Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel screening method for free non-standard amino acids in human plasma samples using AccQ·Tag reagents and LC-MS/MS†
Daniel H. J.
Ng
 *a,
Li Yan
Chan
*a,
Li Yan
Chan
 a,
Laura
Fitzner
b,
Julia Katharina
Keppler
a,
Laura
Fitzner
b,
Julia Katharina
Keppler
 bc,
Shareef M.
Ismail
d,
Simon
Hird
e,
Peter
Hancock
e,
Schwarz
Karin
b and
Demetrowitsch
Tobias
b
bc,
Shareef M.
Ismail
d,
Simon
Hird
e,
Peter
Hancock
e,
Schwarz
Karin
b and
Demetrowitsch
Tobias
b
aInternational Food and Water Research Centre, Waters Pacific Pte Ltd, 1 Science Park Road #01-10, The Capricorn, Singapore Science Park II, Singapore, 117528, Singapore. E-mail: Daniel_ng@waters.com
bDivision of Food Technology, Kiel University, Heinrich-Hecht Platz 10, Kiel, 24118, Germany
cLaboratory of Food Process Engineering, Wageningen University & Research, Bornse Weilanden 9, Wageningen, 6708 WG, the Netherlands
dGlobal Service Education, Waters Pacific Pte Ltd, 1 Science Park Road #01-10, The Capricorn, Singapore Science Park II, Singapore, 117528, Singapore
eFood and Environment Scientific Operations, Waters Corporation, Stamford Avenue, Altrincham Road, Wilmslow, SK9 4AX, UK
First published on 5th January 2023
Abstract
There are at least 500 naturally occurring amino acids, of which only 20 standard proteinogenic amino acids are used universally across all organisms in the synthesis of peptides and proteins. Non-standard amino acids can be incorporated into proteins or are intermediates and products of metabolic pathways. While the analysis of standard amino acids is well-defined, the analysis of non-standard amino acids can be challenging due to the wide range of physicochemical properties, and the lack of both reference standards and information in curated databases to aid compound identification. It has been shown that the use of an AccQ·Tag™ derivatization kit along with LC-MS/MS is an attractive option for the analysis of free standard amino acids in complex samples because it is fast, sensitive, reproducible, and selective. It has been demonstrated that the most abundant quantitative transition for MS/MS analysis of 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) derivatized amino acids corresponds to the fragmentation of the molecule at the 6-aminoquinoline carbonyl group producing a common m/z 171 fragment ion and occurs at similar mass spectrometry collision energy and cone voltages. In this study, the unique properties of AQC derivatized amino acids producing high intensity common fragment ions, along with chromatographic separation of amino acids under generic chromatography conditions, were used to develop a novel screening method for the detection of trace levels of non-standard amino acids in complex matrices. Structural elucidation was carried out by comparing the MS/MS fragment ion mass spectra generated with in silico predicted fragmentation spectra to enable a putative identification, which was confirmed using an appropriate analytical standard. This workflow was applied to screen human plasma samples for bioactive thiol-group modified cysteine amino acids and S-allylmercaptocysteine (SAMC), S-allylcysteine sulfoxide (SACS or alliin) and S-propenylcysteine (S1PC) are reported for the first time to be present in human plasma samples after the administration of garlic supplements.
Introduction
Amino acids are organic molecules that contain a carboxyl group, an amino group, and a side chain R group. There are more than 500 naturally occurring amino acids, of which only 20 standard proteinogenic amino acids are used universally across all organisms in the synthesis of peptides and proteins.1–3 Some non-standard amino acids are formed during post-translational modification of amino acids in peptides and proteins such as phosphorylation, myristoylation and carboxylation.4,5 The wide range of non-standard amino acids that can be incorporated into proteins resulting in new protein folding structures with unique chemical properties has fueled interest in biochemical engineering, and to date over 100 non-standard amino acids have been chemically engineered into proteins.2,6 Other non-standard amino acids are intermediates and products of metabolic pathways important for biological functions. For example, neurotransmitter amino acids such as N-methyl aspartate (NMDA) and γ-aminobutyric acid (GABA) are involved in excitatory and inhibitory neurotransmission.3,7 Therefore, non-standard amino acids are important for a broad range of biological and biochemical activities, and the identification and quantification of non-standard amino acids are required to further understand proteins and amino acid metabolic pathways.Free amino acids have been analyzed through a variety of techniques including capillary electrophoresis, thin-layer chromatography, chemical derivatization followed by high-performance liquid chromatography (HPLC), nuclear magnetic resonance (NMR), gas-chromatography mass spectrometry (GC-MS), and liquid-chromatography mass spectrometry (LC-MS).8 Regardless of the analytical technique applied, the first step is to determine the amount of free amino acids present using analytical standards. While the analysis of standard amino acids is well-defined, the analysis of non-standard amino acids can be challenging due to the wide range of physicochemical properties, and the lack of both analytical standards and information in curated databases to aid compound identification. In addition, developing a consolidated method for the analysis of the wide range of non-standard amino acids in complex biological matrices is difficult due to interference from the matrix, and free non-standard amino acids can be present at trace levels. Obtaining analytical standards can be expensive or time consuming and is an analytical risk, especially when there is uncertainty over the presence of these non-standard amino acids in biological matrices.
It has been shown that the use of AccQ·Tag™ reagents for chemical derivatization along with liquid chromatography (LC) separation is an attractive option in the analysis of free amino acids.9–11 The derivatization of free amino acids involves the reaction of 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) with primary and secondary amines forming stable fluorescent urea derivatives that are retained and separated on a reverse-phase C18 LC column. The use of AQC derivatization along with the latest liquid chromatography (LC) and mass spectrometry (MS) technology has enabled the rapid, specific, and sensitive analysis of free amino acids. Boogers et al. (2008) showed a reduction in analysis time and better limits of detection (LODs) when using ultra-performance liquid chromatography (UPLC™) as compared to HPLC.9 Armenta et al. (2010) coupled a UPLC system with tandem quadrupole mass spectrometry (MS/MS) to quantify free amino acids in complex biological samples and showed a dynamic range between 2 and 4 orders of magnitude, a low LOD in the fmol per μL range, and good technical and biological reproducibility.10 In addition, the use of AQC derivatization has been extended to the analysis of non-standard amino acids, metabolites with amine moieties, and N-glycans.12–14 Due to the flexibility of the detection device coupled to the LC, high sensitivity, good chromatographic separation, and high selectivity, AQC derivatization has been used in a wide range of application areas including the analysis of neurotransmitter amino acids,15 characterizing the nutritional value of food and beverage,16,17 and even soil amino acid analysis.18
After AQC derivatization, the hydrophobic tag allows good chromatographic retention and separation of amino acids with different polarities on a reverse phase C18 column under generic chromatography conditions thereby enabling easy LC method development.9,15,19 Armenta et al. (2010) demonstrated that the most abundant quantitative ion of all the derivatized amino acids for MS/MS analysis corresponds to the fragmentation of the molecule at the 6-aminoquinoline carbonyl group producing a common m/z 171 fragment ion.10 Furthermore, the optimal cone voltages (CVs) and collision energies (CEs) were similar for most amino acids. These properties make AQC chemical derivatization an attractive option for the analysis of non-standard amino acids due to high sensitivity, generic chromatography conditions and easy multiple-reaction monitoring (MRM) method development. As MRM conditions are similar for derivatized amino acids with similar R groups, it is theoretically possible to generate MRMs of AQC derivatized amino acids with standard MS parameters for analytes in situations where reference standards are not available. Further quick confirmatory investigations can be carried out to determine if analytical standards need to be obtained for further identification and quantitative analysis. This screening approach would reduce analytical risk and speed up method development.
An application area that is particularly challenging is the analysis of bioactive thiol-group modified cysteine amino acids in human biofluids after the ingestion of garlic and its supplements. At least 9 thiol-group modified cysteine amino acids such as S-allylcysteine (SAC), S-allylmercaptocysteine (SAMC), S-allylcysteine sulfoxide (SACS, or alliin), S-ethylcysteine (SEC), S-ethylcysteine sulfoxide (SECS), S-methylcysteine (SMC), S-methylcysteine sulfoxide (SMCS), S-propenylcysteine (S1PC) and S-propyl cysteine (SPC) are postulated to be present in garlic and garlic supplements. They form a subset of the total organosulfur compounds, which contribute to the unique taste and smell of garlic, while conferring antimicrobial, anticancer and antioxidant properties.20–23 Many in vivo studies have shown medicinal properties of many of these thiol-group modified cysteine amino acids;21,24–26 however, which of these amino acids are present in human plasma upon administration of garlic supplements is currently unknown. Park et al. (2017) developed a LC-MS/MS method to determine SAC and its metabolites in rat plasma after the administering of SAC or aged garlic extract (AGE).27 After administering the AGE supplement, SAC the predominant thiol-group modified cysteine amino acid was detected at a concentration of approximately 0.4 μg mL−1 a little higher than the lower limit of quantification (LLOQ) of 0.1 μg mL−1. It is expected that other thiol-group modified cysteine amino acids would be present at lower concentrations, and hence an alternative analytical method would be required to detect these other compounds.
In this study, a novel method is developed using a high sensitivity LC-MS/MS system and AQC derivatization to screen trace levels of non-standard amino acids in human plasma samples. In addition, an enhanced MS/MS scanning function (StepWave DS) is used to generate product ions to support structural elucidation. This method reduces analytical risk and facilitates easier method development by providing evidence for the presence of non-standard amino acids and helps to determine which analytical standards are required for further identification and quantitative analysis. To demonstrate the applicability of this method, it is used to screen trace levels of thiol-group modified cysteine amino acids in human plasma samples upon administration of a garlic supplement.
Experimental
Reagents
LC-MS grade acetonitrile (ACN) and water were obtained from Honeywell Burdick & Jackson (Muskegon, MI, USA). LC-MS grade formic acid (FA) and an AccQ·Tag Ultra Derivatization Kit were obtained from Waters (Milford, MA, USA). Ultrapure Milli-Q water was from Millipore (Bedford, MA, USA). Analytical standards L-alanine, L-cysteic acid, L-leucine, L-methionine, L-methionine sulfone, L-norvaline, L-phenylalanine, (±) L-alliin (SACS) and L-serine were obtained from Sigma Aldrich (Saint Louis, MO, USA). S1PC was obtained from MedChemExpress (Monmouth Junction, NJ, USA). SAC was synthesized as previously described by Besada et al. (2005) and SAMC was synthesized as previously described by Wilde et al. (2016).28,29 More details of SAMC synthesis can be found in the ESI.†Plasma samples
Plasma samples were obtained from a larger study group investigating the bioavailability of garlic organosulfur compounds after eating garlic supplements. Participants consisted of non-smoking, 20–29 year old, normal weight (BMI around 19–25 kg m−2) male volunteers. The study was approved by the ethics committee of the Medical Faculty of the Christian-Albrechts-University of Kiel, Germany (A 111/14). Plasma samples used in this study were from a volunteer that had eaten freeze dried garlic powder with SACS as the key compound in an acid-resistant capsule and an approximate allicin yield of 18 mg. The capsules were made using 952.4 mg of milled freeze-dried garlic slices and filled in acid-resistant capsules (DRcaps™). Before ingesting the capsule, participants avoided vegetables and spices of the genus Allium for one week. Blood plasma samples were drawn before eating the supplement and 4 hours after eating the supplement. The plasma samples were precipitated with 80% cooled methanol (−80 °C) in a dilution ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v). The samples were vortexed and incubated for 10 min at −80 °C. The samples were centrifuged at 15
10 (v/v). The samples were vortexed and incubated for 10 min at −80 °C. The samples were centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 10 min, and the supernatant was transferred to new vials and stored at −80 °C until sample treatment.
000 rpm and 4 °C for 10 min, and the supernatant was transferred to new vials and stored at −80 °C until sample treatment.
Optimization of MRM parameters
Free amino acids were derivatized using AccQ·Tag reagents and analyzed on an ACQUITY Ultra Performance LC™ I-Class System (Waters, Milford, MA, USA) coupled with a Xevo™ TQ-XS Tandem Quadrupole Mass Spectrometer (Waters, Manchester, UK) fitted with an electrospray ionization (ESI) source.To optimize MRM parameters, AQC derivatized amino acids were combined infused with a LC flow into the mass spectrometer. Free amino acid standards were derivatized as per Salazar et al. (2012);11 briefly 100 μL of 100 μg mL−1 amino acid standard was mixed with 700 μL of 50 mM ammonium acetate buffer (pH 9.3) and 200 μL of AccQ·Tag reagent, and heated at 55 °C for 10 min. Derivatized amino acids were infused at 30 μL min−1 together with a LC flow (combined infusion) of 0.7 mL min−1 using 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 water
30 water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile containing 0.2% FA. Xevo TQ-XS System parameters were as follows: positive polarity, 150 °C source temperature, 550 °C desolvation temperature, 1000 L h−1 desolvation gas flow, and capillary voltage of 0.7 kV. IntelliStart™ Application within Waters MassLynx™ 4.2 SCN 1007 Software (Waters, Manchester, UK) was used to profile optimal cone voltages and collision energies.
acetonitrile containing 0.2% FA. Xevo TQ-XS System parameters were as follows: positive polarity, 150 °C source temperature, 550 °C desolvation temperature, 1000 L h−1 desolvation gas flow, and capillary voltage of 0.7 kV. IntelliStart™ Application within Waters MassLynx™ 4.2 SCN 1007 Software (Waters, Manchester, UK) was used to profile optimal cone voltages and collision energies.
LC-MS/MS analysis of free amino acids
Free amino acids were derivatized as per manufacturer's instructions; 9 μL of plasma sample and 1 μL of water or 100 ng mL−1 analytical standard were mixed with 70 μL of AccQ·Tag ultra borate buffer and 20 μL of AccQ·Tag reagent and heated at 55 °C for 10 min. Chromatographic separation of AQC derivatized amino acids was carried out using reverse phase liquid chromatography (RP) with an AccQ·Tag ultra C18, 1.7 μm, 2.1 × 100 mm column (Waters, Milford, MA, USA). 1 μL of sample was injected into the column and gradient elution was performed using mobile phase A (water) and mobile phase B (ACN), containing 0.2% FA, at a flow rate of 0.7 mL min−1, column temperature of 55 °C, and gradient conditions as follows: 0–0.54 min, 0.1% B, and curve 6; 5.74 min, 9.1% B, and curve 5; 7.74 min, 21.2% B, and curve 6; 10 min, 59.6% B, and curve 6; 10.01 min, 90% B, and curve 6; 12.50 min, 90% B, and curve 6; 12.50–12.51 min, 0.1% B, and curve 6; 15 min, 0.1% B, and curve 6. To profile the presence of unknown bioactive thiol-group modified cysteine amino acids, theoretical MRMs with the common fragment ion (m/z 171.1) was used. Generic MRM conditions of cone voltage, 30 V, and collision energy, 29 eV were used.Structural elucidation of unknown derivatized amino acids
To aid putative identification of unknown derivatized amino acids, the ScanWave DS function of the Xevo TQ-XS system was used to generate MS/MS product ion mass spectra. This function allows the fragmentation and accumulation of ions, while synchronizing the ejection of the fragment ions with the scanning cycle of the second quadrupole to improve the duty cycle.30 For each ScanWave DS function, the second quadrupole was set to scan between 80 or 100 and 320 m/z at 1000 amu per sec, and the cone voltage was set at 30 V. Four injections of collision energies 10, 15, 20 and 25 eV were acquired, and the raw files were combined.In silico structural elucidation
The chemical structures, chemical formula and molecular weight were generated using ChemDraw™ 18 (PerkinElmer Informatics, Waltham, MA, USA). In silico fragmentation of thiol-group modified cysteine amino acids was carried out with competitive fragmentation modeling for metabolite identification (CFM-ID) (https://cfmid.wishartlab.com/) using the spectra prediction tool.31,32 The simplified molecular input line entry system (SMILES) string for AQC derivatized SAC, SACS, SPC and S1PC is O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NC1
C(NC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC2
CC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC
CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CN
CN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2C
C2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)NC(CSCC
C1)NC(CSCC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)C(O)
C)C(O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, O
O, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NC(C(O)
C(NC(C(O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)CS(CC
O)CS(CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C)
C)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NC1
O)NC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC2
CC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC
CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CN
CN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2C
C2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1, O
C1, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NC2
C(NC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC1
CC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC
CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CN
CN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1C
C1C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)NC(CSCCC)C(O)
C2)NC(CSCCC)C(O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and O
O and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NC(C(O)
C(NC(C(O)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)CS/C
O)CS/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C)NC1
C/C)NC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC2
CC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC
CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CN
CN![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2C
C2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1.
C1.
Results and discussion
Development of theoretical MRMs for screening
It has been shown that the most abundant quantitative ion used for MRM transitions for AQC derivatized amino acids corresponds to the fragmentation of the molecules at the 6-aminoquinoline carbonyl group producing a common m/z 171 fragment ion (common ion) (Fig. S1†).10,11,33 Furthermore Salazar et al. (2012) showed that the optimal CV and CE for this common ion were similar for most of the derivatized amino acids.11 To expand on this observation and apply it towards screening of thiol-group modified cysteine amino acids, a panel of amino acids with similar R-chain groups to target compounds were used to profile the optimal CV and CE to produce the common ion (Fig. 1). One group of amino acids consisted of alanine and its extended side-chain groups such as leucine, norvaline, phenylalanine, and serine. Another group of amino acids consisted of cysteine and its derivatives such as cysteic acid, SAC and SAMC. The last group of amino acids consisted of methionine and its oxidized derivative methione sulfone.The MassLynx Intellistart MRM optimizer selected the common ion as the most abundant product ion in all the amino acids tested, confirming previous reports.10,11,33 At 80% peak height, all derivatized amino acids had a range of at least 40 V for optimal CV and overlapped with the other amino acids tested. A common CV of 30 V was used for screening MRMs (Fig. 1A and B). With regard to CE, all derivatized amino acids had a range of at least 10 eV for optimal CE (Fig. 1C and D). While most of the amino acids had similar optimal CE profiles of approximately 18 to 34 eV at an 80% peak height, SAMC had an optimal CE profile of 26 to 52 eV at 80% peak height. A common CE of 29 eV runs through all the overlapping CE range profiles for all the amino acids and was used for screening MRMs.
The theoretical structures of AQC bioactive derivatized thiol-group modified cysteine amino acids were generated and the molecular weight was calculated (Fig. 2). The derivatized amino acids were SAC (1), SAMC (2), SACS (3), SEC (4), SECS (5), SMC (6), SMCS (7), S1PC (8), and SPC (9). Generic LC conditions were used along with screening MRMs developed using the calculated molecular weight of the singly protonated derivatized amino acids, and common CVs and CEs.
 | ||
| Fig. 2 Theoretical chemical structures of AQC derivatized thiol-group modified cysteine amino acids: SAC (1), SAMC (2), SACS (3), SEC (4), SECS (5), SMC (6), SMCS (7), S1PC (8), and SPC (9). | ||
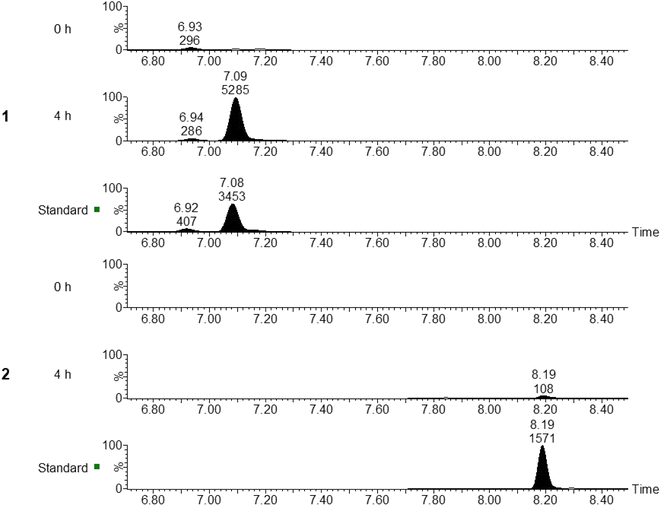
Plasma samples from individuals before and after the administration of the garlic powder supplement were tested for SAC and SAMC. Common ion MRMs for both 1 and 2 were specific as shown by the equivalent comparison against the 10 ng mL−1 analytical standard spiked into blank plasma (Fig. 3). At 4 h after administration of the garlic supplement, 1 and 2 were enriched compared to 0 h, suggesting that other trace level thiol-group modified cysteine amino acids may also be enriched in plasma samples (Fig. 3). To date, SAMC has not been reported to be present in plasma samples after the administration of garlic supplements, and the low levels detected here are probably due to the quick metabolism of SAMC as shown previously by Yang et al. (2018), in which the pharmacokinetic mean residence time (MRT) and half-life (t1/2) were less than 5 min after intravenous injection of SAMC into rats.34
Screening and identification of thiol-group modified cysteine amino acids
Plasma samples were screened for the panel of bioactive thiol-group modified cysteine amino acids with screening MRMs (Fig. 4). Unique chromatographic peaks for 3, 8 and 9 were found in plasma samples after the administration of the garlic supplement, whereas 4, 5, 6 and 7 showed no difference (Fig. 4). A unique chromatographic peak was found for 3 at a retention time of 3.31 min, and two unique chromatographic peaks were found for 8 and 9 at retention times of 7.07–7.10 min and 7.47–7.50 min. These two unique chromatographic peaks could be related to 1, 8 or 9 as 1 and 8 are isomeric compounds, and their 13C isotopic peaks have a similar mass to 9.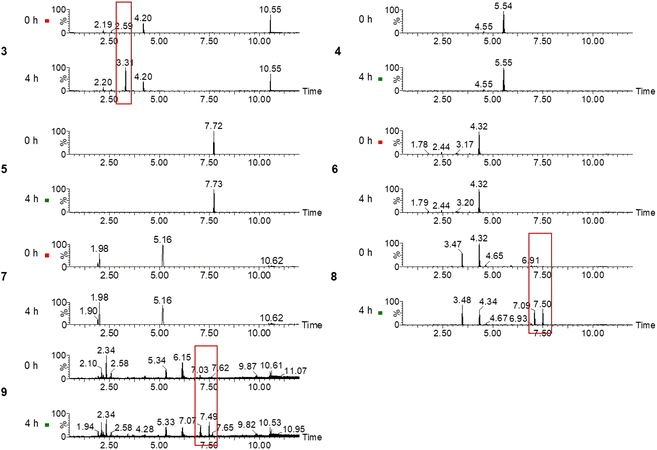 | ||
| Fig. 4 Screening of plasma samples for 3, 4, 5, 6, 7, 8 and 9 at 0 h and 4 h after garlic supplement administration. Unique chromatographic features are highlighted with a red box. | ||
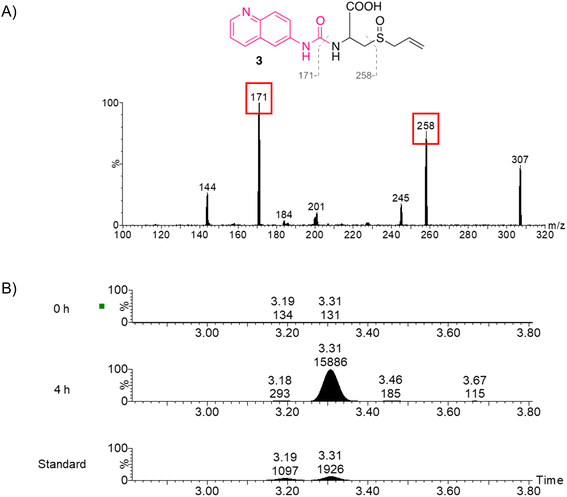
Due to the low intensity of fragment ions, an enhanced MS/MS scanning function (StepWave DS) was used.30 The fragment spectra of these AQC derivatized amino acids are not available in MS/MS databases, and therefore the fragment spectra were compared against competitive fragmentation modeling for metabolite identification (CFM-ID)31,32in silico predicted fragment spectra to provide additional evidence to support the identification before obtaining the analytical standard for confirmation. The unique chromatographic peak of 3 produced a series of fragment spectra (Fig. 5A) and a fragment ion at m/z 258 was consistent with CFM-ID fragment spectra (Fig. S2a†). The enrichment of the molecule 4 h after garlic administration along with the fragment spectra provided evidence for the presence of SACS, and therefore the analytical standard was obtained to confirm the identity. The chromatographic profile of 3 (Fig. 5B) and MS/MS spectra (Fig. S2b†) from the analysis of the enriched sample and the analytical standard were compared, and the identity of SACS was confirmed. Two chromatographic peaks at retention times of 3.19 and 3.31 min were seen in the SACS standard, which corresponds to the two sulfur-centered stereoisomers of the standard, whereas only one chromatographic peak at a retention time of 3.31 min was seen 4 h after garlic supplement administration and is consistent with previous studies, which showed that only the (+) sulfur-centered stereoisomer is present in garlic preparations.35,36 In the study by Park et al. (2015), SAC and SACS were detected in rats that were orally administered 50 mg kg−1 SAC, whereas rats orally administered 1 g kg−1 AGE showed significantly lower levels of SAC and undetectable SACS.27 In this study, for the first time the presence of SACS in human plasma samples after the administration of garlic supplements is shown.
The fragment spectra of the two unique chromatographic peaks at retention times of 7.07–7.10 min and 7.47–7.50 min showed the ion at m/z 171 to be the most abundant fragment ion present and other fragment ions with low intensities (Fig. 6A). Comparing the fragment spectra, the chromatographic peak at 7.07–7.10 min corresponding to 1 had unique fragment ions at m/z 245, 246 and 290. The CFM-ID in silico predicted fragment spectra of 1 and 8 (Fig. S3A†) were mostly similar; however an ion at m/z 292 was expected to be present at higher levels for 1 compared to 8. A similar ion with the loss of 2H atoms due to an additional C–C double bond in the structure giving a m/z of 290 was also predicted by CFM-ID (Fig. S3B†) and this is probably the fragment ion in 1 (Fig. 6A). The CFM-ID predicted the fragment ion at m/z 246 for both 1 and 8, whereas it was only seen experimentally for 1. This unique fragment ion for 1 was confirmed by analyzing the pure SAC standard (data not shown). The chromatographic peak at 7.47–7.50 min is most probably 8 as it was enriched at 4 h after garlic administration, is isomeric to 1, and has a different fragmentation pattern. The analytical standard was obtained to confirm the identity. The chromatographic profile of 8 (Fig. 6B) and fragment spectra (Fig. S3B†) were compared between the enriched sample and the analytical standard, and the identity was confirmed. Amano et al. (2016) had demonstrated the bioavailability of S1PC in rats after oral and intravenous administration of S1PC,37 and the results in this study are in agreement and show for the first time that S1PC is enriched in human plasma samples after the administration of garlic supplements.
The two unique chromatographic peaks for 9 have the same retention time as 1 and 8 (Fig. 4). In addition, the fragment spectra were offset by 1–2 m/z units when compared to 1 and 8 (Fig. S4†) and did not show any fragment matches with that of CFM-ID. Therefore, 9 is most probably the 13C isotopic M + 1 peaks for 1 and 8. As there is insufficient evidence to prove the identity of 9, it is assumed that it is not present in these samples and was not investigated further.
Applicability and future work
Typical LC-MS/MS experiments require either analytical standards or experimental information in curated databases. Experimental information is not available for many non-standard amino acids and obtaining analytical standards can pose an analytical risk as the presence of these analytes in complex biological matrices is unknown. The novel method presented in this study reduces the analytical risk by screening for trace levels of non-standard amino acids in complex biological matrices under generic LC-MS/MS conditions and provides evidence to obtain relevant analytical standards for further studies. In the application area of bioactive thiol-group modified cysteine molecules presented in this study, seven non-standard amino acids were of interest, but only two required analytical standards for further analytical studies. Therefore, this method reduces waste and streamlines method development. The main steps for this method are: (1) profiling common MRM parameters for screening of AQC derivatized amino acids, (2) development of screening MRMs, (3) screening of samples in which amino acids are expected to be present and absent, and finally (4) structural elucidation of unique chromatographic peaks. Gray et al. (2017) and Zhang et al. (2018) have both shown the use of AQC derivatization of standard and non-standard amino acids for typical LC-MS/MS analysis,12,13 therefore this method can be extended to screen for other groups of non-standard amino acids.Bioactive garlic metabolites are an active area of research.20–27 Previous in vivo animal studies focused on the administration of SAC, SACS and S1PC through intraperitoneal or intravenous injection and does not accurately reflect the bioavailability of these compounds upon ingestion of garlic supplements.27,37,38 To the best of our knowledge, this study has shown for the first time the presence of SAMC, SACS and S1PC in human plasma samples after the administration of dietary garlic supplements. While the presence of other bioactive thiol-group modified cysteine amino acids cannot be ruled out, the results presented here indicate that only SAC, SAMC, SACS and S1PC were enriched 4 hours after eating garlic powder supplements (Fig. 4). Other thiol-group modified cysteine amino acids may not be present in the supplement administered or had been fully metabolized at the time point of plasma sampling.
Different LC-MS/MS methods have been developed for the quantification of garlic bioactive thiol-group modified cysteine amino acids.27,34,37,38 Lee et al. (2014) had used mixed-mode reverse phase and cation exchange chromatography and MRM transition of m/z 162 > 145 for the analysis of SAC,35 whereas Park et al. (2017) had used reverse-phase chromatography and MRM transition of m/z 162 > 73 for the analysis of SAC.27 This study presents a novel screening method for a range of thiol-group modified cysteine acids under generic LC conditions and a common fragment ion for MRM transitions. In the studies by Lee et al. (2014) and Park et al. (2017), they showed a LLOQ of 5 ng mL−1 and 100 ng mL−1 respectively for SAC.27,38 In another study, SAC and S1PC were detected at concentration levels around 10 ng mL−1 and SAMC below 10 ng mL−1 (Fig. 3), which suggest that the LLOQ for this method could be even lower. Therefore, further investigations can be carried out into the concentration of thiol-group modified cysteine amino acids in plasma samples after the administration of different types of supplements, the presence of these amino acids in supplements, other biological fluids like urine, and different time points after the administration of the supplement.
In relation to garlic organosulphur compounds, several dimers and trimers such as γ-glutamyl-S-methylcysteine (GSMC), γ-glutamyl-S-allylcysteine (GSAC), and γ-glutamyl-S-1-propenylcysteine (GS1PC) are involved in the synthesis and metabolism of bioactive thiol-group modified cysteine amino acids in garlic.21–23 Furthermore, thiol-group modified cysteine amino acids can also undergo N-acetylation during metabolism.27,37 Therefore, an extension to this study is to screen plasma samples for dimers, trimers, and N-acetylated thiol-group modified cysteine amino acids. Due to the different properties of the molecules, the current method presented herein will need to be assessed and modified for the applicability to these other molecules.
Conclusion
In this study, the unique properties of AQC derivatized amino acids producing a high intensity common fragment ion under generic LC-MS/MS conditions were used to develop a screening method for the detection of trace levels of non-standard amino acids. Structural elucidation was carried out by MS/MS fragment ion scan, followed by comparison of the spectra with CFM-ID in silico predicted fragmentation spectra, and confirmed with analytical standards. This method was applied to screen human plasma samples for bioactive thiol-group modified cysteine amino acids after the administration of garlic supplements. SAMC, SACS and S1PC are reported for the first time to be present in human plasma samples after the administration of garlic supplements. Altogether, this workflow facilitates quick method development and reduces analytical risk by using a generic LC-MS/MS method to screen samples for non-standard amino acids and provides evidence to determine if an analytical standard is required for further confirmatory and quantification investigations.Funding
This study was supported by Waters Corporation and funded by the Federal Ministry for Education and Research in Germany, Food Chain Plus (FoCus, 0315539A).Author contributions
Daniel H. J. Ng: conceptualization, methodology, software, formal analysis, investigation, resources, writing – original draft, writing – review and editing, supervision, and project administration. Li Yan Chan: conceptualization, methodology, software, formal analysis, investigation, resources, writing – review and editing, and visualization. Laura Fitzner: resources and writing – review and editing. Julia Katharina Keppler: resources and writing – review and editing. Shareef M. Ismail: conceptualization and writing – review and editing. Simon Hird: writing – review and editing. Peter Hancock: supervision, project administration, and writing – review and editing. Schwarz Karin: resources, project administration, funding acquisition, writing – review and editing. Demetrowitsch Tobias: conceptualization, resources, project administration, and writing – review and editing.Conflicts of interest
The authors declared no competing interest.Abbreviations
| ACN | Acetonitrile |
| AGE | Aged garlic extract |
| AQC | 6-Aminoquinolyl-N-hydroxysuccinimidyl carbamate |
| CE | Collision energy |
| CFM-ID | Competitive fragmentation modeling for metabolite identification |
| CV | Cone voltages |
| FA | Formic acid |
| GABA | γ-Aminobutyric acid |
| GC-MS | Gas-chromatography mass spectrometry |
| GS1PC | γ-Glutamyl-S-1-propenylcysteine |
| GSAC | γ-Glutamyl-S-allylcysteine |
| GSMC | γ-Glutamyl-S-methylcysteine |
| HPLC | High-performance liquid chromatography |
| LC | Liquid chromatography |
| LC-MS | Liquid-chromatography mass spectrometry |
| LLOQ | Lower limit of quantification |
| LOD | Limit of detection |
| MRM | Multiple-reaction monitoring |
| MS | Mass spectrometry |
| MS/MS | Tandem quadrupole mass spectrometry |
| NMDA | N-Methyl aspartate |
| NMR | Nuclear magnetic resonance |
| RP | Reverse phase liquid chromatography |
| S1PC | S-Propenyl cysteine |
| SAC | S-Allylcysteine |
| SACS | S-Allyl cysteine sulfoxide |
| SAMC | S-Allylmercaptocysteine |
| SEC | S-Ethyl cysteine |
| SECS | S-Ethyl cysteine sulfoxide |
| SMC | S-Methyl cysteine |
| SMCS | S-Methyl cysteine sulfoxide |
| SMILES | Simplified molecular input line entry system |
| SPC | S-Propyl cysteine |
| UPLC | Ultra-performance liquid chromatography |
Acknowledgements
We thank the volunteers and Division of Food Technology Kiel University for the human plasma samples provided.References
- Y. Lu and S. Freeland, Genome Biol., 2006, 7, 102 CrossRef PubMed.
- T. S. Young and P. G. Schultz, J. Biol. Chem., 2010, 285, 11039–11044 CrossRef PubMed.
- I. Wagner and H. Musso, Angew. Chem. Int., Ed. Engl., 1983, 22, 816–828 CrossRef.
- G. A. Khoury, R. C. Baliban and C. A. Floudas, Sci. Rep., 2011, 1, 90 CrossRef PubMed.
- R. G. Krishna and F. Wold, in Methods in Protein Sequence Analysis, ed. K. Imahori and F. Sakiyama, Springer US, Boston, MA, 1993, pp. 167–172 Search PubMed.
- S. H. Hong, Y.-C. Kwon and M. C. Jewett, Front. Chem., 2014, 2, 34 Search PubMed.
- R. Dalangin, A. Kim and R. E. Campbell, Int. J. Mol. Sci., 2020, 21, 6197 CrossRef PubMed.
- W. Xu, C. Zhong, C. Zou, B. Wang and N. Zhang, Amino Acids, 2020, 52, 1071–1088 CrossRef.
- I. Boogers, W. Plugge, Y. Q. Stokkermans and A. L. L. Duchateau, J. Chromatogr. A, 2008, 1189, 406–409 CrossRef PubMed.
- J. M. Armenta, D. F. Cortes, J. M. Pisciotta, J. L. Shuman, K. Blakeslee, D. Rasoloson, O. Ogunbiyi, D. J. Sullivan and V. Shulaev, Anal. Chem., 2010, 82, 548–558 CrossRef PubMed.
- C. Salazar, J. M. Armenta, D. F. Cortés and V. Shulaev, Methods Mol. Biol., 2012, 828, 13–28 CrossRef PubMed.
- N. Gray, R. Zia, A. King, V. C. Patel, J. Wendon, M. J. W. McPhail, M. Coen, R. S. Plumb, I. D. Wilson and J. K. Nicholson, Anal. Chem., 2017, 89, 2478–2487 CrossRef PubMed.
- Q. Zhang, H. Xu, R. Liu, P. Gao, X. Yang, P. Li, X. Wang, X. Wang, Y. Zhang, K. Bi and Q. Li, Anal. Chem., 2018, 90, 11941–11948 CrossRef.
- Y. Wu, Q. Sha, J. Du, C. Wang, L. Zhang, B. Liu, Y. Lin and X. Liu, J. Chromatogr. A, 2018, 1535, 114–122 CrossRef PubMed.
- H. Liu, M. C. Sañuda-Peña, J. D. Harvey-White, S. Kalra and S. A. Cohen, J. Chromatogr. A, 1998, 828, 383–395 CrossRef PubMed.
- L. Gwatidzo, B. M. Botha and R. I. McCrindle, Food Chem., 2013, 141, 2163–2169 CrossRef PubMed.
- L. X. Loh, D. H. J. Ng, M. Toh, Y. Lu and S. Q. Liu, J. Agric. Food Chem., 2021, 69, 14024–14036 CrossRef PubMed.
- S. Hou, H. He, W. Zhang, H. Xie and X. Zhang, Talanta, 2009, 80, 440–447 CrossRef.
- S. A. Cohen and D. P. Michaud, Anal. Biochem., 1993, 211, 279–287 CrossRef.
- P. Ahmad, S. S. Alvi and M. S. Khan, Natural Bio-Active Compounds: Volume 1: Production and Applications, Springer Singapore, Singapore, 2019, vol. 20, pp. 561–585 Search PubMed.
- H. Amagase, J. Nutr., 2006, 136, 716S–725S CrossRef PubMed.
- M. Iciek, I. Kwiecień and L. Włodek, Environ. Mol. Mutagen., 2009, 50, 247–265 CrossRef.
- Y. Kodera, M. Ushijima, H. Amano, J. Suzuki and T. Matsutomo, Molecules, 2017, 22, 570 CrossRef.
- C. G. Sheela and K. T. Augusti, Indian J. Exp. Biol., 1992, 30, 523–526 Search PubMed.
- K. T. Augusti and C. G. Sheela, Experientia, 1996, 52, 115–119 CrossRef PubMed.
- H. P. Koch and L. D Lawson, Garlic: the Science and Therapeutic Application of Allium Sativum L. And Related Species, Williams &Wilkins, Baltimore, 1996, vol. 12, pp. 37–108 Search PubMed.
- T. Park, J.-H. Oh, J. H. Lee, S. C. Park, Y. P. Jang and Y.-J. Lee, Planta Med., 2017, 83, 1351–1360 CrossRef PubMed.
- P. Besada, L. Mamedova, C. J. Thomas, S. Costanzi and K. A. Jacobson, Org. Biomol. Chem., 2005, 3, 2016–2025 RSC.
- S. C. Wilde, J. K. Keppler, K. Palani and K. Schwarz, Food Chem., 2016, 197, 1015–1021 CrossRef PubMed.
- M. Twohig, P. Alden, G. Fujimoto, D. Kenny and R. S. Plumb, Waters Application Note, 2008, p. 720002828EN Search PubMed.
- F. Allen, A. Pon, M. Wilson, R. Greiner and D. Wishart, Nucleic Acids Res., 2014, 42, W94–W99 CrossRef CAS PubMed.
- F. Wang, J. Liigand, S. Tian, A. David, R. Greiner and D. S. Wishart, Anal. Chem., 2021, 93, 11692–11700 CrossRef CAS PubMed.
- S. Karakawa, K. Shimbo, N. Yamada, T. Mizukoshi, H. Miyano, M. Mita, W. Lindner and K. Hamase, J. Pharm. Biomed. Anal., 2015, 115, 123–129 CrossRef CAS PubMed.
- M. Yang, Z. Dong, X. Jiang, Z. Zhao, J. Zhang, X. Cao and D. Zhang, J. Chromatogr. Sci., 2018, 56, 396–402 CAS.
- B. Dethier, M. Laloux, E. Hanon, K. Nott, S. Heuskin and J.-P. Wathelet, Talanta, 2012, 101, 447–452 CrossRef.
- E. Block, Sci. Am., 1985, 252, 114–121 CrossRef PubMed.
- H. Amano, D. Kazamori and K. Itoh, Xenobiotica, 2016, 46, 1017–1025 CrossRef PubMed.
- S. Lee, N. In Chang, M. Yoo, J. Hoon Choi and D. Shin, J. Chromatogr. Sci., 2015, 53, 54–59 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ay01588a |
| This journal is © The Royal Society of Chemistry 2023 |