 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Gold nanoparticle probes for colorimetric detection of plasma galectin-3: a simple and rapid approach†
Maria
António
 a,
Tânia
Lima
bc,
Rita
Ferreira
de,
Margarida
Fardilha
b,
José Mesquita
Bastos
bf,
Rui
Vitorino
beg and
Ana L.
Daniel-da-Silva
a,
Tânia
Lima
bc,
Rita
Ferreira
de,
Margarida
Fardilha
b,
José Mesquita
Bastos
bf,
Rui
Vitorino
beg and
Ana L.
Daniel-da-Silva
 *a
*a
aCICECO-Aveiro Institute of Materials, Department of Chemistry, University, of Aveiro, Aveiro 3810-193, Portugal. E-mail: ana.luisa@ua.pt
biBiMED-Institute of Biomedicine, Department of Medical Sciences, University of Aveiro, Aveiro, 3810-193, Portugal
cCancer Biology and Epigenetics Group, Research Center of Portuguese Oncology Institute of Porto (GEBC CI-IPOP) & Porto Comprehensive Cancer Center (P.CCC), Porto 4200-072, Portugal
dQOPNA, Department of Chemistry, University of Aveiro, Aveiro, 3810-193, Portugal
eLAQV-REQUIMTE, Chemistry Department, University of Aveiro, Aveiro, 3810-193, Portugal
fCardiology Department, Hospital Infante D. Pedro, Centro Hospitalar do Baixo Vouga, Aveiro, Portugal
gUnIC@RISE, Department of Surgery and Physiology, Cardiovascular R&D Center, Faculty of Medicine of the University of Porto, Alameda Professor Hernâni Monteiro, Porto 4200-319, Portugal
First published on 10th May 2023
Abstract
Galectin-3 (Gal-3) is a carbohydrate-binding protein associated with the development and progress of heart failure. Here, we report the first colorimetric and low-cost approach for detecting and quantifying Gal-3 using gold nanoparticles (AuNPs) bioconjugated with Gal-3 antibody. The interaction of Gal-3 with the resulting nanoprobes led to a linear response of the absorbance ratio A750nm/A526nm to Gal-3 concentration, accompanied by a change in color intensity. The assay showed a linear optical response even in complex samples, such as saliva and fetal bovine serum (FBS), up to a concentration of 200 μg L−1. The limit of detection (LOD) followed the trend LODPBS (10.0 μg L−1) < LODsaliva (22.6 μg L−1) < LODFBS (25.3 μg L−1). The potential applicability of this method to the analysis of human plasma samples was also demonstrated. Compared to conventional detection techniques, this colorimetric assay provides faster results (∼1 h) and is more cost-effective due to the use of simple and unexpensive equipment. This assay represents an exciting solution for the rapid screening of high risk for rapid progression of heart failure in patient samples (Gal-3 > 25.9 μg L−1).
Introduction
Galectin-3 (Gal-3) is a carbohydrate-binding protein1 and belongs to the lectin family that binds β-galactosides.2 This protein can be found in tumor cells,3,4 immunological cells,5 tissues and organs such as the heart.6 Gal-3 is associated with the development and progression of heart failure (HF). Upregulated Gal-3 levels lead to increased cardiac fibroblast production, which increases collagen production and contributes to ventricular dysfunction. Increased Gal-3 levels have been found in hypertrophied hearts of patients with aortic stenosis7 and in patients with acute8 and chronic HF.9 This protein is considered an independent biomarker of income HF10 and a predictor of 60 days mortality in HF patients.8 High Gal-3 levels observed in individuals without cardiovascular disease are associated with HF hospitalization, coronary artery disease, ischemic stroke, and all-cause mortality.11 Gal-3 levels > 25.9 μg L−1 predict that a patient is prone to rapid progression of HF, with a high risk of hospitalization or death.12 This risk is moderate for levels in the range of 17.8–25.9 μg L−1. Below 17.8 μg L−1, which was established as the upper limit for a normal Gal-3 level, the risk is low.12,13 Salivary Gal-3 level was correlated with serum Gal-3 level.14 Salivary Gal-3 level was found to be useful for diagnosis of HF and prediction of primary outcome in HF patients. HF patients had elevated salivary Gal-3 levels (mean: 282.0 μg L−1) compared with healthy controls (mean: 601.3 μg L−1). A cutoff value for salivary Gal-3 levels was established (>384.7 μg L−1).14 In addition, HF patients with salivary Gal-3 levels >172.58 μg L−1 were found to have a higher cumulative risk for the primary outcome than HF patients with lower Gal-3 levels.15In recent years, several analytical methods have been proposed for the determination of Gal-3 content based on electrochemical and localized surface plasmon resonance detection.16–19 Most of these methods are sandwich-type immunoassays17,18 that are expensive because of the need for two specific antibodies. The synthesis of the materials used is also complex, as several steps are required to obtain the final probes, which also increases the time and cost of their fabrication.17,18 In addition, these assays require the use of expensive equipment.17–19 Therefore, simple, inexpensive, and sensitive methods are urgently needed for Gal-3 detection, and colorimetric biosensors are an attractive alternative because they can be observed visually and are easy to use.20,21 Nanomaterials are widely used in biosensing, especially noble metals such as gold nanoparticles (AuNPs).22,23 AuNPs can be prepared by simple and reproducible chemical methods,24 exhibit tunable morphology and particle size,25 and are biocompatible26 and negligibly toxic. These properties make AuNPs suitable for bioapplications. In addition, AuNPs are chemically stable,25 exhibit a high surface-to-volume ratio and molar absorption coefficient, and their surface area can be modified by simple techniques.27,28 Moreover, their application to various detection methods can improve the conductivity and sensitivity of the materials.27 AuNPs exhibit a characteristic optical property in the optical spectrum, localized surface plasmon resonance (LSPR). This phenomenon is observed when the electromagnetic field of the incident light has a specific frequency that is in phase with the oscillation of the electrons on the surface of the nanoparticle.29 Numerous approaches have been used to develop LSPR-based detection strategies. One exciting approach is a colorimetric assay based on LSPR coupling30,31 in which the LSPR band red-shifts based on changes in interparticle distance. The resulting color change can be monitored with UV-VIS spectroscopy inexpensive equipment or detected by the naked eye, making colorimetric assays very attractive for point-of-care testing. However, colorimetric methods often have limited sensitivity32 but modification of the AuNPs surface determines the sensitivity and specificity of the sensor.33,34 To our knowledge, colorimetric sensors for detection of Gal-3 have not been reported previously.
In this study, we developed for the first time a method for colorimetric detection of Gal-3 using modified AuNPs. In this approach, Gal-3 interacted with antibody-bioconjugated Au nanoprobes and promoted the formation of nanoparticle agglomerates that showed a linear optical response to Gal-3 concentration. The intensity of the colloid color decreased at high Gal-3 concentrations and was visible to the human eye. The performance of the colorimetric assay in buffer, saliva, and fetal bovine serum was investigated. Detection of Gal-3 in human plasma samples is also reported.
Materials and methods
Chemicals
The chemicals tetrachloroauric(III) acid trihydrate (HAuCl4·3H2O, ≥ 99.9%), sodium citrate tribasic dihydrate (Na3C6H5O7·2H2O, ≥ 99.0%), 11-mercaptoundecanoic acid (MUDA, C11H22O2S, 95%), phosphate-buffered saline (PBS) pH 7.4, potassium phosphate dibasic (K2HPO4, 98%), trizma hydrochloride (C4H11NO3·HCl, 99%), Tween 20 (C58H114O2), urea (CH4ON2, 99%), albumin from bovine serum (98%), fetal bovine serum, α-amylase from porcine pancreas (≥10 units per mg) and N-hydroxysuccinimide (NHS, C4H5NO3, 98%) were purchased from Sigma-Aldrich. The 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC, C8H17N3·HCl, > 98%) was obtained from Alfa Aesar. The monoclonal antibody of Gal-3 from mouse (B2C10, sc-32790) was acquired from Santa Cruz Biotechnology. The human recombinant Gal-3 (98%) was purchased from RayBiotech. The 6-protein mixture was acquired from AB Sciex, the integrin beta 3 antibody – IgG from porcine (>90.0%) was purchased from ThermoFisher Scientific, the glycine (C2H5NO2, 99%) was purchased from Riedel and the L(+) ascorbic acid for analysis (C6H8O6) was acquired from Carlo Erba Reagents. Sodium chloride (NaCl, >99.5%) was acquired in VWR Chemicals The nitrocellulose membranes (Amersham Protran NC 0.45), horseradish peroxidase-conjugated anti-mouse antibody, and enhanced chemiluminescence kit (ECL Select western blotting detection reagent, RPN2235) from Amersham Pharmacia Biotech were purchased in Cytiva. All the reactants were used without further purification.Synthesis of Au nanoparticles
The Turkevich method was used to synthesize gold nanoparticles with spheroidal shape. The following procedure was used with some modifications.35 Briefly, an aqueous solution of HAuCl4 (100 mL, 1 mM) was left to heat until boiling, under vigorous stirring and reflux. Then, a sodium citrate solution (10 mL, 38.8 mM) was added. The reaction remained for 1 h and the solution color shifted from light yellow to reddish-purple. After that, the solution was allowed to cool until room temperature under stirring.Functionalization of Au nanoparticles with MUDA
Au nanoparticles were functionalized with MUDA following procedures adapted from the literature.36,37 Briefly, MUDA solution was firstly prepared by dispersing MUDA (10 mg) in NaOH aqueous solution (1 mL, 0.01 M), followed by the addition of 4 mL of Milli-Q (M.Q.) water. Then 5 mL of MUDA solution was added to 3.82 mL of Au colloid as previously prepared, in a molar ratio 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (MUDA
1 (MUDA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Au) and left to react for 24 h at 25 °C under magnetic stirring (800 rpm). The resulting AuNPs@MUDA were centrifuged at 9500 rpm for 15 min, twice, and resuspended in an equivalent volume of M.Q. water.
Au) and left to react for 24 h at 25 °C under magnetic stirring (800 rpm). The resulting AuNPs@MUDA were centrifuged at 9500 rpm for 15 min, twice, and resuspended in an equivalent volume of M.Q. water.
Conjugation of AuNPs@MUDA with the antibody of Gal-3
The antibody-conjugated Au nanoparticles (AuNPs@MUDA@Ab) were prepared according to the following procedures from the literature17,37 with several alterations. EDC solution (10 μL, 5 mM) was added to 1 mL of AuNPs@MUDA, followed by NHS solution (10 μL, 7.5 mM). The nanoparticles were left to react for 30 min at 25 °C under orbital stirring (325 rpm). After that, the EDC/NHS activated AuNPs@MUDA were centrifuged at 9500 rpm for 15 min and resuspended in the same volume of K2HPO4 (2.5 mM, pH 7.4). The antibody of Gal-3 (Ab) (25 μL, 200 μg mL−1) was added and left to react for 60 min at 25 °C under orbital stirring (325 rpm). Then, 10 μL of 1% BSA 0.1 M PBS solution was added to AuNPs@MUDA@Ab and left to react for 10 min to block non-specific interactions. The resulting AuNPs@MUDA@Ab@BSA nanoparticles were centrifuged at 6500 rpm for 11 min. The supernatant was centrifuged at 8000 rpm for another 11 min. The recovered nanoparticles were resuspended in 800 μL of PBS (0.01 M, pH 8).Preparation of Gal-3 solution series
The Gal-3 was acquired in a lyophilized form and resuspended in 0.5 M phosphate-buffered saline (PBS) solution at pH = 7.2. Gal-3 stock solutions with a concentration of 10 mg L−1 were prepared by dilution in 0.5 M PBS pH 7.4 and kept in the freezer at −20 °C until use. The Gal-3 solution series were prepared through dilution of Gal-3 stock solution in 0.01 M PBS pH 7.4. The concentration of the prepared solutions ranged from 0 to 320 μg L−1.Procedure for detecting Gal-3 using AuNPs@MUDA@Ab@BSA
In a typical procedure, 30 μL of Gal-3 solution series (0–320 μg L−1) were added to 200 μL of AuNPs@MUDA@Ab@BSA. The resulting mixture was stirred in an orbital shaker incubator (325 rpm) for 60 min at 25 °C and then analyzed through UV-VIS spectroscopy (200 μL sample, in a microplate reader) to determine the absorbance ratio (A750/A526), where A750 and A526 are the absorbance values at 750 nm and 526 nm, respectively.Specificity for Gal-3 detection
To study the specificity of the AuNPs@MUDA@Ab@BSA, the potential interferents BSA, α-amylase, IgG, ascorbic acid, urea and glycine were added to the colloid in the absence of Gal-3, at a concentration of 1.53 nM. The absorbance ratio was evaluated using UV-VIS spectroscopy as described previously and compared to the A750/A526 produced by Gal-3 at an identical concentration (40 μg L−1).Stability of the probes
After synthesis, the AuNPs@MUDA@Ab@BSA probes were stored in the refrigerator at 4 °C. The absorbance ratio of the probes was evaluated after a storage period of 5 h, 3 days, 5 days, 7 days, 11 days and 15 days in the absence and presence of Gal-3 (40 μg L−1 concentration).Salivary and plasma samples
The saliva was collected after breakfast (1 hour) from a healthy individual and placed in Eppendorf microtubes that were kept ice bath until centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 20 min, 4 °C). The supernatant was recovered and kept at −20 °C until used. The plasma samples were obtained from the Cardiology Service of the Baixo Vouga Hospital Center from patients who were admitted for acute myocardial infarction with non-obstructive coronary heart disease and their respective controls. After collection, the plasma samples were kept at −80 °C until use.
000g, 20 min, 4 °C). The supernatant was recovered and kept at −20 °C until used. The plasma samples were obtained from the Cardiology Service of the Baixo Vouga Hospital Center from patients who were admitted for acute myocardial infarction with non-obstructive coronary heart disease and their respective controls. After collection, the plasma samples were kept at −80 °C until use.
Calibration curves for Gal-3 in different complex matrices
The method for Gal-3 detection was employed in different biofluids and a mixture of proteins besides the PBS buffer. Different concentrations of Gal-3 (0–320 μg L−1) were spiked into a diluted 6 protein mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100), saliva (1
100), saliva (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10), and fetal bovine serum (FBS) (1
10), and fetal bovine serum (FBS) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100). The 6 proteins mixture was resuspended in Milli Q water, the saliva and the FBS were thawed in an ice bath and diluted in Milli-Q water. The procedure for Gal-3 detection was described previously.
100). The 6 proteins mixture was resuspended in Milli Q water, the saliva and the FBS were thawed in an ice bath and diluted in Milli-Q water. The procedure for Gal-3 detection was described previously.
Gal-3 detection in plasma samples
The plasma samples of 2 patients with acute myocardial infarction with non-obstructive coronary heart disease and 2 controls were diluted in M.Q. water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) and kept in an ice bath until analysis. A volume of 30 μL of the diluted sample was added to 200 μL of the AuNPs@MUDA@Ab@BSA colloid. Then, the resulting solution was left to react and analyzed through UV-VIS spectroscopy as described previously.
10) and kept in an ice bath until analysis. A volume of 30 μL of the diluted sample was added to 200 μL of the AuNPs@MUDA@Ab@BSA colloid. Then, the resulting solution was left to react and analyzed through UV-VIS spectroscopy as described previously.
Western blot analysis
The plasma samples were analyzed by western blot to observe the presence of Gal-3. The plasma proteins were separated on 12% SDS-PAGE using 20 μg of protein from each sample. After, the proteins were spotted on nitrocellulose membranes through electroblotting, followed by Ponceau staining to promote the efficacy of protein transference from the gel to the membrane and as a loading control. The membranes were blocked with 5% nonfat milk in 1 × Tris-buffered saline – TBS (25 mM Tris–HCl, pH 7.4, 150 mM NaCl) containing 0.1% Tween 20 – TBS-T to avoid unspecific protein binding sites. Then, the membranes were incubated with Ab diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 for 1 hour at RT, the unreacted Ab was washed with TBS-T and the horseradish peroxidase-conjugated mouse antibody was added and left to react for 1 h at RT. The membranes were washed with TBS-T and 1× TBS. The resulting immunoreactive bands were marked using an enhanced chemiluminescence kit and detected by the ChemiDocTM touch imaging system. Image Lab software (version 6.1) was used to evaluate the protein abundance through densitometric analysis. The protein abundance was normalized to Ponceau S staining intensity and loading control.
500 for 1 hour at RT, the unreacted Ab was washed with TBS-T and the horseradish peroxidase-conjugated mouse antibody was added and left to react for 1 h at RT. The membranes were washed with TBS-T and 1× TBS. The resulting immunoreactive bands were marked using an enhanced chemiluminescence kit and detected by the ChemiDocTM touch imaging system. Image Lab software (version 6.1) was used to evaluate the protein abundance through densitometric analysis. The protein abundance was normalized to Ponceau S staining intensity and loading control.
Instrumentation
The LSPR band and the absorbance ratio (A750/A526) of AuNPs were measured in a Multiskan GO microplate UV-VIS spectrophotometer from Thermo Fisher Scientific operating at RT using fast mode and 1 nm of bandwidth. The samples (200 μL) were placed on a UV-Star 96-wells microplate from Greiner. The AuNPs and AuNPs@MUDA were diluted in M.Q. water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) for UV-VIS analysis, while AuNPs@MUDA@AB@BSA were analyzed as prepared. Zeta potential measurements were employed to evaluate the surface charge of AuNPs by electrophoretic light scattering of aqueous solutions. These measurements were performed in a Zetasizer Nano ZS from Malvern Instruments, supplied with a 633 nm HeNe laser and a scattering detector at 173°. Dynamic light scattering (DLS) was utilized to assess the hydrodynamic diameter of AuNPs using the same Zetasizer Nano ZS. The pH measurements, zeta potential, and DLS analysis were performed without diluting the colloid. Scanning transmission electron microscopy (STEM) was used to analyze the morphology and size of AuNPs in a Hitachi HD-2700 STEM microscope operated at 200 kV. The samples were prepared through the evaporation of the solvent of the colloid on carbon-coated copper grids. The TEM images were explored by the ImageJ software version 1.46 to create the AuNPs size histogram. The attenuated total reflection – Fourier transform infrared (ATR-FTIR) spectroscopy was used to evaluate the presence of MUDA and antibodies on the surface of AuNPs. The ATR-FTIR spectra were obtained using a Bruker Optics Tensor 27 spectrometer combined with a horizontal ATR cell, at 256 scans with a resolution of 4 cm−1. The sample was prepared by the deposition of 2 to 3 drops of the Au colloid, PBS buffer, and Ab Gal-3 solution onto the crystal surface of the FTIR. The colloids and solutions were dried under N2 gas to obtain a film that was analyzed. An orbital shaker incubator (IKA KS 4000 i) was used to prepare AuNPs@MUDA@Ab@BSA and during the procedure for detecting Gal-3 at 25 °C and under 325 rpm. Inductively coupled plasma mass spectrometry (ICP-MS) was employed to determine the concentration of Au in the AuNPs. The sample for analysis was 250 μL of as-synthesized AuNPs. The digestion was performed using aqua regia by adding 10 μL of HNO3 (65%) and 30 μL of HCl (37%) to 250 μL of AuNPs colloid. The digestion was performed at room temperature for periods longer than 48 h. The proteins present in the plasma samples were separated using a Mini-Protean system from Bio-Rad Laboratories operating at 200 V for about 1 hour. The proteins separated in gels were transferred to membranes using the Trans-Blot Turbo system from Bio-Rad Laboratories in mixed proteins mode with a transference time of 7 min. A ChemiDocTM Touch Imaging System from Bio-Rad Laboratories was used to detect the immunoreactive bands resulting from Gal-3 recognition with Ab and images acquisition.
3) for UV-VIS analysis, while AuNPs@MUDA@AB@BSA were analyzed as prepared. Zeta potential measurements were employed to evaluate the surface charge of AuNPs by electrophoretic light scattering of aqueous solutions. These measurements were performed in a Zetasizer Nano ZS from Malvern Instruments, supplied with a 633 nm HeNe laser and a scattering detector at 173°. Dynamic light scattering (DLS) was utilized to assess the hydrodynamic diameter of AuNPs using the same Zetasizer Nano ZS. The pH measurements, zeta potential, and DLS analysis were performed without diluting the colloid. Scanning transmission electron microscopy (STEM) was used to analyze the morphology and size of AuNPs in a Hitachi HD-2700 STEM microscope operated at 200 kV. The samples were prepared through the evaporation of the solvent of the colloid on carbon-coated copper grids. The TEM images were explored by the ImageJ software version 1.46 to create the AuNPs size histogram. The attenuated total reflection – Fourier transform infrared (ATR-FTIR) spectroscopy was used to evaluate the presence of MUDA and antibodies on the surface of AuNPs. The ATR-FTIR spectra were obtained using a Bruker Optics Tensor 27 spectrometer combined with a horizontal ATR cell, at 256 scans with a resolution of 4 cm−1. The sample was prepared by the deposition of 2 to 3 drops of the Au colloid, PBS buffer, and Ab Gal-3 solution onto the crystal surface of the FTIR. The colloids and solutions were dried under N2 gas to obtain a film that was analyzed. An orbital shaker incubator (IKA KS 4000 i) was used to prepare AuNPs@MUDA@Ab@BSA and during the procedure for detecting Gal-3 at 25 °C and under 325 rpm. Inductively coupled plasma mass spectrometry (ICP-MS) was employed to determine the concentration of Au in the AuNPs. The sample for analysis was 250 μL of as-synthesized AuNPs. The digestion was performed using aqua regia by adding 10 μL of HNO3 (65%) and 30 μL of HCl (37%) to 250 μL of AuNPs colloid. The digestion was performed at room temperature for periods longer than 48 h. The proteins present in the plasma samples were separated using a Mini-Protean system from Bio-Rad Laboratories operating at 200 V for about 1 hour. The proteins separated in gels were transferred to membranes using the Trans-Blot Turbo system from Bio-Rad Laboratories in mixed proteins mode with a transference time of 7 min. A ChemiDocTM Touch Imaging System from Bio-Rad Laboratories was used to detect the immunoreactive bands resulting from Gal-3 recognition with Ab and images acquisition.
Statistical analysis
The limit of detection (LOD) and limit of quantification (LOQ) were determined from the linear correlation between A750/A526 values and Gal-3 concentrations. The LOD was determined according to eqn (1)–(4) and LOQ from eqn (5), adapted from ref. 38 and 39, as presented. The xi, yi, m, and n are the concentrations of Gal-3, values of A750/A526, slope, and number of points of the calibration curve, respectively.| yB = ∑yi − (m∑xi) | (1) |
| ycalc = (mxi) + yB | (2) |
 | (3) |
| LOD = 3(Sy/x/m) | (4) |
| LOQ = 10(Sy/x/m) | (5) |
Results and discussion
Preparation of AuNPs@MUDA@Ab@BSA probes
AuNPs@MUDA@Ab@BSA nanoprobes were developed to detect Gal-3 by modifying gold nanoparticles. Several aspects were considered in the development of the probe, including the optical properties of AuNPs, the covalent binding of Ab to AuNPs achieved by surface modification with MUDA and carbodiimide chemistry, and the blocking of the nanoprobe surface with BSA to prevent the nonspecific adsorption of other biomolecules (Fig. 1(A)). Citrate-capped gold nanoparticles were synthesized by the Turkevich method. The obtained AuNPs were nearly spherical and had an average size of 14.9 ± 2.7 nm, as determined by TEM (Fig. S1, ESI†). The histogram of particle size is shown in Fig. 1(B). To bioconjugate the Gal-3 antibody (Ab) with the gold nanoparticles, the AuNPs were modified with MUDA (AuNPs@MUDA). Then, bovine serum albumin (BSA) was added to the AuNPs@MUDA@Ab to block the non-specific adsorption of other proteins, resulting in AuNPs@MUDA@Ab@BSA. The TEM image of the synthesized AuNPs@MUDA@Ab@BSA probes is shown in Fig. 1(B). In Fig. 1(C), the optical spectrum of AuNPs shows a localized surface plasmon resonance (LSPR) band at 523 nm with an absorbance of 0.986. The Au concentration in the form of nanoparticles (ICP-MS) was 0.853 mM and the AuNP concentration was estimated to be 8.36 nM, assuming a spherical morphology and a fcc crystal structure.40 The LSPR band shifted to 526 nm with an absorbance of 0.709 after modification with MUDA (Fig. 1(C)). After modification with Ab and BSA, the LSPR band remained at 526 nm with an absorbance of 0.765.Zeta potential (ZP) measurements were performed to determine the surface charge of the AuNPs. The AuNPs were found to be negatively charged (−59.3 ± 1.7 mV; pH 5.8) due to citrate capping. Modification with MUDA decreased the ZP (−73.1 ± 2.6 mV; pH 7.2) due to the carboxylate functional groups. The ZP increased after conjugation with Ab (−29.9 ± 0.4 mV; pH 7.0) as a result of the binding of Ab to the carboxylic acid groups. The surface charge was not affected by modification with BSA (ZP = −30.5 ± 1.2 mV; pH 7.0). The changes in hydrodynamic diameter (HD) confirmed the surface modification (Table S1, ESI†). The HD of the citrate-capped AuNPs was 24.7 ± 0.5 nm with a polydispersity index (PDI) of 0.356. After functionalization with MUDA and Ab conjugation, the HD increased to 65.5 ± 0.4 nm (PDI = 0.454) and 111.3 ± 0.6 nm (PDI = 0.301), respectively. The HD of the final probes AuNPs@MUDA@Ab@BSA was 103.6 ± 0.1 nm with a PDI of 0.290, indicating moderate polydispersity.41
ATR-FTIR spectroscopy analysis confirmed MUDA and Ab modification at the surface of AuNPs (Fig. S2(A), ESI†). The AuNPs covered with citrate showed vibrational bands of citrate at 1389 cm−1 and 1569 cm−1, corresponding to C–O–H stretching vibration and symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, respectively.42 After functionalization with MUDA, new vibrational bands appeared at 2922 cm−1 and 2846 cm−1, which were assigned to the C–H stretching vibrations of the carbon backbone chain of MUDA.36 Conjugation of Ab on the surface of AuNPs@MUDA effectively changed the observed vibrational bands. The presence of characteristic IgG vibrational bands at 1355 cm−1 and 1257 cm−1 and 1654 cm−1 and 1559 cm−1 of the formation of amide bands from carbodiimide coupling of AuNPs@MUDA and Ab43 in Fig. S2(B) of the ESI† indicates that the modification of AuNPs@MUDA with Ab was successful.
O stretching vibration, respectively.42 After functionalization with MUDA, new vibrational bands appeared at 2922 cm−1 and 2846 cm−1, which were assigned to the C–H stretching vibrations of the carbon backbone chain of MUDA.36 Conjugation of Ab on the surface of AuNPs@MUDA effectively changed the observed vibrational bands. The presence of characteristic IgG vibrational bands at 1355 cm−1 and 1257 cm−1 and 1654 cm−1 and 1559 cm−1 of the formation of amide bands from carbodiimide coupling of AuNPs@MUDA and Ab43 in Fig. S2(B) of the ESI† indicates that the modification of AuNPs@MUDA with Ab was successful.
Detection of Gal-3
The developed nanoprobe contain monoclonal antibodies that bind Gal-3 to a single epitope. Previous studies suggested that an increase in Gal-3 concentration would cause a red shift in LSPR due to changes in the refractive index (RI) of the surrounding medium triggered by Gal-3 binding.44 However, we did not detect any band shift. Instead, the decrease in the absorbance of the LSPR band at 526 nm (A526) and minor changes in the absorbance of a weak band at 750 nm (A750) (Fig. 2) were observed when the Gal-3 concentration increased. In a previous work,45 antibody-modified AuNPs exhibited a similar decrease in ALSPR, which was attributed to the formation and precipitation of large aggregates resulting from antibody–antigen interactions. Given that Gal-3 can form homodimers in the absence of carbohydrate-binding ligands,46 we hypothesized that Gal-3 dimers can promote interparticle bridging by binding to two Au probes, leading to the formation of aggregates at high Gal-3 concentrations. Furthermore, the minor variations of A750 could be related with the agglutination of the probes. The formation of nanoparticle agglomerates was confirmed by analyzing the hydrodynamic diameter (HD) of the nanoprobes after incubation of Gal-3 at a concentration of 0, 40, and 160 μg L−1 (Table S2, ESI†). The HD of the probes at 0 μg L−1 Gal-3 in PBS was similar to the HD (121.9 ± 1.4 nm) of the nanoprobes. However, after incubation with Gal-3 at 40 and 160 μg L−1, the HD values increased to 332.9 ± 4.6 nm and 422.6 ± 3.8 nm, respectively, consistent with the formation of agglomerates by interaction with Gal-3. The PDI value also increased with the concentration of Gal-3. For concentrations of 0, 40, and 160 μg L−1, the PDI values were 0.310, 0.518, and 0.708, respectively, indicating the polydisperse size distribution of the probes after interaction with Gal-3 41. The absorbance ratio (AR), which is the ratio between A750 and A526, was calculated and correlated with the Gal-3 concentration. | ||
| Fig. 2 (Bottom) UV-VIS spectra of AuNPs@MUDA@Ab@BSA probes in response to the presence of Gal-3 at variable concentration (0–160 μg L−1) in PBS (0.01 M, pH 7.4). (Top) Photograph of the trials. | ||
The amount of antibody (Ab) used in the bioconjugation of the Au probes and the incubation time with Gal-3 are key parameters that were optimized to achieve higher sensitivity. It was found that 5 μg Ab at a Gal-3 concentration of 80 μg L−1 gave a higher AR value (0.320) than 0.5 μg (0.199) and 2.5 μg (0.232) (Fig. S3, ESI†). Subsequent experiments were performed using 25 μL of 200 μg mL−1 Ab (5 μg) to modify 1 mL of AuNPs@MUDA. The effect of incubation time with Gal-3 (15, 30, and 60 min) in AR was examined for initial Gal-3 concentrations of 40 and 160 μg L−1 (Fig. S4 of ESI†). For the lowest Gal-3 concentration (40 μg L−1), the effect of incubation time in AR was meaningless. However, at higher Gal-3 concentration (160 μg L−1), AR increased with incubation time of 0.258, 0.285, 0.421 for 15, 30 and 60 min, respectively. Subsequently, the incubation time was fixed at 60 minutes.
The Gal-3 was detected in PBS (0.01 M, pH = 7.4) at optimized conditions for Ab amount and incubation time. The resulting absorption spectra are shown in Fig. 2. The intensity of the colloid color decreased for Gal-3 ≥ 20 μg L−1, as shown in Fig. 2 (top). A linear relationship between the AR and Gal-3 concentration was observed in the range from 0 to 160 μg L−1 (Fig. 3).
 | ||
| Fig. 3 Calibration curve for AuNPs@MUDA@Ab@BSA in Gal-3 concentrations in PBS (grey), spiked into diluted saliva (black), a diluted mixture of 6 proteins (green) and diluted FBS (blue). | ||
A correlation coefficient of 0.9921 was observed and LOD of 10.0 μg L−1 was obtained. The present method was also tested for the detection of Gal-3 in more complex matrices, in a mixture of 6 proteins (BSA, β-galactosidase, α-lactalbumin, β-lactoglobulin, lysozyme, and apotransferrin), saliva, and FBS diluted 100-, 10-, and 100-fold, respectively. The linear relationship between AR and Gal-3 concentration was observed despite increasing matrix complexity. The calibration curves are shown in Fig. 3. It was found that the working range of the calibration curve increased with the complexity of the matrix. In buffer, the working range was up to 160 μg L−1 Gal-3, while in the 6-protein mixture and saliva, the highest concentration was 180 μg L−1. In diluted FBS, the working range extended to 200 μg L−1. However, the increase in matrix complexity resulted in a decrease in the slope of the calibration curve compared to the detection in buffer. Since the slope is related to the sensitivity of method,47 the sensitivity in FBS decreased by about half compared to PBS. In saliva, this decrease was less pronounced. However, the correlation coefficient remained almost unchanged when matrix complexity increased. The LOD followed the trend LODPBS (10.0 μg L−1) < LODsaliva (22.6 μg L−1) < LOD6 protein (23.5 μg L−1) < LODFBS (25.3 μg L−1). The detection range of Gal-3 in saliva and FBS was 24.6–180 μg L−1 and 26.9–200 μg L−1, respectively. The coefficient of variation (CV) varied from 7.20 to 10.7% in buffer, 2.16 to 8.53% in saliva, 2.57 to 8.82% in a mixture of 6 proteins, and 1.45 to 9.16% in FBS. The recovery rate (%) was determined for Gal-3 in biofluids and ranged from 99.1–120.3% in saliva, 91.0–118.1% in a mixture of 6 proteins, and 83.0–112.7% in FBS.
Our developed system shows great potential for Gal-3 detection as it offers a wider working range in FBS (26.9–200 μg L−1) and shorter time to result with lower cost compared to commercially available ELISA kits with a narrower working range (0.49–30.0 μg L−1) and longer time to result (2 h 30 min). Other complex immunoassays reported in literature have shown lower limits of detection but narrower detection ranges18 and longer time to result.17 Additionally, other novel Gal-3 detection methods have been described but their analytical performance is uncertain without defined LOQ16 and LOD19 values (as shown in Table 1).
| Method/Nanomaterial | LOD | Detection range | Time to result | Cost | Ref. |
|---|---|---|---|---|---|
| a NM: not mentioned; ND: not defined. | |||||
| ELISA | 0.29 μg L−1 | 0.47–30.0 μg L−1 | ∼2 h 30 min | ∼560 euros (kit for 96 tests) | 48 |
| AuNPs/MB/MSN composite | 0.17 fg mL−1 | 0.5–500 ng L−1 | ∼2 h | NMa | 17 |
| g-C3N4@AuNPs Ti-MOF@COFs | 0.025 pg mL−1 | 0.0001–20 ng L−1 | ∼45 min | NMa | 18 |
| AuNPs@Fc-Lac | 4.16 mg L−1 | ND–13 mg L−1 | ∼1 h 20 min | NMa | 16 |
| MIPs 2-aminophenol | ND | 0.5–5000 ng L−1 | ∼20 min | NMa | 19 |
| Modified AuNPs | 25.3 μg L−1 | 26.9–200 μg L−1 | ∼1 h 10 min | ∼200 euros (96 tests) | This work |
Gal-3-antibody interaction
The dissociation constant (kd) was estimated to evaluate the strength of the Gal-3-antibody interaction. For this calculation, the specific binding – one site equation (equation S1, ESI†) was used applying the AR plotted against Gal-3 concentration in the range 0–320 μg L−1 (Fig. S5 of ESI†). The kd was found to be 0.388 nM, 0.170 nM, and 0.512 nM in buffer, diluted saliva, and FBS, respectively. The values of kd indicate high affinity of the antibody to the Gal-3 (∼10−10 M) similar to others in the literature using probes (∼10−10 M)49 or aptamers (peptide-specific) (∼10−5 M).50Specificity and stability studies
The AR of the probes in the presence of the potentially interfering species BSA, α-amylase, IgG, ascorbic acid, urea, and glycine (1.53 nM concentration) was measured and compared to the effect of the Gal-3 at an identical concentration (Fig. 4). The AR values due to the interfering species were markedly lower than the AR due to the Gal-3, and similar to the results observed for the blank. These results indicate that the methods allow for the specific detection of Gal-3. | ||
| Fig. 4 AR of AuNPs@MUDA@Ab@BSA in the presence of Gal-3 and different biomolecules at the concentration of 1.53 nM. Dashed line indicates the result of a blank test. | ||
The stability of the AuNPs@MUDA@Ab@BSA stored at 4 °C was investigated. The absorbance ratio of the nanoparticles was evaluated after 5 h, 3 days, 5 days, 7 days, 11 days, and 15 days of their synthesis. The nanoparticles were analyzed in the absence and presence of 40 μg L−1 of galectin-3 revealing good stability until 11 days (Fig. S6 of ESI†).
Detection in plasma samples
The developed nanoprobes were able to detect Gal-3 in complex matrices. The level of Gal-3 in saliva, serum, or plasma is relevant to the diagnosis and prognosis of HF. Determination of Gal-3 level in saliva of healthy controls and patients with HF was desirable, using the presented strategy. However, saliva collection is not observed in the clinical context. Therefore, nanoprobes were used to determine the level of Gal-3 in plasma. Plasma from 2 myocardial infarction patients (P) and 2 controls (C) was analyzed by the developed colorimetric method. The following Gal-3 concentrations were determined for the different plasma samples: P0 = 280.5 μg L−1, P1 = 260.3 μg L−1, C3 = 233.6 μg L−1, and C2 = 143.9 μg L−1. For comparison purposes, the plasma samples were also analyzed by western blot technique using the same antibody against Gal-3. Fig. 5 shows that the intensity of the Gal-3 30 kDa band labeled by the antibody varies between samples. Stronger bands correspond to samples with a higher concentration of Gal-3 (Fig. 5). According to the adj. vol. ratio (Table S3, ESI†), the concentration of Gal-3 varied as follows P0 > P1 > C3 > C2. This trend is consistent with the values of Gal-3 concentration obtained by the colorimetric method. In addition, the relationship between the adj. vol. of P0 and the other samples was similar to the relationship between the Gal-3 concentration of P0 and the other samples, as shown in Table S3 of the ESI.† These results indicate the applicability of the developed method in human plasma.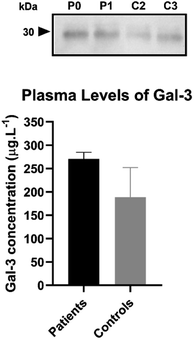 | ||
| Fig. 5 Plasma protein levels of Gal-3 in myocardial infarction patients (P0, P1) and controls (C2, C3) using the present method and western blot. | ||
In addition, the difference between Gal-3 concentrations in patients and controls was small. This could be due to the presence of other cardiovascular diseases in the controls, since they were controls with respect to the absence of acute myocardial infarction.
Conclusions
In summary, we have developed a novel method for the colorimetric detection and quantification of Gal-3 using AuNPs@MUDA@Ab@BSA probes. This is the first reported method for colorimetric detection of Gal-3. Our results show that the changes in absorbance ratio monitored by UV-VIS spectroscopy follow a linear correlation with Gal-3 concentration in PBS (0.01 M, pH = 7.4), dilute protein mixture, saliva, and FBS, with a working range of 10.0–200 μg L−1. The LOD of our method follows the trend LODPBS (10.0 μg L−1) < LODSaliva (22.6 μg L−1) < LOD6 protein (23.5 μg L−1) < LODFBS (25.3 μg L−1). The potential applicability of the developed method in human plasma samples was also demonstrated. The developed method offers several advantages over existing commercial ELISA kits. It has a wider working range, faster turnaround time, and reduces detection costs to almost half. In addition, our method can be performed with routine laboratory equipment (UV-VIS spectrometer), which makes it more accessible for near-patient testing. However, further studies are needed to validate the applicability of our method on a larger scale and in different biofluids.Ethical statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards and approved by the Ethics Committee of Centro Hospitalar do Baixo Vouga, (ref 2404-2022). Informed consent was obtained from all subjects involved in the study.Author contributions
Maria António: conceptualization, methodology, validation, formal analysis, writing-original article, writing-review and edition. Tânia Lima: methodology, validation, formal analysis, writing-review and edition. Rita Ferreira, Margarida Fardilha, José Mesquita Bastos: writing-review and edition. Rui Vitorino and Ana L. Daniel-da-Silva: conceptualization, investigation, supervision, writing-original article, writing-review and edition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Portuguese Foundation for Science and Technology (FCT), FCT/MEC (PIDDAC), European Union, QREN, FEDER, and COMPETE for the project CICECO-Aveiro Institute of Materials, UIDB/50011/2020 & UIDP/50011/2020 & LA/P/0006/2020, the project iBiMED (UIDB/04501/2020, POCI-01-0145-FEDER-007628), UnIC (UID/IC/00051/2019) and the project LAQV- REQUIMTE (UIDB/50006/2020). M. A. thanks FCT for the PhD grants (SFRH/BD/136881/2018; COVID/BD/152911/2023). T. L. thanks FCT for the PhD grant (SFRH/BD/136904/2018). A. L. D.-d.-S. acknowledges FCT for the contract (IF/00405/2014). R. V. thanks the FCT for the Investigator Grant (IF/00286/2015). COST Action CA21153Network for implementing multiomics approaches in atherosclerotic cardiovascular disease prevention and research. The graphical abstract was built using images from Servier Medical Art. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/).References
- A. Krześlak and A. Lipińska, Cell. Mol. Biol. Lett., 2004, 9, 305–328 Search PubMed.
- S. H. Barondes, D. N. W. Cooper, M. A. Gitt and H. Leffler, J. Biol. Chem., 1994, 269, 20807–20810 CrossRef CAS.
- R. S. Bresalier, P.-S. Yan, J. C. Byrd, R. Lotan and A. Raz, Cancer, 1997, 80, 776–787 CrossRef CAS.
- P. Nangia-Makker, R. Sarvis, D. W. Visscher, J. Bailey-Penrod, A. Raz and F. H. Sarkar, Breast Cancer Res. Treat., 1998, 49, 171–183 CrossRef CAS PubMed.
- H. Sano, D. K. Hsu, J. R. Apgar, L. Yu, B. B. Sharma, I. Kuwabara, S. Izui and F. T. Liu, J. Clin. Invest., 2003, 112, 389–397 CrossRef CAS.
- H. Kim, J. Lee, J. W. Hyun, J. W. Park, H. gu Joo and T. Shin, Cell Biol. Int., 2007, 31, 655–662 CrossRef CAS PubMed.
- U. C. Sharma, S. Pokharel, T. J. Van Brakel, J. H. Van Berlo, J. P. M. Cleutjens, B. Schroen, S. André, H. J. G. M. Crijns, H. J. Gabius, J. Maessen and Y. M. Pinto, Circulation, 2004, 110, 3121–3128 CrossRef CAS PubMed.
- R. R. van Kimmenade, J. L. Januzzi, P. T. Ellinor, U. C. Sharma, J. A. Bakker, A. F. Low, A. Martinez, H. J. Crijns, C. A. MacRae, P. P. Menheere and Y. M. Pinto, J. Am. Coll. Cardiol., 2006, 48, 1217–1224 CrossRef CAS PubMed.
- D. J. A. Lok, P. Van Der Meer, P. W. B. A. De La Porte, E. Lipsic, J. Van Wijngaarden, H. L. Hillege and D. J. Van Veldhuisen, Clin. Res. Cardiol., 2010, 99, 323–328 CrossRef CAS.
- R. A. De Boer, D. J. A. Lok, T. Jaarsma, P. Van Der Meer, A. A. Voors, H. L. Hillege and D. J. Van Veldhuisen, Ann. Med., 2011, 43, 60–68 CrossRef CAS.
- D. Aguilar, C. Sun, R. C. Hoogeveen, V. Nambi, E. Selvin, K. Matsushita, A. Saeed, J. W. McEvoy, A. M. Shah, S. D. Solomon, E. Boerwinkle and C. M. Ballantyne, J. Am. Heart Assoc., 2020, 9, e015405 CrossRef PubMed.
- P. A. Mccullough, A. Olobatoke and T. E. Vanhecke, Rev. Cardiovasc. Med., 2011, 12, 200–210 CrossRef PubMed.
- T. Mueller, M. Egger, I. Leitner, C. Gabriel, M. Haltmayer and B. Dieplinger, Clin. Chim. Acta, 2016, 456, 19–23 CrossRef CAS PubMed.
- X. Zhang, Y. Wan, R. Chata, A. Brazzale, J. J. Atherton, K. Kostner, G. Dimeski and C. Punyadeera, J. Clin. Pathol., 2016, 69, 1100–1104 CrossRef CAS.
- X. Zhang, N. Karunathilaka, S. Senanayake, V. N. Subramaniam, W. Chan, K. Kostner, J. Fraser, J. J. Atherton and C. Punyadeera, Clin. Res. Cardiol., 2020, 109, 685–692 CrossRef CAS PubMed.
- M. C. Martos-Maldonado, I. Quesada-Soriano, L. García-Fuentes and A. Vargas-Berenguel, Nanomaterials, 2020, 10, 203 CrossRef CAS PubMed.
- H. Liu, Y. Cheng, Y. Chen, H. Xiao, Y. Sui, Q. Xie, R. Liu and X. Yang, J. Electroanal. Chem., 2020, 861, 113952 CrossRef CAS.
- M. L. Yola and N. Atar, Nanoscale, 2020, 12, 19824–19832 RSC.
- S. M. V. Cerqueira, R. Fernandes, F. T. C. Moreira and M. G. F. Sales, Microchem. J., 2021, 164, 105992 CrossRef CAS.
- H. Aldewachi, T. Chalati, M. N. Woodroofe, N. Bricklebank, B. Sharrack and P. Gardiner, Nanoscale, 2018, 10, 18–33 RSC.
- H. Wang, H. Rao, M. Luo, X. Xue, Z. Xue and X. Lu, Coord. Chem. Rev., 2019, 398, 113003 CrossRef CAS.
- E. B. Aydın, M. Aydın and M. K. Sezgintürk, Sens. Actuators, B, 2021, 345, 130355 CrossRef PubMed.
- X. Ou, Y. Liu, M. Zhang, L. Hua and S. Zhan, Microchim. Acta, 2021, 188, 304 CrossRef CAS PubMed.
- P. Ghosh, G. Han, M. De, C. K. Kim and V. M. Rotello, Adv. Drug Delivery Rev., 2008, 60, 1307–1315 CrossRef CAS PubMed.
- H. Robenek, Colloidal Gold: Principles, Methods, and Applications, Academic Press, Inc., New York, 1989, vol. 12 Search PubMed.
- R. Shukla, V. Bansal, M. Chaudhary, A. Basu, R. R. Bhonde and M. Sastry, Langmuir, 2005, 21, 10644–10654 CrossRef CAS PubMed.
- H. Li and L. J. Rothberg, J. Am. Chem. Soc., 2004, 126, 10958–10961 CrossRef CAS PubMed.
- W. Zhou, X. Gao, D. Liu and X. Chen, Chem. Rev., 2015, 115, 10575–10636 CrossRef CAS PubMed.
- G. C. Pavassiliou, Prog. Solid State Chem., 1980, 12, 185–271 CrossRef.
- X. Ma, X. Kou, Y. Xu, D. Yang and P. Miao, Nanoscale Adv., 2019, 1, 486–489 RSC.
- M. António, R. Ferreira, R. Vitorino and A. L. Daniel-da-Silva, Talanta, 2020, 214, 120868 CrossRef PubMed.
- L. Guo, J. A. Jackman, H. H. Yang, P. Chen, N. J. Cho and D. H. Kim, Nano Today, 2015, 10, 213–239 CrossRef CAS.
- Y. Chen, Y. Xianyu and X. Jiang, Acc. Chem. Res., 2017, 50, 310–319 CrossRef CAS PubMed.
- J. E. Kim, J. H. Choi, M. Colas, D. H. Kim and H. Lee, Biosens. Bioelectron., 2016, 80, 543–559 CrossRef CAS PubMed.
- K. C. Grabar, R. G. Freeman, M. B. Hommer and M. J. Natan, Anal. Chem., 1995, 67, 735–743 CrossRef CAS.
- L. Meng, J. H. Yin, Y. Yuan and N. Xu, RSC Adv., 2018, 8, 9327–9333 RSC.
- J. P. Oliveira, A. R. Prado, W. J. Keijok, P. W. P. Antunes, E. R. Yapuchura and M. C. C. Guimarães, Sci. Rep., 2019, 9, 1–8 CrossRef CAS PubMed.
- J. H. Williamson, Can. J. Phys., 1968, 46, 1845–1847 CrossRef.
- A. Shrivastava and V. Gupta, Chron. Young Sci., 2011, 2, 21 CrossRef.
- A. Rahman, A. K. Rahman, W. Ghann, H. Kang and J. Uddin, Int. J. Biosens. Bioelectron., 2018, 4, 169–174 Search PubMed.
- S. Bhattacharjee, J. Controlled Release, 2016, 235, 337–351 CrossRef CAS PubMed.
- R. Kurrey, M. K. Deb, K. Shrivas, B. R. Khalkho, J. Nirmalkar, D. Sinha and S. Jha, Anal. Bioanal. Chem., 2019, 411, 6943–6957 CrossRef CAS PubMed.
- S. K. Vashist, E. M. Schneider, E. Lam, S. Hrapovic and J. H. T. Luong, Sci. Rep., 2014, 4, 1–7 Search PubMed.
- P. Englebienne, Analyst, 1998, 123, 1599–1603 RSC.
- B. Veigas, A. Matias, T. Calmeiro, E. Fortunato, A. R. Fernandes and P. V. Baptista, Analyst, 2019, 144, 3613–3619 RSC.
- Y. S. Li, X. T. Li, L. G. Yu, L. Wang, Z. Y. Shi and X. L. Guo, Int. J. Biol. Macromol., 2020, 142, 463–473 CrossRef CAS PubMed.
- L. A. Currie, Pure Appl. Chem., 1995, 67, 1699–1723 CrossRef CAS.
- T. F. S. Invitrogen, Human Galectin-3 ELISA Kit, https://www.thermofisher.com/elisa/product/Galectin-3-Human-ELISA-Kit/BMS279-4, accessed 3 March 2022 Search PubMed.
- M. Stasenko, E. Smith, O. Yeku, K. J. Park, I. Laster, K. Lee, S. Walderich, E. Spriggs, B. Rueda, B. Weigelt, D. Zamarin, T. D. Rao and D. R. Spriggs, Sci. Rep., 2021, 11, 1–13 CrossRef PubMed.
- M. Hashimoto, T. Miki, I. V. Chang, H. Tsutsumi and H. Mihara, Bioorg. Med. Chem. Lett., 2021, 37, 127835 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ay00347g |
| This journal is © The Royal Society of Chemistry 2023 |

