Open Access Article
Open Access ArticleNew synthetic red- and orange-emitting luciferases to upgrade in vitro and 3D cell biosensing†
Maria Maddalena
Calabretta
ab,
Denise
Gregucci
ab and
Elisa
Michelini
 *abc
*abc
aDepartment of Chemistry “Giacomo Ciamician”, University of Bologna, Via Selmi 2, 40126, Bologna, Italy. E-mail: elisa.michelini8@unibo.it
bCenter for Applied Biomedical Research (CRBA), Azienda Ospedaliero-Universitaria Policlinico S. Orso-la-Malpighi, 40138 Bologna, Italy
cHealth Sciences and Technologies Interdepartmental Center for Industrial Research (HSTICIR), University of Bologna, 40126, Bologna, Italy
First published on 28th September 2023
Abstract
Bioluminescence (BL), i.e., the emission of light in living organisms, has become an indispensable tool for a plethora of applications including bioassays, biosensors, and in vivo imaging. Current efforts are focused on the obtainment of new luciferases having optimized properties, such as improved thermostability at 37 °C, pH-insensitive emission, high quantum yield, extended kinetics and red-shifted emission. To address these issues we have obtained two new synthetic luciferases, an orange and a red-emitting luciferase, which were designed to achieve high sensitivity (BoLuc) and multiplexing capability (BrLuc) for in vitro and in vivo biosensing using as a starting template a recently developed thermostable synthetic luciferase (BgLuc). Both luciferases were characterized in terms of emission behaviour and thermal and pH stability showing promising features as reporter proteins and BL probes. As proof-of-principle application, an inflammation assay based on Human Embryonic Kidney (HEK293T) 3D cell cultures was developed using either the orange or the red-emitting mutant. The assay provided good analytical performance, with limits of detection for Tumor Necrosis Factor (TNFα) of 0.06 and 0.12 ng mL−1 for BoLuc and BrLuc, respectively. Moreover, since these luciferases require the same substrate, D-luciferin, they can be easily implemented in dual-color assays with a significant reduction of total cost per assay.
Introduction
Bioluminescence (BL), i.e., the emission of light in living organisms, has become an indispensable tool for a plethora of applications including bioassays, biosensors, and in vivo imaging.1 BL, like chemiluminescence, does not require any light source and is characterized by a high signal to noise ratio without the main issues related to fluorescence detection, such as photobleaching and autofluorescence.2Several BL proteins, including luciferases from different species and photoproteins such as aequorin and obelin, have been investigated and used as reporter proteins in drug screening assays, to monitor target pathway activation, or to track cell location over time in small laboratory animals.2–4 Cells can be genetically engineered to express a BL reporter under the regulation of constitutive promoters or inducible transcription elements to respond to different physical and biochemical stimuli. BL imaging provides additional advantages when compared to other approaches, such as the possibility to perform longitudinal non-invasive monitoring of patho-physiological events with almost zero background. As concerns BL imaging, the main limitations which cause insufficient sensitivity are mostly connected to low photon emission and tissue absorption. These issues hinder applications to deep imaging and large animal models.5
An interesting work was performed in which the growth of a gliobastoma tumor was monitored in mice with both BL imaging and magnetic resonance imaging (MRI). Unexpectedly, a discrepancy was observed between the two imaging modalities with no correlation between light emission and tumor volume at day 10 in a GL261-luc-GFP mouse model. This divergence was explained by possible instability in luciferase expression.6 In addition, BL provided an invaluable toolbox for upgrading predictive in vitro 3D cell models not only for the early stages of drug discovery, but also in perspective of replacing animal models in compliance with the 3Rs principle of reduction, replacement and refinement.7
Of all the proteins employed in BL assays and BL imaging, the wild-type North American firefly (Photinus pyralis) luciferase (PpyLuc) is certainly the most investigated. PpyLuc, a 550 aa protein with a size of about 62 kDa, catalyzes a multi-step reaction in which the substrate, D-luciferin (D-LH2), is adenylated and oxidated with the production of an electronically exited state oxyluciferin which emits yellow-green light (λmax 562 nm) when returning to the ground state.8 The requirement of adenosine triphosphate (ATP) as co-factor made this enzyme a valuable tool in all applications in which ATP detection is required, for example for hygiene monitoring or quantification of intracellular ATP levels.9 Other luciferases were also explored, such as those isolated from marine species, requiring coelenterazine as substrate and producing blue-shifted light without requiring any cofactor except molecular oxygen.10,11 More recently the BL system of mushroom Neonothopanus nambi was studied12 and successfully expressed in bacteria, yeast, Xenopus laevis embryos, and human cell lines.13
Since wild-type PpyLuc is a heat-sensitive enzyme, which rapidly loses its catalytic activity at 37 °C, several efforts were aimed at improving its stability at higher temperatures and at different pH starting from the pioneering work of White et al.14 A chimeric enzyme having the N-domain of PpyLuc joined to the C-domain of Luciola italica luciferase was obtained with additional mutations conveying thermostability.9De novo enzyme design has been explored leading to a small thermostable luciferase (13.9 kDa) having a melting temperature higher than 95 °C.15 In the last decades research has been mainly focused on the one hand on the obtainment of new luciferases16 and, on the other hand, on the development of highly sensitive photodetectors to improve light collection efficiency and enable the implementation of BL into portable biosensors.17 Among the main drivers for the obtainment of improved luciferases there are several factors, depending on the final application; a non-exhaustive list of these factors includes: (i) improved thermostability at 37 °C, (ii) pH-insensitive emission, (iii) high quantum yield, (iv) extended kinetics and (v) red-shifted emission. Most of these issues have been addressed by mutating the wild-type enzyme, by developing new substrates, and by conjugating the luciferase to fluorescent molecules to exploit the Bioluminescence Resonance Energy Transfer (BRET) for red-shifting the emission spectrum.18 Several D-luciferin and coelenterazine analogues were also obtained to improve light emission or change the emission wavelength and a multicolor toolbox of D-LH2 analogues has been published.19,20 The effectiveness of the BRET-based approach has been exemplified in a recent work by Afshari et al. who immobilized the Nanoluc protein on the surface of silver sulfide quantum dots, obtaining the emission of NIR-II photons with a signal to noise ratio about 2 times higher than that obtained with fluorescence modality in mice tumor models.21 In most of red-emitting luciferases the emission peak is narrow and intensity much lower than the green-emitting counterparts. This narrow emission is highly advantageous for multicolor applications,22 in which spectral resolution enables to separate the signals from red and green-emitting luciferases reducing the interference, but reflects a lower photon production. Thus, such narrow emitters are not suitable for applications in which high sensitivity is required, for instance in cancer imaging.23
Despite the undoubted advantages of these strategies, the majority of these applications are restricted to a small community of field experts and a few of these luciferases entered the market. A possible explanation could be related both to the low intensity of red emitting probes which account for reduced sensitivity and to the high cost of substrates (e.g., furimazine for NanoLuc reporter).
Therefore, researchers who are not directly involved in the obtainment of such probes prefer to adopt commercial solutions. In an attempt to address this need we have developed two new synthetic luciferases, an orange and a red-emitting luciferases, which were on purpose designed to achieve high sensitivity (BoLuc) and multiplexing capability (BrLuc) for in vitro and in vivo biosensing using as a starting template a recently developed thermostable synthetic luciferase (BgLuc).24 These luciferases require the same substrate, D-luciferin, thus reducing total cost per assay, showing promising features as reporter proteins and BL probes. As proof-of-principle application, an inflammation assay based on Human Embryonic Kidney (HEK293T) 3D cell cultures was developed using the orange and the red-emitting mutant as reporter protein.
Experimental details
Chemicals and reagents
E. coli BL21 competent cells were from Agilent Technologies (Santa Clara, CA, USA). E. coli JM109 competent cells for plasmid propagation, Select Agar and LB (Lennox L Broth), and the SOC medium (tryptone 20 g L−1, yeast extract 5 g L−1, NaCl 5.0 M 2 mL L−1, KCl 1.0 M 2.5 mL L−1, MgCl2 1.0 M 10 mL L−1, MgSO4 1.0 M 10 mL L−1, D-glucose 1.0 M 20 mL L−1) were from Sigma (St Louis, MO, USA). Human embryonic kidney (HEK293T) cells were from ATCC (American Type Culture Collection [ATCC], Manassas, VA, USA) and all reagents for cell cultures were from Carlo Erba Reagents (Cornaredo, Milano, Italy). Beetle luciferin potassium salt (D-luciferin), Bright-Glo substrate, the kits for plasmid extraction and the mammalian expression plasmid pGL4.32[luc2P NF-kB-RE Hygro] were from Promega (Madison, WI). The enzymes required for cloning were from Thermo Fisher Scientific (Waltham, MA, USA). Protino Ni-IDA Resin and 14 mL Protino Columns for protein extraction were from Macherey-Nagel GmbH and Co. KG (Düren, Germany). The new P. pyralis luciferase mutants BoLuc and BrLuc genes were synthesized by Eurofins Genomics (Ebersberg, Germany) and cloned into pcDNA3.1 (+) vector and pQE-30 UA plasmid (Qiagen) by standard molecular cloning procedures.Plasmid construction and mutations of BoLuc and BrLuc variants
The sequences of the P. pyralis luciferases mutants BoLuc and BrLuc were synthesized by Eurofin MWG Operon (Ebersberg, Germany).The two sequences were cloned into pcDNA3.1 (+) vector backbone (Invitrogen, Waltham, Massachusetts, USA) and pQE-30 UA plasmid (Qiagen) by mean of a blunt ligation, obtaining plasmids pQEBoLuc, pQEBrLuc, pCDNA-BoLuc, and pcDNA-BrLuc. All constructs were verified by DNA sequencing. The BoLuc (bright orange luciferase) contained the following mutations F14R, L35Q, V182K, I232K, F465R, Y33N, T214A, A215L, F295L, E354K, V241I, G246A, F250S, S284T, N119G, N50D. While BrLuc had the same mutations except V241I, G246A, F250S (Fig. S1†).
Protein expression and purification
Plasmids pQE-BoLuc and pQE-BRLuc were transformed in JM109 competent cells for luciferase expression following the protocol previously described.24 Briefly transformed E. coli 250 mL cultures were grown in LB medium with 50 μg mL−1 ampicillin at 37 °C with shaking until an OD600 nm of 0.6 was reached. Cultures were induced with 0.1 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) and incubated with shaking for 5 h at 30 °C. Bacterial cells were pelleted by centrifugation at 3000g, 4 °C for 20 min. Cell-lysis extraction buffer solution was prepared with 10 μL of lysozyme (10 mg mL−1 in PBS) and 1 μL of serine protease, PMSF (phenylmethylsulfonyl fluoride) 100 mM in EtOH per 1 mL of B-PER Reagent. The washing solution was LEW Buffer, 300 mM NaCl and 50 mM NaH2PO4·H2O at pH 8.0. Elution Buffer was LEW Buffer plus 250 mM imidazole solution, adjusted to pH 8.0 with NaOH. The bacterial pellet was then resuspended on ice using 2 mL of cell-lysis extraction buffer for 20 min. A 4 mL volume of LEW Buffer was added to the cell lysate and resuspended at room temperature for 10 min. After ultracentrifugation at 4500g, at 4 °C for 1 h, the clear supernatant was collected to proceed with the purification using Protino Ni-IDA resins for purification of His-tag proteins according to the manufacturer's instructions. Protein concentration was assessed by a Bradford assay with bovine serum albumin as standard. The activity of the purified proteins was evaluated using a luminometer (Thermo Scientific Varioskan LUX Multimode microplate reader) on 10 μL-aliquots of eluted protein, and 10 μL of Bright-Glo Luciferase Assay System (Promega).In vitro characterization of luciferase mutants
For thermal inactivation studies BoLuc and BrLuc aliquots (0.6 mg mL−1) were incubated at and 25 °C, 37 °C and 45 °C in 0.2 mL of Lew buffer (pH 7.8). Aliquots (5 μL) were taken every 10 min till 3 hours and then after 5, 15 hours and overnight incubation to track enzyme inactivation. Emission spectra were obtained in a white 384-well microplate after 30 min incubation at 25 °C, 37 °C and 45 °C with a Thermo Scientific Varioskan LUX Multimode Microplate Reader using 10 μL of the purified luciferases with 10 μL of Bright-Glo substrate. BL emission spectra were obtained at pH 5.0, 7.0 and 8.0 as previously described.25Heat inactivation studies
BoLuc luciferase (0.6 mg) and BrLuc luciferase (0.6 mg) were incubated at 37 °C in 0.2 mL of 25 mM glycylglycine Lew buffer (pH 8.0). To track enzyme inactivation 5 μL-aliquots of BoLuc and BrLuc luciferases were taken at regular intervals (every 10 min and after an overnight incubation). BL signals were obtained after the addition of the Bright-Glo substrate (5 μL).Determination of kinetic parameters
To calculate the apparent Michaelis–Menten (Km) for ATP, BoLuc and BrLuc luciferase activities were determined in presence of the enzyme (2.4 μg), excess of D-LH2 substrate (1.0 mM) with varying concentrations of ATP (from 2.0 × 10−6 to 2.0 × 101 mM).All measurements were repeated at least three times. GraphPad Prism v8.3.0 software (GraphPad Software, La Jolla, CA, USA) was used to calculate apparent Km values, fitting the data to the Michaelis–Menten equation.
Characterization in Hek293T cell models
Hek293T cells were grown routinely in 5% CO2 in air in minimum essential medium with Earle's salts (DMEM) supplemented with 10% (v/v) fetal bovine serum, 2 mM L-glutamine, 0.1 mM nonessential amino acids, MEM vitamins, and antibiotic/antimycotic solution. Cells, previously seeded in a 96-well optical-bottom black plate at a density of 2.0 × 104 cells per well, were transiently transfected with pcDNA3.1-BoLuc or pcDNA3.1-BrLuc expression vectors using the FUGENE® HD transfection reagent at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and incubated under standard conditions for 24 h at 37 °C and 5% CO2. Emission kinetics (20 min with 200 ms integration time) were obtained with the luminometer Thermo Scientific Varioskan LUX Multimode Microplate Reader after injection of 60 μL of D-luciferin citrate solution 1.0 mM pH 5.0. Emission spectra were recorded from 450 to 800 nm, at 2 nm intervals with 1000 ms integration time. HEK293T cells transfected with pcDNA3.1-BgLuc in the same experimental conditions were used to compare the results. To obtain the BL image by smartphone acquisition, one day before transfection cells were plated on a 24-well plate at a density of 8 × 104 cells per well and transfected with 0.5 μg pCDNA3_BgLuc, or 0.5 μg pcDNA3-BoLuc, or 0.5 μg pCDNA3-BrLuc expression vectors using a FuGENE®HD
3, and incubated under standard conditions for 24 h at 37 °C and 5% CO2. Emission kinetics (20 min with 200 ms integration time) were obtained with the luminometer Thermo Scientific Varioskan LUX Multimode Microplate Reader after injection of 60 μL of D-luciferin citrate solution 1.0 mM pH 5.0. Emission spectra were recorded from 450 to 800 nm, at 2 nm intervals with 1000 ms integration time. HEK293T cells transfected with pcDNA3.1-BgLuc in the same experimental conditions were used to compare the results. To obtain the BL image by smartphone acquisition, one day before transfection cells were plated on a 24-well plate at a density of 8 × 104 cells per well and transfected with 0.5 μg pCDNA3_BgLuc, or 0.5 μg pcDNA3-BoLuc, or 0.5 μg pCDNA3-BrLuc expression vectors using a FuGENE®HD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA ratio of 3
DNA ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and incubated at 37 °C with 5% CO2 for 72 h. Then, cells gently detached, were counted and transferred at the same concentration of 5.0 × 103 cells per well in a black 384-well small volume plate. Green, orange and red-emitting cells were imaged after addition of 5 μL BrightGlo substrate with the smartphone camera integrating BL signals for 30 s with ISO 3200. All transfections were performed in triplicate and repeated at least three times.
1 and incubated at 37 °C with 5% CO2 for 72 h. Then, cells gently detached, were counted and transferred at the same concentration of 5.0 × 103 cells per well in a black 384-well small volume plate. Green, orange and red-emitting cells were imaged after addition of 5 μL BrightGlo substrate with the smartphone camera integrating BL signals for 30 s with ISO 3200. All transfections were performed in triplicate and repeated at least three times.
Inflammation assay
To monitor NF-kB pathway activation, one-day spheroids were obtained after seeding of 2.0 × 104 cells per well in 96-well microspace round bottom cell culture plates (Corning® Elplasia® Plates) as described previously26 and transiently transfected with plasmid encoding for BoLuc and BrLuc under the regulation of NF-kB responsive element (pGL4.32[NF-κB-RE]-BoLuc and pGL4.32[NF-κB-RE]-BrLuc). 24 h post-transfection, spheroids were treated in triplicate with 50 μL of TNFα solutions in culture medium (0.1–20 ng mL−1) or with 50 μL of culture medium as a control. After 5 h incubation at 37 °C, 50 μL Bright-Glo substrate was added to each well. The half maximal effective concentration (EC50), which is the concentration of the inducer (TNFα) which produces 50% of the maximum possible response, was calculated using the following equation:Y = bottom + (top − bottom)/(1 + 10((log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EC50 − X) × hillslope)) EC50 − X) × hillslope)) |
All the experiments were performed in triplicate and repeated at least three times.
Results and discussion
Design of BoLuc and BrLuc luciferases
Prompted by the good stability properties of a recently developed luciferase variant (BgLuc) we decided to use this mutant as the starting point for obtaining red-shifted variants for multiplexed and in vivo applications. Since haemoglobin and other biomolecules reduce light transmission at wavelengths below 600 nm in mammalian tissues, red emission enables to reduce both absorption and scattering.27 The most prominent mutation which accounts for a red-shifted emission is the well-described S284T.28 The reasons for this red-shifted emission are still debated and several mechanisms were proposed, a recent comprehensive review underlines the co-existence of multiple forms of oxyluciferin.29 Although S284T mutation provided a significant red-shifting, most likely caused by changes in the microenvironment, with an emission maximum (λmax) of 616 nm at pH 7.8 corresponding to a 60 nm red shift as compared to the wild-type enzyme λmax, it caused a severe loss of activity, retaining only 22% of the specific activity of the unmodified luciferase.30 Therefore, the starting point for developing the red-emitting mutants was the highly thermostable and pH-resistant synthetic luciferase BgLuc.24 This mutant contains 15 mutations which provide advantageous properties in terms of stability and brightness such as mutations replacing solvent-exposed hydrophobic amino acids with hydrophilic residues, a strategy which showed successful not only for luciferase but also with other enzymes, such as acetylcholinesterase.31The mutation S284T was introduced in the human codon optimized version of BgLuc, leading to a very bright orange emitting luciferase (BoLuc-Bright orange Luciferase). This result was ascribed to the three mutations V241I/G246A/F250S that were initially introduced into the BgLuc which have been reported to increase light output and have modest blue-shifted emission (λmax 548 nm).32 These mutations counteract the presence of S284T mutation, resulting in an orange luciferase. Therefore, these mutations were removed in the Brluc variant, resulting into a more pronounced red shifted emission (Fig. S1†). The two new luciferases, BoLuc and BrLuc, were first characterized in vitro, in terms of thermal and pH stability and emission properties, and then in vitro with 3D mammalian cell-based assays.
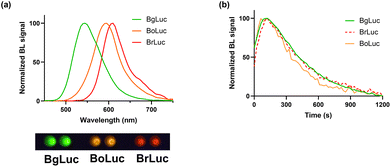
The emission spectra and kinetics of the purified BoLuc and BrLuc were obtained in the presence of saturating concentrations of substrate and co-factors, either Bright-Glo commercial substrate or 1.0 mM D-LH2, 2.0 mM Mg-ATP, as described in Materials and Methods section. Both BoLuc and BrLuc showed flash type emission kinetics with a peak after 10 s and a signal half-life of 25 s and 30 s, respectively (Fig. 1b). As concerns the emission spectra, BoLuc showed a maximum at 595 nm and half bandwidth of 75 nm while BrLuc was characterized by a more pronounced red shifting with a maximum at 615 nm and half bandwidth of 60 nm with Bright-Glo substrate.
As concerns kinetic parameters, while the orange mutant showed a very low Km for ATP (8.2 ± 0.2 μM), the red variant had a marked decreased affinity for ATP (Km of 196 ± 11 μM), this finding is commonly reported for red mutants. As concerns the turnover, both the mutants showed Kcat in the order of 108 cps per M, as the BgLuc, which appear higher than those reported for the wild-type P. pyralis luciferase by others,33 however it must be considered that different measurement conditions were used (Table S1†).
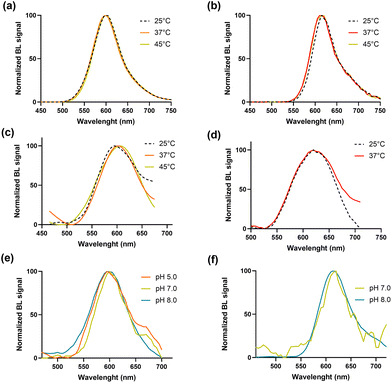
To explore thermal stability of the two luciferases heat inactivation studies were conducted by keeping aliquots of the purified proteins at 25, 37 and 45 °C for different periods of time (from 10 min to 8 h) (Fig. 1c). The thermal stability was better than that of the BgLuc variant (half-life of 2.5 h at 37 °C) and outperformed the wild-type luciferase (half-life of 0.26 h at 37 °C),24 showing a remarkable 71% and 68% of remaining activity after 15 h at 37 °C for BrLuc and BoLuc, respectively. The emission behaviour at different pH was also evaluated. The intensity of the emissions at different pH was very different, for example at pH 5.0 the intensity of BoLuc was 70% of the intensity obtained at pH 7.0 (Fig. S2a†). Both BoLuc and BrLuc showed a pH independent emission peak (Fig. 2). This is a crucial property for multiplexed assays and biosensors since spectral resolution of the two signals could be impaired by partial overlapping of the emissions, as confirmed by using the pH-sensitive luciferases in which red shifted emission is observed at lower pH.33
Conversely, BrLuc emission intensity was highly affected at lower pH (Table 1), with almost negligible signal at pH 5.0 (Fig. S2b†). As reported in Table 1 the broadening of the emission spectrum observed with the BgLuc variant using D-LH2 substrate instead of the optimized commercial BrigthGlo formulation was not observed with the new mutants.
| D-LH2 | BL emission pH 5.0 | BL emission pH 7.0 | BL emission pH 8.0 | |||
|---|---|---|---|---|---|---|
| λ max (nm) | Half bandwidth (nm) | λ max (nm) | Half bandwidth (nm) | λ max (nm) | Half bandwidth (nm) | |
| BgLuc | 548 | 124 | 550 | 94 | 550 | 97 |
| BoLuc | 595 | 78 | 600 | 62 | 595 | 78 |
| BrLuc | n.d. | n.d. | 620 | 61 | 615 | 72 |
After incubation of the enzymes at 25, 37 and 45 °C for 30 min the spectra were measured either with 1.0 mM D-LH2 and 2.0 mM ATP or the commercial Bright-Glo substrate (Fig. 2 and Table 2). The relative intensity of BoLuc at 25 °C was 80% of the intensity obtained at 37° and 45 °C (Fig. S3a†), while those obtained for BrLuc at 25 °C and 37 °C were about 88% of the intensity obtained at 45 °C (Fig. S3b†). Despite no significant differences were observed in λmax of BoLuc and BrLuc at different temperature with the commercial Bright-Glo substrate, a shift of about 5 nm and 10 nm was detected at 37° and 45 °C for BoLuc luciferase when compared to that obtained at 25 °C using 1.0 mM D-LH2 and 2.0 mM ATP. Concerning the BrLuc luciferase the shift of 10 nm was observed only at 37 °C, while at 45 °C a negligible signal was obtained.
| Luciferase mutant | BL emission (25 °C) | BL emission (37 °C) | BL emission (45 °C) | |||
|---|---|---|---|---|---|---|
| λ max (nm) | Half bandwidth (nm) | λ max (nm) | Half bandwidth (nm) | λ max (nm) | Half bandwidth (nm) | |
| a Numbers in brackets refer to results obtained with 1.0 mM D-LH2 and 2.0 mM ATP. b Not detectable due to low signal intensities. | ||||||
| BgLuc | 552 (552)a | 70 (85)a | 552 (552)a | 76 (78)a | 556 (556)a | 82 (90)a |
| BoLuc | 600 (595)a | 75 (117)a | 600 (600)a | 70 (90)a | 600 (605)a | 75 (97)a |
| BrLuc | 615 (615)a | 60 (90)a | 615 (625)a | 65 (100)a | 620 (n.d.)b | 65 (n.d)b |
Characterization of BoLuc and BrLuc expression in mammalian cell models
Prompted by the encouraging results obtained with the purified proteins, the cDNAs encoding for the two luciferases were cloned into pcDNA3 backbone and transfected in Hek293T cell lines. BoLuc and BrLuc were expressed in Hek293T 2D and 3D cell cultures and characterized in terms of emission spectra and kinetics. A preliminary study was performed to assess the BL emission in living cells using complementary metal–oxide semiconductor (CMOS) smartphone-integrated camera. As shown in Fig. 3a, the signal obtained with 5.0 × 103 Hek293T cells (in 10 μL volume) expressing either BgLuc, BoLuc or BrLuc was imaged with a Oneplus 6 (Fig. 3a). Then, spectra were obtained in cell monolayers and 3D cell models using HEK293T transiently transfected with BoLuc and BrLuc (Fig. 3a).When compared to BgLuc, which showed a half bandwidth of 72 nm, BoLuc and BrLuc presented a half bandwidth of 70 nm and 57 nm, respectively. The narrow peak of BrLuc represents an advantage for multiplexed assays although it reflects minor light output, as reported for several other red emitting luciferases which showed a reduced photon yield.34–36 In addition, despite the lower light output of BrLuc makes it less advantageous than BgLuc and BoLuc for biosensing applications, its narrow emission peak in the red region represents an advantage for multiplexed biosensing and in vivo imaging.
Therefore, for applications with 3D models and in vivo imaging, in which spectral unmixing is not required BoLuc could be a more suitable candidate. For this reason, we investigated the performance of BoLuc and BrLuc as reporter proteins for 3D cell bioassays.
3D cell bioassay for inflammation
The interesting properties of these two mutants, especially in terms of stability at 37 °C and red-shifted signals, prompted the implementation into a 3D cell-based assay for inflammation.TNF-α is considered a key modulator of systematic inflammation and represents a biomarker to monitor inflammatory response in several diseases. Immunoassays are widely employed with a few assays suitable for the detection of TNF-α in clinical laboratories (limit of detection-LOD 0.1–10.0 pg mL−1).37
One day old Hek293T spheroids, previously transfected with a reporter construct in which either BoLuc or BrLuc luciferase is placed under the control of the NF-kB response element, were incubated with TNF-α (concentration range 0–20 ng mL−1) for 5 h at 37 °C. Dose–response curves are shown in Fig. 4, showing a LOD, calculated as the TNFα concentration corresponding to the blank plus three times the standard deviation, of 0.06 ng mL−1 and 0.12 ng mL−1 for BoLuc and BrLuc, respectively and EC50 of 1.9 ng mL−1 for both.
As expected, the higher photon yield of BoLuc enabled to lower the limit of quantification (LOQ), calculated as the TNFα concentration corresponding to the blank plus ten times the standard deviation (0.08 vs. 0.22 ng mL−1), however still not competitive with commercial immunoassays used in clinical settings which have LODs in the range 0.1–10.0 pg mL−1.
While in our assay BoLuc provided improved analytical performance, BrLuc should be recommended for dual assays based on spectral resolution in combination with blue-shifted reporter proteins or for in vivo applications. Further studies will be required to assess its suitability for in vivo animal models.
Conclusions
Capitalizing on past efforts in the search for new luciferases, we have developed two new synthetic luciferases, an orange and a red-emitting luciferase. The novel luciferases, obtained by introducing mutations onto the previously reported thermostable mutant BgLuc, were characterized in vitro at different pH and temperature, showing good features, especially in terms of thermal stability. Prompted by these encouraging results the two proteins were expressed in Hek293T 3D cell culture models to explore their suitability as reporter proteins. As proof-of-principle application, an inflammation assay based on Hek293T 3D cell cultures was developed using either the orange or the red-emitting mutant; both assays showed excellent analytical performance. Moreover, since these luciferases require the same substrate, D-luciferin, they can be easily implemented in dual-color assays with a significant reduction of total cost per assay.It must be pointed out that the orange-emitting luciferase had a broader emission, especially in the presence of D-luciferin substrate, thus being suitable for applications demanding high sensitivity. Conversely, the redder mutant was characterized by a very narrow emission, with a half bandwidth of 57 nm when expressed in Hek293T 3D cell cultures, thus it should provide superior performance in in vivo biosensing and multiplexed applications.
Author contributions
Conceptualization, E. M.; methodology, M. M. C., D. G., and E. M.; software, E. M., and M. M. C.; validation, M. M. C., D. G.; formal analysis, M. M. C.; investigation, M. M. C, D. G. and E. M.; resources, M. M. C. and E. M.; data curation, M. M. C.; writing – original draft preparation, E. M.; writing – review and editing, M. M. C. and E. M.; visualization, M. M. C. and E. M.; supervision, E. M.; funding acquisition, E. M. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported in part by the PRIMA program, project Fedkito, supported by the European Union. This study was, in part, carried out within the Agritech National Research Center and received funding from the European Union Next-Generation EU National Recovery and Resilience Plan (NRRP), Mission 04 Component 2, investment 1.4—D.D. 1032 17/06/2022, CN00000022 and investment 1.5—NextGenerationEU, call for tender no. 3277, dated 30/12/2021 and award number: 0001052, dated 23/06/2022. This manuscript reflects only the authors’ views and opinions; neither the European Union nor the European Commission can be considered responsible for them. We acknowledge Ph.D. programs on green topics (PON “Research and Innovation” 2014–2020) funded by FSE REACT-EU.References
- B. Baljinnyam, M. Ronzetti and A. Simeonov, Expert Opin. Drug Discovery, 2023, 18, 25–35 CrossRef CAS PubMed.
- E. E. Bashmakova, V. V. Krasitskaya, A. N. Kudryavtsev, V. G. Grigorenko and L. A. Frank, Photochem. Photobiol., 2017, 93, 548–552 CrossRef CAS PubMed.
- S. D'Alessandro, G. Camarda, Y. Corbett, G. Siciliano, S. Parapini, L. Cevenini, E. Michelini, A. Roda, D. Leroy, D. Taramelli and P. Alano, J. Antimicrob. Chemother., 2016, 71, 1148–1158 CrossRef.
- S. J. Williams and J. A. Prescher, Acc. Chem. Res., 2019, 52, 3039–3050 CrossRef CAS PubMed.
- S. Liu, Y. Su, M. Z. Lin and J. A. Ronald, ACS Chem. Biol., 2021, 16, 2707–2718 CrossRef CAS PubMed.
- M. Bausart, E. Bozzato, N. Joudiou, X. Koutsoumpou, B. Manshian, V. Préat and B. Gallez, Cancers, 2023, 15, 1919 CrossRef CAS PubMed.
- E. Michelini, L. Cevenini, M. M. Calabretta, D. Calabria and A. Roda, Anal. Bioanal. Chem., 2014, 406, 5531–5539 CrossRef CAS.
- B. R. Branchini, T. L. Southworth, D. M. Fontaine, D. Kohrt, F. S. Welcome, C. M. Florentine, E. R. Henricks, D. B. DeBartolo, E. Michelini, L. Cevenini, A. Roda and M. J. Grossel, Anal. Biochem., 2017, 534, 36–39 CrossRef CAS PubMed.
- B. R. Branchini, T. L. Southworth, D. M. Fontaine, D. Kohrt, M. Talukder, E. Michelini, L. Cevenini, A. Roda and M. J. Grossel, Anal. Biochem., 2015, 484, 148–153 CrossRef CAS PubMed.
- S. V. Markova, M. D. Larionova and E. S. Vysotski, Photochem. Photobiol., 2019, 95, 705–721 CrossRef CAS PubMed.
- P. V. Natashin, L. P. Burakova, M. I. Kovaleva, M. B. Shevtsov, D. A. Dmitrieva, E. V. Eremeeva, S. V. Markova, A. V. Mishin, V. I. Borshchevskiy and E. S. Vysotski, Int. J. Mol. Sci., 2023, 24, 6869 CrossRef CAS PubMed.
- A. A. Kotlobay, K. S. Sarkisyan, Y. A. Mokrushina, M. Marcet-Houben, E. O. Serebrovskaya, N. M. Markina, L. Gonzalez Somermeyer, A. Y. Gorokhovatsky, A. Vvedensky, K. V. Purtov, V. N. Petushkov, N. S. Rodionova, T. V. Chepurnyh, L. I. Fakhranurova, E. B. Guglya, R. Ziganshin, A. S. Tsarkova, Z. M. Kaskova, V. Shender, M. Abakumov, T. O. Abakumova, I. S. Povolotskaya, F. M. Eroshkin, A. G. Zaraisky, A. S. Mishin, S. V. Dolgov, T. Y. Mitiouchkina, E. P. Kopantzev, H. E. Waldenmaier, A. G. Oliveira, Y. Oba, E. Barsova, E. A. Bogdanova, T. Gabaldón, C. V. Stevani, S. Lukyanov, I. V. Smirnov, J. I. Gitelson, F. A. Kondrashov and I. V. Yampolsky, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 12728–12732 CrossRef CAS PubMed.
- A. A. Kotlobay, M. A. Dubinnyi, K. V. Purtov, E. B. Guglya, N. S. Rodionova, V. N. Petushkov, Y. V. Bolt, V. S. Kublitski, Z. M. Kaskova, R. H. Ziganshin, Y. V. Nelyubina, P. V. Dorovatovskii, I. E. Eliseev, B. R. Branchini, G. Bourenkov, I. A. Ivanov, Y. Oba, I. V. Yampolsky and A. S. Tsarkova, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 18911–18916 CrossRef CAS PubMed.
- P. J. White, D. J. Squirrell, P. Arnaud, C. R. Lowe and J. A. H. Murray, Biochem. J., 1996, 319, 343–350 CrossRef CAS PubMed.
- A. H.-W. Yeh, C. Norn, Y. Kipnis, D. Tischer, S. J. Pellock, D. Evans, P. Ma, G. R. Lee, J. Z. Zhang, I. Anishchenko, B. Coventry, L. Cao, J. Dauparas, S. Halabiya, M. DeWitt, L. Carter, K. N. Houk and D. Baker, Nature, 2023, 614, 774–780 CrossRef CAS PubMed.
- E. S. Vysotski, Int. J. Mol. Sci., 2022, 24, 281 CrossRef.
- M. M. Calabretta, L. Montali, A. Lopreside, F. Fragapane, F. Iacoangeli, A. Roda, V. Bocci, M. D'Elia and E. Michelini, Anal. Chem., 2021, 93, 7388–7393 CrossRef CAS PubMed.
- G. Borghei and E. A. H. Hall, Analyst, 2014, 139, 4185–4192 RSC.
- G. Kamiya, N. Kitada, T. Furuta, T. Hirano, S. A. Maki and S.-B. Kim, Int. J. Mol. Sci., 2023, 24, 1420 CrossRef CAS PubMed.
- Y. Sun, J. Liu, P. Wang, J. Zhang and W. Guo, Angew. Chem., Int. Ed., 2012, 51, 8428–8430 CrossRef CAS PubMed.
- M. J. Afshari, C. Li, J. Zeng, J. Cui, S. Wu and M. Gao, ACS Nano, 2022, 16, 16824–16832 CrossRef CAS PubMed.
- B. R. Branchini, D. M. Ablamsky, J. M. Rosenman, L. Uzasci, T. L. Southworth and M. Zimmer, Biochemistry, 2007, 46, 13847–13855 CrossRef CAS PubMed.
- V. R. Viviani, G. F. Pelentir and V. R. Bevilaqua, Biosensors, 2022, 12, 400 CrossRef CAS PubMed.
- M. M. Calabretta, D. Gregucci, H. Martínez-Pérez-Cejuela and E. Michelini, Biosensors, 2022, 12, 742 CrossRef CAS PubMed.
- H. Martínez-Pérez-Cejuela, D. Gregucci, M. M. Calabretta, E. F. Simó-Alfonso, J. M. Herrero-Martínez and E. Michelini, Anal. Chem., 2023, 95, 2540–2547 CrossRef.
- L. Cevenini, M. M. Calabretta, A. Lopreside, B. R. Branchini, T. L. Southworth, E. Michelini and A. Roda, Photochem. Photobiol., 2017, 93, 531–535 CrossRef CAS.
- M. Endo and T. Ozawa, Int. J. Mol. Sci., 2020, 21, 6538 CrossRef CAS.
- B. R. Branchini, T. L. Southworth, N. F. Khattak, E. Michelini and A. Roda, Anal. Biochem., 2005, 345, 140–148 CrossRef CAS PubMed.
- M. B. Al-Handawi, S. Polavaram, A. Kurlevskaya, P. Commins, S. Schramm, C. Carrasco-López, N. M. Lui, K. M. Solntsev, S. P. Laptenok, I. Navizet and P. Naumov, Chem. Rev., 2022, 122, 13207–13234 CrossRef CAS PubMed.
- H. Caysa, R. Jacob, N. Müther, B. Branchini, M. Messerle and A. Söling, Photochem. Photobiol. Sci., 2009, 8, 52–56 CrossRef CAS PubMed.
- C. Strub, C. Alies, A. Lougarre, C. Ladurantie, J. Czaplicki and D. Fournier, BMC Biochem., 2004, 5, 9 CrossRef PubMed.
- B. R. Branchini, D. M. Ablamsky, J. M. Rosenman, L. Uzasci, T. L. Southworth and M. Zimmer, Biochemistry, 2007, 46, 13847–13855 CrossRef CAS PubMed.
- T. Pozzo, F. Akter, Y. Nomura, A. Y. Louie and Y. Yokobayashi, ACS Omega, 2018, 3, 2628–2633 CrossRef CAS PubMed.
- Y. Liang, J. Biomed. Opt., 2012, 17, 016004 CrossRef PubMed.
- B. R. Branchini, T. L. Southworth, N. F. Khattak, E. Michelini and A. Roda, Anal. Biochem., 2005, 345, 140–148 CrossRef CAS.
- H. Caysa, R. Jacob, N. Müther, B. Branchini, M. Messerle and A. Söling, Photochem. Photobiol. Sci., 2009, 8, 52–56 CrossRef CAS PubMed.
- A. Valaperti, Z. Li, M. Vonow-Eisenring and E. Probst-Müller, J. Pharm. Biomed. Anal., 2020, 179, 113010 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an01251d |
| This journal is © The Royal Society of Chemistry 2023 |