An allosteric DNA switch–mediated catalytic DNA circuit for ratiometric and sensitive nucleic acid detection†
Xiaofan
Zhang
a,
Shunjun
Xie
a,
Xue
Chen
a,
Li
Wang
*b,
Fang
Li
*a and
Shufeng
Liu
 *b
*b
aCollege of Marine Science and Biological Engineering, Qingdao University of Science and Technology, 53 Zhengzhou Road, Qingdao 266042, China. E-mail: fangli@qust.edu.cn
bCollege of Chemistry and Chemical Engineering, Yantai University, 30 Qingquan Road, Yantai, 264005, China. E-mail: wangliytu@126.com; sliu@ytu.edu.cn
First published on 29th November 2022
Abstract
Herein, a new allosteric DNA switch–mediated catalytic DNA circuit reaction strategy has been proposed for ratiometric and sensitive nucleic acid detection. The sensing system was based on two DNA hybrid probes, each of which was constructed by annealing a reconfigurable DNA hairpin with single-stranded DNA. Upon target recognition by the first DNA hybrid probe, a reconfigurable DNA switch was liberated, triggering a toehold-mediated strand displacement reaction (TSDR) with the second DNA hybrid probe, which was accompanied by the release of another reconfigurable DNA switch. This released allosteric DNA switch could further interact with the first hybrid DNA probe via the TSDR strategy to form a reciprocal strand displacement network between the two DNA hybrid probes. Theoretically, this reciprocal strand displacement reaction would continue till the complete consumption of the reaction substrates. Thus, it provides a new signal amplification method leading toward target recognition. More interestingly, it creates a ratiometric signal response mode for target recognition, which involves the fluorescence increment of one fluorophore (Cy5) and concurrent decrement of another fluorophore (Cy3) accompanied by the target-triggered reciprocal strand displacement reaction. This process could achieve a low detection limit of about 0.1 pM toward the target nucleic acid and selective discrimination toward different mismatched targets. It could also be applied for detection in a serum sample. Thus, the developed catalytic DNA circuit reaction strategy together with ratiometric signal readout provides a new avenue for programmable, reliable and sensitive detection of nucleic acids and might also pave the way for developing more advanced DNA circuits or biosensors.
Introduction
Nucleic acids, which are responsible for genetic information storage, protein synthesis and many important physiological and pathological activities, can be used as biomarkers for individual discrimination and diagnostic applications as well as building units in DNA nanotechnology and information processing.1,2 The sensitive and accurate analysis of specific nucleic acid sequences or nucleic acid–related analytes is highly demanded in applications such as pathogen identification, disease diagnosis and genotyping.3–6 To achieve the sensitive detection of specific nucleic acid sequences, sequence or signal amplification methods are often necessary before measurement to overcome the intrinsic limitations of the method or instrument itself. The polymerase chain reaction (PCR) amplification technique is considered the gold standard for nucleic acid analysis.7,8 However, its requirements of specialized equipment, reagents, relatively complicated temperature control and trained personnel are not easily available for point-of-care applications. Moreover, non-specific amplification is difficult to avoid and increases the probability of false positive output. To address these issues, isothermal nucleic acid amplification strategies, such as rolling circle amplification (RCA), loop-mediated isothermal amplification (LAMP), helicase-dependent amplification (HDA), recombinase polymerase amplification (RPA), and strand displacement amplification (SDA), have been developed for nucleic acid analysis.9–13 However, these methods also suffer from complexity, time consumption and high cost. Moreover, the activity of protein enzymes in these methods might be easily influenced by environmental factors, limiting their wide application in clinical analysis.Compared with the above-mentioned strategies, the enzyme-free nucleic acid assembly–based signal amplification strategy is very attractive for bioanalysis owing to its advantages such as low cost, high specificity, and programmable operation.14–18 The toehold-mediated strand displacement reaction (TSDR) is generally used to harness nucleic acid assemblies for the development of DNA nanotechnology and DNA circuits applied in signal transduction, magnification or biocomputation.19–27 The typical catalytic DNA circuit–based signal amplification strategies mediated via TSDR include the so-called catalytic hairpin DNA assembly (CHA) and hybridization chain reactions (HCR).28–33 They usually take advantage of the metastable nucleic acid hairpins that act as assembly units, and thus their thermodynamic tendency for stable assembly even in the absence of the targets poses unfavorable challenges to the sensing performance. Further, very careful design of the assembly units and thorough optimization of the experimental conditions are usually needed for the analysis of different targets, which limits their application generality to some extent. Thus, the development of more simple and robust signal amplification or transduction strategies with no involvement of the metastable assembly units might provide enormous opportunities for application in bioanalysis and DNA circuit design.
Herein, an allosteric DNA switch–mediated catalytic DNA circuit strategy was developed as a signal magnifier for sensitive target analysis. The sensing system consisted of two DNA hybrid probes, each of which was obtained by annealing a reconfigurable DNA hairpin with a single-stranded DNA. Since the DNA hairpin was caged inside the DNA hybrid probe, the toehold domain was positioned away from the branch migration domain, due to which the toehold-mediated strand displacement reaction (TSDR) activity was blocked. In the presence of target DNA, it could displace the first DNA switch from the first DNA hybrid probe via a TSDR reaction. The released DNA switch reconfigured into a hairpin-like DNA structure, inducing the toehold and branch migration domains to move to adjacent positions, recovering the TSDR activity.34,35 Then, its reaction with the second DNA hybrid probe via TSDR released the second DNA switch, which could further interact with the first hybrid probe via TSDR to circularly liberate the first DNA switch. Thus, the reciprocal strand displacement reaction network between two hybrid probes could afford signal magnification toward the initial target recognition event. Moreover, the designed DNA hybrid probes created the opportunity for ratiometric signal response toward the target. The ratiometric method shows potential in reliable bioanalysis owing to the presence of two or more concurrent signal responses, thus providing signal calibration toward target recognition.36–40 In the current system, the first hybrid probe was labeled with a fluorophore (Cy5) and a quencher (BHQ) in the respective strands for fluorescence quenching. The second hybrid probe was labeled with only a fluorophore (Cy3). These two fluorophores (Cy5 and Cy3) shared the same quencher, namely BHQ. Following the target DNA–triggered reciprocal strand displacement process between the two hybrid probes, the fluorescence response of Cy5 was enhanced, but the fluorescence response of Cy3 was synchronously attenuated, showing ratiometric signal response for target analysis.
Experimental section
Chemicals and reagents
All the oligonucleotides were HPLC-purified and synthesized by Sangon Biotech. Co., Ltd (Shanghai, China), and their base sequences are shown in Table S1.† Tris(hydroxymethyl)aminomethane was obtained from Sigma-Aldrich (St. Louis, MO, USA). Acryl/bis 40% solution (29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), N,N,N′,N′-tetramethylethylenediamine (TEMED), ammonium persulfate (APS) and ethidium bromide (EB) were obtained from Sangon Biotech. Co., Ltd (Shanghai, China). All other chemicals were obtained from Shanghai Chemical Reagents Co., Ltd (Shanghai, China). Deionized water (18.2 MΩ cm) collected from a Milli-Q water purification system was used in all experiments (Millipore Corp., Bedford, MA, USA).
1), N,N,N′,N′-tetramethylethylenediamine (TEMED), ammonium persulfate (APS) and ethidium bromide (EB) were obtained from Sangon Biotech. Co., Ltd (Shanghai, China). All other chemicals were obtained from Shanghai Chemical Reagents Co., Ltd (Shanghai, China). Deionized water (18.2 MΩ cm) collected from a Milli-Q water purification system was used in all experiments (Millipore Corp., Bedford, MA, USA).
Preparation of the probes AB and CD
Probe AB was obtained by annealing the same concentration (each 10 μM) of strand A and strand B in TE buffer (10 mM Tris–HCl, 1 mM EDTA, 12.5 mM MgCl2, pH 7.8), which was first heated to 95 °C for 5 minutes and then cooled to room temperature. Probe CD was obtained by annealing the strands of C and D in the same way. The obtained probes AB and CD were stored in the dark in a refrigerator (4 °C) before use.Target DNA detection by the sensing system
Target DNA detection by the allosteric DNA switch–mediated catalytic DNA circuit strategy was tested by mixing 2.5 μM probe AB (8 μL) and 2.5 μM probe CD (8 μL) with different concentrations of the target in TE buffer (10 mM Tris–HCl, 1 mM EDTA, 12.5 mM MgCl2, pH 7.8) in a total sample volume of 100 μL. Except for the probe concentration optimization experiments, 200 nM of probes AB and CD were always used in the testing sample. Before fluorescence measurement, the mixture was incubated at 25 °C for 180 min (other temperatures such as 20, 30 and 35 °C were also studied for optimization). The above assay procedure was also applied for target DNA detection in the diluted serum sample.In the case of the allosteric DNA switch–mediated strand displacement strategy (herein, no cyclic amplification step was involved), the above procedure was applied except that probe CD was replaced by the same concentration of probe CD2, which was obtained by annealing strand C and strand D2.
Recovery experiments were carried out according to the same procedure with three different concentrations of target DNA (50 pM, 500 pM and 5 nM) spiked in 10% diluted sheep serum.
Gel electrophoresis characterization
The samples were mixed with the 6× loading buffer (Sangon Biotech. Co., Ltd, Shanghai, China) and run on a 17% native polyacrylamide gel in 1× TAE buffer. The gel electrophoresis experiments were operated at 180 V for 3 min followed by 80 min at 135 V, and then, the gels were stained using the EB dye. The gel electrophoresis images were captured by an FR-980 gel image analysis system (Shanghai, China).Fluorescence measurements
Fluorescence spectroscopy was performed on a Hitachi F-2700 spectrometer (Hitachi, Japan). The corresponding parameters were set as follows: the scan rate was 1500 nm min−1, 24 photomultiplier voltage was 700 V, the excitation wavelength was 540 nm for Cy3 and 635 nm for Cy5, and the slits of excitation and emission were 5 nm/5 nm for Cy3 and 5 nm/10 nm for Cy5, respectively.Results and discussion
Detection principle of the allosteric DNA switch–mediated catalytic DNA circuit strategy
The design principle of the proposed allosteric DNA switch–mediated catalytic DNA circuit reaction strategy is schematically illustrated in Scheme 1. The sensing system consisted of two DNA hybrid probes (probes AB and CD). These two probes were obtained by annealing the corresponding strands (strands A and B for probe AB, strands C and D for probe CD). Among them, the strands B and D could be reconfigured into the DNA stem-loop structures as the allosteric DNA switches were liberated from probes AB and CD, respectively. For the allosteric DNA switches of B and D, the toehold and branch migration regions were brought adjacent to each other, and then their toehold-mediated strand displacement reactions (TSDR) with the respective probes CD and AB could automatically occur. However, the probes AB and CD could be not directly and reciprocally communicated between them since the toehold and branch migration regions in the caged strands of B and D were kept away for blocking the TSDR activity. In probe AB, the fluorophore (Cy5) and quencher (BHQ) were labeled at the 5′ terminus of strand A and the internal position of strand B, respectively. They were located at the opposite base positions on the respective strands, inducing Cy5 fluorescence quenching by BHQ. In probe CD, another fluorophore (Cy3) was labeled at the corresponding position in strand C but no quencher was attached to strand D. Thus, in the initial state of the sensing system with probes AB and CD, only the fluorescence response of Cy3 from probe CD would occur. Upon addition of the target DNA, it would hybridize with strand A in probe AB via a TSDR process to liberate the caged strand B. Then, the fluorescence response of Cy5 in strand A would be activated, and its fluorescence intensity is related to the initial target recognition event. The released strand B would then be reconfigured into a stem-loop structure, bringing the toehold and branch migration domains adjacent to each other. Thus, the TSDR reaction between the allosteric switch B and probe CD would proceed, resulting in the formation of the BC hybrid and the release of strand D. After the formation of the BC hybrid, the Cy3 group on strand C remains adjacent to the quencher BHQ at the opposite position of strand B, leading to the fluorescence quenching of Cy3. The released strand D would be also reconfigured into a stem-loop structure, bringing the corresponding toehold and branch migration domains close for the successive TSDR process with probe AB. The sequence encompassed by the toehold and branch migration domains in switch D is designed to be consistent with the target DNA sequence. Thus, the released switch D would be used as a target DNA substitute and its further interaction with probe AB via a TSDR reaction induces the formation of an AD hybrid and the release of strand B. The released strand B would repeat the above cyclic process and constitute the reciprocal strand displacement reaction network between probe AB and CD. Accompanying the above process, the fluorescence response of Cy5 could be enhanced continuously, while the fluorescence response of Cy3 will attenuate accordingly, contributing to the amplified and ratiometric signal response of the system toward the target DNA.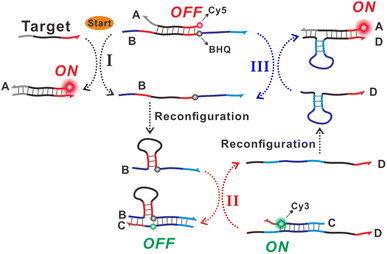 | ||
| Scheme 1 Schematic illustration of the allosteric DNA switch–mediated catalytic DNA circuit strategy for ratiometric nucleic acid sensing. | ||
Detection feasibility of the constructed sensing system toward the target
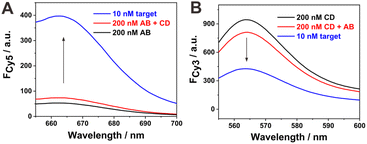
The nucleic acid detection feasibility of the proposed sensing strategy was first verified by the fluorescence method. Firstly, the fluorescence responses of the individual probes (probe AB or CD) were recorded, as shown in Fig. 1A and B (black curves). Only a very low Cy5 fluorescence response for probe AB could be observed owing to the fluorescence quenching of Cy5 by BHQ. The sole probe CD showed high fluorescence of Cy3. After mixing probes AB and CD at the same concentration, a slight increase in Cy5 fluorescence and a decrease in Cy3 fluorescence could be seen (Fig. 1A and B, red curves). Compared with the fluorescence response before mixing, the change in the background fluorescence response, especially the decrease in Cy3 fluorescence, might be attributed to the fluorescence quenching of Cy3 to some extent caused by the approaching or spontaneous association of the two hybrid probes. The further introduction of target DNA could induce a distinct increase in Cy5 fluorescence response and a concurrent decrease in Cy3 fluorescence (Fig. 1A and B, blue curves). Thus, the fabricated sensing system showed detection feasibility toward the target nucleic acid, and the opposite trends of the two signal responses suggested a ratiometric mode for target nucleic acid analysis.The whole sensing strategy could be subdivided into three cascade steps. Then, each step was individually characterized to support the reaction principle. The corresponding results are shown in Fig. S1.† Step 1 was based on the TSDR between target DNA and probe AB. It can be seen from Fig. S1A† that the mixing of probe AB with the target DNA induced the fluorescence recovery of Cy5, suggesting the release of strand B from probe AB. Step II was based on the TSDR between the allosteric switch B and probe CD. Then, we directly mixed strand B and probe CD for fluorescence characterization (Fig. S1B†). After the addition of strand B, the fluorescence response of Cy3 decreased significantly, suggesting the formation of the BC hybrid via the TSDR reaction between switch B and probe CD. In the BC hybrid, the fluorescence of Cy3 in strand C was effectively quenched by the adjacent BHQ quencher in strand B. Step III was similar to step I except that the target DNA was substituted by switch D. The addition of strand D also led to a significant enhancement in Cy5 fluorescence (Fig. S1C†), indicating that switch D could also hybridize with probe AB via a TSDR reaction process. The sensing system was also characterized by gel electrophoresis (Fig. S2†). Lanes 2–5 represent the bands of probe AB, probe CD, hybrid BC and hybrid AD, respectively. After the mixing of probe AB and CD, only the bands related to probe AB and CD could be seen, but no new bands appeared, suggesting no evident reaction between them in the absence of target DNA (lane 6). However, in the presence of target DNA, the bands of AB and CD became dim, and new bright bands appeared corresponding to the hybrid of target DNA and strand A, the immediate product of switches B and D (lane 8).
Optimization of the experimental conditions
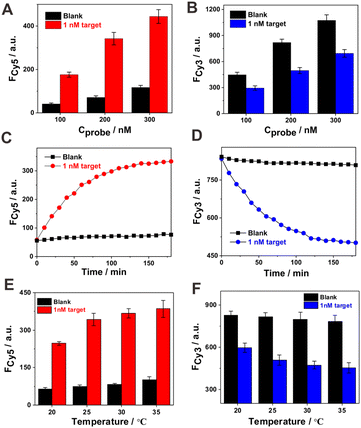
The concentration of probes AB and CD might affect the detection performance to some extent. Three different concentrations of probes AB and CD were studied for the signal response toward 1 nM target DNA. It can be seen from Fig. 2A and B that 200 nM of probes AB and CD demonstrated a better signal response (the increment of Cy5 and decrement of Cy3 fluorescence signals) toward 1 nM target DNA compared with a lower (100 nM) or higher (300 nM) concentration. The low concentration of probes limited the signal response toward the target DNA, and the high concentration of probes induced a relatively high background signal. The time-dependent fluorescence response toward 1 nM target DNA was monitored. As shown in Fig. 2C (Cy5) and 2D (Cy3), evident increase in the Cy5 fluorescence signal and decrease in the Cy3 signal could be observed during the whole reaction process in the case of target DNA. Moreover, a relatively rapid fluorescence signal change occurred at the initial stage (within 1 h), and then, the trend slowed down till signal saturation. However, in the absence of target DNA, only very weak fluorescence response changes could be observed for Cy5 and Cy3 during the whole reaction process. In this study, we employed a relatively long reaction time of 180 min to ensure sufficient target DNA–induced signal output. However, a short reaction time of less than 60 min was found to be effective for signal tracking toward the target recognition event. The reaction temperature was then optimized by considering the signal response and background level at four different temperatures (20, 25, 30, and 35 °C). It can be seen from Fig. 2E and F that the developed sensing system could detect target DNA effectively in the studied temperature range. The elevation in reaction temperature could induce an increase in signal response, but the background signal level was also elevated. At a lower reaction temperature of 20 °C, the signal response was inhibited to some extent. Although the fluorescence signal change induced by the target at 30 °C was slightly larger than that at 25 °C, the signal response-to-background ratio between them was comparable. Thus, we took the more friendly temperature 25 °C as the optimal temperature for the following experiments.The detection performance of the sensing system
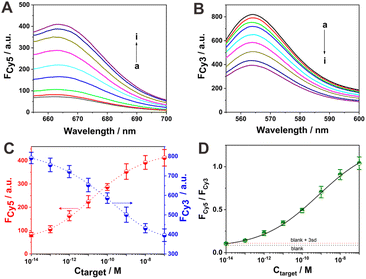
Under the optimized sensing conditions, we then explored the detection performance of the sensing system toward the target DNA. The fluorescence spectra of the sensing system at different concentrations of target DNA are shown in Fig. 3A and B. With the increment in target DNA concentration from 10 fM to 100 nM, the fluorescence response of Cy5 enhanced continuously and that of Cy3 decreased accordingly, indicating the dependence of these two signal response behaviors on target concentration. The calibration curves of the fluorescence responses of Cy5 and Cy3 versus the target amount are displayed in Fig. 3C. Such a dual-signal reporting system with opposite response trends toward target DNA provides an effective means for the ratiometric analysis of target DNA. Furthermore, the calibration curve of the fluorescence ratio of Cy5 to Cy3 (FCy5/FCy3) versus different concentrations of target DNA was obtained, as shown in Fig. 3D. At a low target DNA concentration of 0.1 pM, all the signal responses, including the Cy5 fluorescence increase, Cy3 fluorescence decrease and the ratio between, them could be easily discriminated. The detection limit for the target was determined according to the typical 3σ method. The ratiometric response of Cy5 to Cy3 at 0.1 pM target exceeded the calculated value (blank response plus three times its standard deviation; the standard deviation for the blank solution was based on five repetitive experiments). Thus, the detection limit of the current catalytic DNA circuit reaction strategy was determined as 0.1 pM, which is superior to many reported enzyme-free nucleic acid sensing strategies (Table S2†). The relative standard deviations (RSD) at three concentrations of the target (10 pM, 1 nM and 100 nM) were obtained based on the ratiometric response (FCy5/FCy3) as 7.88%, 9.50% and 7.02%, respectively, from three repetitive experiments, suggesting the relatively good detection reproducibility of the current strategy for target analysis. Furthermore, its ratiometric feature is attractive for reliable nucleic acid analysis.In order to confirm the signal amplification ability of the current strategy, a control experiment was designed for comparison with strand D2 as the substitute for D. As schematically illustrated in Fig. S3,† the designed sensing system underwent two cascade TSDR processes: one between target DNA and probe AB, and the other between the released switch B and probe CD2 (probe CD2 was obtained by annealing strand C with strand D2). However, the D2 strand released from probe CD2 could not further participate in the signal amplification reaction with probe AB. Thus, this can be considered a simple DNA switch–mediated strand displacement strategy. The designed control system was also used for the ratiometric detection of target DNA. However, owing to no signal amplification, the fluorescence responses in the presence of 1 nM target DNA (fluorescence increment of Cy5 and descent of Cy3) were distinctly lower compared with that of the allosteric DNA switch–mediated catalytic DNA circuit system. Moreover, a relatively slow signal response toward target DNA was observed in this control system (Fig. S4†). The detection performance of this control sensing system toward the same target DNA was also studied for further comparison. The target DNA–dependent fluorescence response behaviors of Cy5 and Cy3 are revealed in Fig. S5A and S5B,† respectively. Particularly, the fluorescence response of Cy5 increased and that of Cy3 decreased accordingly with the increment of target DNA. The relationships of the fluorescence intensities (Cy5 and Cy3) and the ratio between them with the target DNA concentration are shown in Fig. S5C and S5D,† respectively. It could only achieve target DNA detection with a low limit of about 10 pM, which is inferior to that of the allosteric DNA switch–mediated catalytic DNA circuit strategy by about two orders of magnitude. This further verified the signal amplification ability of the proposed allosteric DNA switch–mediated catalytic DNA circuit strategy.
Detection selectivity of the proposed sensing strategy
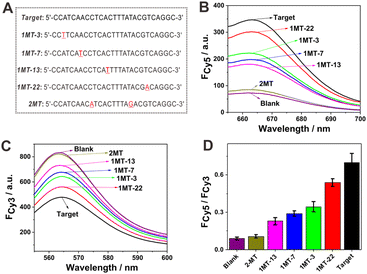
The detection selectivity of the proposed sensing strategy was then examined by using mismatched DNA sequences as analytes. A two-base mismatched DNA sequence (2MT) and four one-base mismatched DNA sequences (1MT-3, 1MT-7, 1MT-13 and 1MT-22) were chosen. In the names of the one-base mismatched DNA sequences, the numbers indicate the position of the mismatched base in the DNA sequence. For example, the number 3 in 1MT-3 indicates that the third base from the 5′-terminus is mismatched. The mismatched base of 1MT-3 was located at the toehold region and that of 1MT-7 was at the joint between the toehold and branch migration regions. The mismatched bases of 1MT-13 and 1MT-22 were chosen at the branch migration region. The mismatched DNA sequences are shown in Fig. 4A, and the mismatched bases are underlined and marked in red. The corresponding fluorescence spectra of Cy5 and Cy3 in the mismatched DNA sequences are shown in Fig. 4B and C, respectively. In addition, the Cy5 to Cy3 fluorescence ratios of the different DNA sequences are shown in Fig. 4D. As expected, the 2MT sequence hardly induced any fluorescence signal change. In the case of 1MT, the fluorescence signal responses, including the fluorescence increase of Cy5, fluorescence decrease of Cy3, and the ratio of Cy5 to Cy3, were lower than those of the complementary target DNA. It was also found that the mismatched base positions exerted different impacts on the fluorescence responses. It has been known that the mismatched base posited at the toehold domain, especially at the joint between the toehold and branch migration domains, can evidently inhibit the TSDR process.41,42 This effect was less evident when the mismatched base was positioned at the branch migration region. It could be seen that 1MT-22 showed a minimal impact on the fluorescence response among the studied 1MT sequences. In this case, the mismatched base was distant from the toehold region, and hence its TSDR process with probe AB would be less affected. The fluorescence responses of 1MT-3, 1MT-7 and 1MT-13 were evidently lower than that of the complementary target DNA. In the cases of 1MT-3 and 1MT-7, the mismatched bases positioned at the toehold region could obviously weaken their TSDR with probe AB. The mechanism underlying the lower fluorescence response of 1MT-13 is not currently clear. In 1MT-13, the mismatched base situated at the branch migration domain of TMSD is also related to the binding domain for the loop sequence of switch B. Thus, it can be speculated that the mismatched base of 1MT-13 would not only limit the TSDR reaction process but also affect the reconfiguration process of DNA switch B to some extent. These experiments demonstrated the excellent ability of the current allosteric DNA switch–mediated catalytic DNA circuit reaction strategy to discriminate mismatched targets.Application potential of the sensing system in serum samples
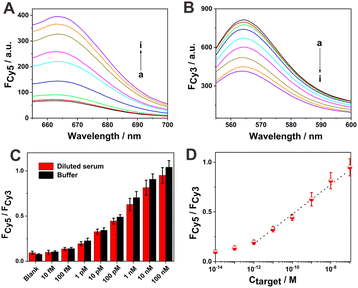
The application potential of the current sensing system in complex biological matrices was studied by assessing its target DNA detection ability in diluted sheep serum. The corresponding fluorescence spectra of Cy5 and Cy3 at different target DNA concentrations are shown in Fig. 5A and B, respectively. Again, the observed simultaneous fluorescence increment of Cy5 and decrement of Cy3 upon increasing target DNA concentrations suggest the reliable detection capability of the current sensing strategy in relatively complicated biological samples, such as diluted serum. The fluorescence ratios of Cy5 to Cy3 at different concentrations of target DNA in the serum system were basically comparable with those in the buffer system (Fig. 5C). This indicates that the serum matrix could not induce evident interferences toward target DNA detection. The calibration curve of the fluorescence ratio of Cy5 to Cy3 versus target DNA concentration is shown in Fig. 5D. A low detection limit of about 0.1 pM for target DNA could also be achieved in diluted serum, suggesting again the good target DNA detection ability of the current sensing system even in the relatively complicated biological matrix. A linear relationship between the fluorescence response (FCy5/FCy3) and target DNA concentration could be plotted in the range of 1 pM to 100 nM. The regression equation was expressed as FCy5/FCy3 = 1.967 + 0.149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(Ctarget/M) with a regression coefficient of 0.9892. According to this calibration curve, the recovery experiment was then conducted with spiked target DNA in the diluted serum (Table S3†). Relatively satisfactory recovery rates of 83–119% with relative standard deviations of 6.9–14.3% could be obtained, further revealing the application potential of this strategy in relatively complicated biological matrices.
log(Ctarget/M) with a regression coefficient of 0.9892. According to this calibration curve, the recovery experiment was then conducted with spiked target DNA in the diluted serum (Table S3†). Relatively satisfactory recovery rates of 83–119% with relative standard deviations of 6.9–14.3% could be obtained, further revealing the application potential of this strategy in relatively complicated biological matrices.
Conclusions
Herein, a new allosteric DNA switch–mediated catalytic DNA circuit strategy was developed for ratiometric and sensitive nucleic acid analysis. It was based on two designed DNA hybrid probes, and a reciprocal strand displacement reaction network between them could be realized by target-triggered allosteric DNA switch cascades for signal amplification. The ratiometric signal responses (the simultaneous signal increment in Cy5 and the decrement of Cy3 signals) provide an attractive means for reliable analysis of nucleic acids. The achieved detection limit toward target DNA was as low as 0.1 pM, which is superior to most of the reported enzyme-free nucleic acid sensing strategies. It also demonstrated selective detection toward different mismatched targets and applicative potential in relatively complex biological samples, such as diluted serum. The proposed sensing strategy displays the features of facile probe design, programmable operation, free of protein enzymes, and ratiometric and sensitive response. It thus provides an attractive means for the analysis of nucleic acids and might pave the way for developing more advanced DNA circuits and various biosensors.Conflicts of interest
The authors declared that they have no commercial or financial conflicts of interest.Acknowledgements
We acknowledge the support of the Natural Science Foundation of Shandong Province (ZR2022MB035, JQ201704), the National Natural Science Foundation of China (21475072), and the Key Research and Development Program of Shandong Province of China (2016GSF201208).Notes and references
- H. Schwarzenbach, D. S. Hoon and K. Pantel, Nat. Rev. Cancer, 2011, 11, 426–437 CrossRef
.
- R. Breaker, Nature, 2004, 432, 838–845 CrossRef
.
- K. Yang and J. C. Chaput, J. Am. Chem. Soc., 2021, 143, 8957–8961 CrossRef
.
- J. Das, I. Ivanov, L. Montermini, J. Rak, E. H. Sargent and S. O. Kelley, Nat. Chem., 2015, 7, 569–575 CrossRef PubMed
.
- X. Wang, X. Chen, R. Zhou, P. Hu, K. Huang and P. Chen, Anal. Chem., 2021, 93, 3889–3897 CrossRef
.
- R. Dai, P. Hu, X. Wang, S. Wang, X. Song, K. Huang and P. Chen, Analyst, 2019, 144, 4407–4412 RSC
.
- B. Peeters, S. Safdar, D. Daems, P. Goos, D. Spasic and J. Lammertyn, Anal. Chem., 2020, 92, 10783–10791 CrossRef PubMed
.
- R. Schoske, P. M. Vallone, C. M. Ruitberg and J. M. Butler, Anal. Bioanal. Chem., 2003, 375, 333–343 CrossRef
.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef PubMed
.
- H. Shi, X. Mao, X. Chen, Z. Wang, K. Wang and X. Zhu, Biosens. Bioelectron., 2017, 91, 136–142 CrossRef PubMed
.
- H. Yuan, Y. Chao, S. Li, M. Y. H. Tang, Y. Huang, Y. Che, A. S. T. Wong, T. Zhang and H. C. Shum, Anal. Chem., 2018, 90, 13173–13177 CrossRef
.
- M. Vincent, Y. Xu and H. Kong, EMBO Rep., 2004, 5, 795–800 CrossRef CAS PubMed
.
- S. Lutz, P. Weber, M. Focke, B. Faltin, J. Hoffmann, C. Muller, D. Mark, G. Roth, P. Munday, N. Armes, O. Piepenburg, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 887–893 RSC
.
- N. Srinivas, J. Parkin, G. Seelig, E. Winfree and D. Soloveichik, Science, 2017, 358, eaal2052 CrossRef
.
- H. Wang, C. Li, X. Liu, X. Zhou and F. Wang, Chem. Sci., 2018, 9, 5842–5849 RSC
.
- W. Li, L. Wang and W. Jiang, Chem. Commun., 2017, 53, 5527–5530 RSC
.
- P. Chen, Y. Wang, Y. He, K. Huang, X. Wang, R. Zhou, T. Liu, R. Qu, J. Zhou, W. Peng, M. Li, Y. Bai, J. Chen, J. Huang, J. Geng, Y. Xie, W. Hu and B. Ying, ACS Nano, 2021, 15, 11634–11643 CrossRef CAS
.
- P. Hu, X. Wang, L. Wei, R. Dai, X. Yuan, K. Huang and P. Chen, J. Mater. Chem. B, 2019, 7, 4778–4783 RSC
.
- F. C. Simmel, B. Yurke and H. R. Singh, Chem. Rev., 2019, 119, 6326–6369 CrossRef CAS PubMed
.
- P. Irmisch, T. E. Ouldridge and R. Seidel, J. Am. Chem. Soc., 2020, 142, 11451–11463 CrossRef CAS
.
- P. Hu, M. Li, X. Wei, B. Yang, Y. Li, C. Y. Li and J. Du, Anal. Chem., 2018, 90, 9751–9760 CrossRef
.
- A. J. Genot, D. Y. Zhang, J. Bath and A. J. Turberfield, J. Am. Chem. Soc., 2011, 133, 2177–2182 CrossRef PubMed
.
- G. Seelig, D. Soloveichik, D. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef PubMed
.
- J. Chen, L. Tang, X. Chu and J. Jiang, Analyst, 2017, 142, 3048–3061 RSC
.
- X. Wang, Z. Luo, Q. Xie, Z. Huang, M. Wu and Y. Duan, Anal. Chim. Acta, 2020, 1139, 138–145 CrossRef PubMed
.
- W. Meng, R. A. Muscat, M. L. McKee, P. J. Milnes, A. H. El-Sagheer, J. Bath, B. G. Davis, T. Brown, R. K. O'Reilly and A. J. Turberfield, Nat. Chem., 2016, 8, 542–548 CrossRef PubMed
.
- C. Gu, X. Kong, X. Liu, P. Gai and F. Li, Anal. Chem., 2019, 91, 8697–8704 CrossRef PubMed
.
- Z. Wu, H. Fan, N. S. R. Satyavolu, W. Wang, R. Lake, J. H. Jiang and Y. Lu, Angew. Chem., Int. Ed., 2017, 56, 8721–8725 CrossRef
.
- R. Y. Zhang, S. H. Luo, X. M. Lin, X. M. Hu, Y. Zhang, X. H. Zhang, C. M. Wu, L. Zheng and Q. Wang, Anal. Chim. Acta, 2021, 1157, 338396 CrossRef PubMed
.
- J. Shang, C. Li, F. Li, Q. Wang, B. Yuan and F. Wang, Anal. Chem., 2021, 93, 2403–2410 CrossRef PubMed
.
- G. Xu, H. Zhao, J. Reboud and J. M. Cooper, ACS Nano, 2018, 12, 7213–7219 CrossRef CAS PubMed
.
- Y. Zhao, J. Xiang, H. Cheng, X. Liu and F. Li, Biosens. Bioelectron., 2021, 194, 113581 CrossRef CAS PubMed
.
- P. Chen, Y. Bai, S. Tang, N. Wang, Y. He, K. Huang, J. Huang, B. Ying and Y. Cao, Nano Lett., 2022, 22, 1710–1717 CrossRef CAS PubMed
.
- W. Lai, L. Ren, Q. Tang, X. Qu, J. Li, L. Wang, L. Li, C. Fan and H. Pei, ACS Nano, 2018, 12, 7093–7099 CrossRef CAS
.
- Y. Guo, D. Yao, B. Zheng, X. Sun, X. Zhou, B. Wei, S. Xiao, M. He, C. Li and H. Liang, ACS Nano, 2020, 14, 8317–8327 CrossRef CAS PubMed
.
- X. J. Yang, K. Zhang, T. T. Zhang, J. J. Xu and H. Y. Chen, Anal. Chem., 2017, 89, 4216–4222 CrossRef CAS PubMed
.
- Z. M. Ying, Z. Wu, B. Tu, W. Tan and J. H. Jiang, J. Am. Chem. Soc., 2017, 139, 9779–9782 CrossRef CAS PubMed
.
- S. Wang, S. Lu, J. Zhao and X. Yang, ACS Appl. Mater. Interfaces, 2019, 11, 25066–25073 CrossRef CAS
.
- M. Liu, R. Shen, H. Li, Y. Jia, P. I. Mak and R. P. Martins, J. Mater. Chem. C, 2022, 10, 655–664 RSC
.
- L. Yang, X. Yin, B. An and F. Li, Anal. Chem., 2021, 93, 1709–1716 CrossRef CAS
.
- R. R. Machinek, T. E. Ouldridge, N. E. Haley, J. Bath and A. J. Turberfield, Nat. Commun., 2014, 5, 5324 CrossRef CAS PubMed
.
- Z. Zhang, D. Zeng, H. Ma, G. Feng, J. Hu, L. He, C. Li and C. Fan, Small, 2010, 6, 1854–1858 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: DNA sequences (Table S1); additional experimental characterizations (Fig. S1–S5), performance comparison (Table S2) and recovery experiment (Table S3). See DOI: https://doi.org/10.1039/d2ay01751b |
| This journal is © The Royal Society of Chemistry 2023 |