Open Access Article
Open Access ArticleDevelopment of an MIP based electrochemical sensor for TGF-β1 detection and its application in liquid biopsy†
Giulia
Siciliano
 *a,
Maria Serena
Chiriacò
a,
Francesco
Ferrara
a,
Antonio
Turco
*a,
Maria Serena
Chiriacò
a,
Francesco
Ferrara
a,
Antonio
Turco
 a,
Luciano
Velardi
b,
Maria Assunta
Signore
b,
Marco
Esposito
a,
Luciano
Velardi
b,
Maria Assunta
Signore
b,
Marco
Esposito
 a,
Giuseppe
Gigli
a and
Elisabetta
Primiceri
*a
a,
Giuseppe
Gigli
a and
Elisabetta
Primiceri
*a
aInstitute of Nanotechnology, CNR-Nanotec, via per Monteroni, 73100, Lecce, Italy. E-mail: giulia.siciliano@nanotec.cnr.it; elisabetta.primiceri@nanotec.cnr.it
bInstitute for Microelectronics and Microsystems, CNR-IMM, via per Monteroni, 73100, Lecce, Italy
First published on 2nd August 2023
Abstract
Oral cancer is one of the most common types of cancer in Europe and its large diffusion requires, together with prevention, the development of low-cost and reliable portable platforms for its diagnosis, with features of high selectivity and sensitivity. In this study, the development and characterization of a molecularly imprinted polymer (MIP)-based electrochemical sensor for TGF-β1 detection are reported. The optimized biosensor is a potential tool for the early screening of oral cancer. A biomimetic surface has been obtained by electropolymerization of o-phenylenediamine (o-PD) on platinum electrodes, in the presence of TGF-β1 as a template molecule. MIP synthesis, template removal and TGF-β1 rebinding have been monitored by Differential Pulse Voltammetry (DPV). Atomic Force Microscopy (AFM) has been performed to investigate and characterize the surface morphology and the influence of the washing step on MIP and NIP (non-imprinted polymer as the control) while the thickness of the polymer layer has been measured by Scanning Transmission Electron Microscopy (STEM) analysis. The MIP sensor performance has been tested in both buffer solution and saliva samples with TGF-β1, showing a linear response in the considered range (from 20 ng ml−1 down to 0.5 ng ml−1), an outstanding LOD of 0.09 ng mL−1 and affinity and selectivity to TGF-β1 also in the presence of interfering molecules. The sensor was used also for the detection of target molecules in spiked saliva samples with good recovery results suggesting the possibility of the use of the proposed system for large scale fast screening in oral cancer diagnosis.
Introduction
Molecular recognition is the underlying principle of most biological processes. Bioreceptors for target analyte recognition (such as antibody/antigen interaction and enzymatic catalysis) play a key role in the development of numerous powerful analytical platforms even if their physical and chemical stability can limit some applications. For this reason, there is a steadily growing interest in mimicking natural recognition systems using synthetic analogues with high affinity and specificity. Artificial receptors can offer improved stability, cost-effectiveness and a rapid fabrication process1 overcoming limitations of natural receptors such as low preservation temperature and short shelf life. In this respect, molecularly imprinted polymers (MIPs) offer valuable opportunities for biosensing purposes providing cavities in the polymer matrix with high affinity for a chosen target molecule used as a template. Notably, MIPs can offer greater long-term storage stability, low-cost production, potential reusability, resistance to microbial spoilage and custom synthesis of selective receptors without the need to inoculate laboratory animals, as well as facile integration with transducers.2 Among different strategies for MIP preparation, bulk polymerization is most frequently used,3 producing monolithic structures, which are then sieved to obtain the final product. On the other hand, electro-polymerization is a simple way to prepare MIPs directly on the surface of a transducer by controlling the thickness of the MIP layer by the amount of charge passed. This approach is particularly attractive for the fabrication of small devices for clinical diagnostics, environmental control and pharmaceutical industries. Moreover, the ease of measurement, the cheapness, the portability and the possibility to determine and monitor a wide range of analytes with a good limit of detection by electrochemical techniques have made electrochemical transducers very popular.4–8 Several works reported the use of MIPs based on electro-polymerization of o-phenylenediamine (o-PD) for the detection of several small template molecules, such as dopamine,9 glucose,10DL-phenylalanine,11 sorbitol,12 and TnT,13 while few works reported the use of MIPs for large molecules, including proteins,13,14 because of the difficulty in removing high molecular weight molecules from the polymeric matrix.Transforming growth factor β (TGF-β) includes three forms (TGF-β1, 2 and 3) which are expressed by all cells in human blood and play a key role in many processes such as cell growth, differentiation, apoptosis and immune responses. TGF-β signaling plays multiple roles in cancer, since it exhibits a tumor-suppressive activity in normal cells but a tumor-promoting role in malignant cells.15–17 The expression of TGF-β has been studied in many types of cancer, including breast, colorectal, prostate, and liver cancer.18,19 It was demonstrated that in early-stage tumors, the levels of TGF-β are associated with a favorable prognosis, since the TGF-β pathway promotes cell cycle arrest and apoptosis.20,21 In contrast, in advanced tumors, the TGF-β pathway promotes tumor progression and metastasis20,21 by stimulating invasion, cancer cell motility, and cell stemness.16 Regarding the role of TGF-β1 in oral cancer, Chen et al. demonstrated that patients affected by locally advanced oral cancer overexpressed TGF-β1.22 Together with IL-10, TNF-alpha, and VEGF, TGF-β regulatory cytokine is secreted by the stromal cells in a tumor environment and it has been found both in serum and in saliva secretion.23 Polz-Dacewicz et al., considering a cohort of around 80 patients, found that the concentration of serum TGF-β in patients was higher than that in healthy controls (11.3 ng ml−1vs. 7.8 ng ml−1) and the gap was higher in saliva, where the levels were of 24.1 ng ml−1 and 14.8 ng ml−1 for patients and controls, respectively.24 It is not difficult to predict that a minimally invasive assay like saliva sampling could represent a powerful means to broaden the application of early screening of oral cancer.25–27 Nowadays, the determination of TGF-β1 is mostly based on ELISA colorimetric kits that exploit sandwich-immunoassay for its quantification in the range from hundreds to thousands of pg mL−1. However, this approach has several shortcomings, such as not being rapid or suitable for near-to-bed diagnosis. The use of electrochemical methods for the determination of TGF-β1 can overcome these limitations, making them competitive in terms of LOD and selectivity.
In this work, we demonstrate the high performances of the first TGF-β1 MIP electrochemical sensor tested on a saliva sample as a screening tool for oral cancer. In particular the design and synthesis of the artificial macromolecular receptor for TGF-β1 based on molecular imprinting were achieved by electro-polymerization of the functional monomer, o-phenylenediamine (o-PD), in the presence of TGF-β1 as the target molecule. PoPD diamine was chosen as it is an insulating polymer able to grow in a self-limiting manner, producing a very thin layer to facilitate protein template removal. The MIP is obtained using three process steps: polymerization, template removal and rebinding. The analytical performance of the developed sensor was evaluated both in buffer solution and in a complex matrix such as saliva. In order to demonstrate the high affinity and specificity of the synthetic receptor for the template molecule, the rebinding test was performed also in the presence of another protein as an interfering molecule, namely interleukine-10 chosen for its chemical structure being similar to that of TGF-β1. The overall results about the sensor performance indicate that this approach is particularly attractive and promising for the detection of TGF-β1 to quickly diagnose oral cancer. The use of artificial receptors based on MIP as an alternative to antibodies has been exploited here for the first time in a saliva sample without requiring any labeling steps and with only simple pre-treatment.28,29 For these reasons, the developed sensor is suitable for point of care diagnostics since it allows very fast and simple analysis in a single step. The integration of MIPs into miniaturized devices, combined with their long shelf-life and no need for refrigerated storage, makes them innovative and powerful tools for developing easy and portable devices for liquid biopsy with the aim to increase the life expectancy of patients.
Materials and methods
Materials
o-Phenylenediamine (o-PD, ≥98%, Sigma-Aldrich, USA), potassium ferricyanide (K3[Fe(CN)6], 99%, E. Merck, Germany), potassium ferrocyanide (K4[Fe(CN)6], 99%, E. Merck, Germany) and TGFβ-1 (powder, Sigma-Aldrich, USA) were used as received. The stock solution of TGF-β1 (1 μg mL−1) was prepared in phosphate buffer solution at pH 7.4 and stored at −20 °C if not used. The standard stock solution of o-PD (0.1 mg mL−1) was prepared in acetate buffer solution (pH 5.2). Artificial saliva NeutraSal® (powder, dibasic sodium phosphate 0.010 mg, monobasic sodium phosphate 0.010 mg, calcium chloride 0.050 mg, sodium chloride 0.450 mg, sodium bicarbonate 0.016 mg, silicon dioxide 0.002 mg, Wisconsin Pharmacal Company, WI) was prepared by dissolving a foil sachet in 30 mL of milliQ water. All solvents, purchased from Sigma-Aldrich, are of the highest purity available. All aqueous solutions were prepared by using water obtained from a Milli-Q Gradient A-10 system (Millipore, 18.2 MΩ cm, organic carbon content ≤4 μg L−1).Electrochemical measurements
Electrochemical measurements were performed using an Autolab potentiostat (PGSTAT 204, Metrohm). Platinum (Pt) interdigitated microelectrodes, obtained by standard lithography fabrication, composed of two Pt connection tracks and patterned on a glass substrate were used as working and counter electrodes, while an Ag/AgCl electrode was used as the reference. All measurements were performed at room temperature (22 °C).TGF-β1 MIP fabrication
Electrosynthesis of a poly(ortho-phenylenediamine) (Po-PD) film was performed by cyclic voltammetry (CV) (5 scans) in the potential range −0.2–0.8 V vs. Ag/AgCl at a scan rate of 50 mV s−1 in a solution of acetate buffer (0.5 M, pH 5.2) containing 0.1 mg mL−1o-PD. Before polymerization, TGF-β1 was added in the o-PD solution as a template molecule at a concentration of 1 μg mL−1. After polymerization, the modified electrode was washed with different solutions to explore the best conditions for the template removal. The electrochemical stepwise process of MIP fabrication was investigated by the Differential Pulse Voltammetry (DPV) technique in the potential range of −0.2–0.8 V at a scan rate of 100 mV s−1. The control electrode was modified with a non-imprinted polymer (NIP), without TGF-β1 being added as a template. Modified electrodes were stored at room temperature (22 °C).Template removal and rebinding
Various solutions were explored for template removal from the polymer film. In particular, two solutions were tested. Firstly, modified electrodes were washed for 15 minutes with a solution based on acetic acid 5% in water, followed by washing with water. For comparison, another washing solution based on ethanol–water (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) and NaOH 0.25 M at 50 °C for 15 minutes was tested. The characterization of imprinted electrodes was carried out by the DPV technique in the presence of 10 mM K3[Fe(CN)6]/K4[Fe(CN)6], (1
1 v/v) and NaOH 0.25 M at 50 °C for 15 minutes was tested. The characterization of imprinted electrodes was carried out by the DPV technique in the presence of 10 mM K3[Fe(CN)6]/K4[Fe(CN)6], (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at room temperature (22 °C). The binding properties of the MIP modified electrodes were investigated by incubating them with spiked PBS solution at different concentrations of TGF-β1 for 1 h. The electrodes were then washed with PBS to remove the excess protein not specifically adsorbed to the sensor surface. The NIP receptor was used as a control. Devices were characterized by electrochemical measurements in the presence of 10 mM K3[Fe(CN)6]/K4[Fe(CN)6], (1
1) at room temperature (22 °C). The binding properties of the MIP modified electrodes were investigated by incubating them with spiked PBS solution at different concentrations of TGF-β1 for 1 h. The electrodes were then washed with PBS to remove the excess protein not specifically adsorbed to the sensor surface. The NIP receptor was used as a control. Devices were characterized by electrochemical measurements in the presence of 10 mM K3[Fe(CN)6]/K4[Fe(CN)6], (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at room temperature. The MIP sensor was tested also in a complex matrix such as artificial saliva, by incubating MIP modified electrodes with artificial saliva solutions containing different concentrations of TGF-β1 for 1 h.
1) at room temperature. The MIP sensor was tested also in a complex matrix such as artificial saliva, by incubating MIP modified electrodes with artificial saliva solutions containing different concentrations of TGF-β1 for 1 h.
Morphological characterization
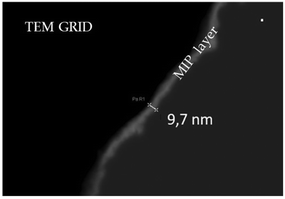
The surface morphology and the roughness of the samples were investigated by Atomic Force Microscopy (AFM) analyses. AFM topography images were acquired over 1 × 1 μm2 scan areas (resolution of 256 × 256 points) using a Nanosurf CoreAFM instrument operating in tapping mode, at room temperature and in air environment. A monolithic silicon probe tip with gold reflective coating (TAP300GD-G) for high frequency, non-contact and tapping mode operation was used at a typical resonance frequency of 300 kHz, with a constant force of 40 N m−1. The roughness values were determined using the CoreAFM 3.10 software, whereas the 3D images were obtained using the Gwyddion 2.45 software.To measure the polymer thickness, Scanning Transmission Electron Microscopy (STEM) characterization was performed by means of a Merlin Zeiss microscope on a TEM grid covered with a PoPD layer synthesized under the same conditions as those for the sensor devices. In order to obtain Z contrast sensitiveness from the images, the STEM was configured in high-angle annular dark-field mode (HAADF).
Results and discussion
Synthesis and characterization of the TGF-β1 MIP receptor
A schematic diagram of MIP sensor preparation is shown in Scheme 1. The MIP-film was synthesized by cyclic voltammetric deposition (5 scans) of o-PD in the presence of TGF-β1 on single-use platinum microelectrodes patterned on glass substrates (Fig. S1†). A typical cyclic voltammogram recorded during the electro-polymerization of o-PD in the presence of TGF-β1 on platinum electrodes is shown in Fig. 1A. A significant decrease in the anodic peak was observed from the 1st to the 5th cycle, which corresponds to an irreversible monomer oxidation on the electrode surface during continuous cycling and illustrates the formation of a non-conductive polymer film on the electrode surface. Different trends were observed in the cyclic voltammogram evolution in the presence of TGF-β1 (MIP) and in its absence (NIP) (Fig. 1B). Despite the fact that NIP electro-polymerization follows the standard behaviour already reported in the literature, MIP deposition evidenced substantial differences. Moreover, during MIP electro-polymerization we observed that an oxidation current loop formed at the positive end of the potential scan. The observed current loops could be due to the strong interaction between TGF-β1, monomers and polymeric chains formed during electro-polymerization. In fact, it was also demonstrated that TGF-β1 could be adsorbed on the positively charged polymer due to electrostatic interactions.30 Therefore, TGF-β1 molecules can act as counterions that participate in the polymerization process as the balancing species.31 This could partially inhibit the electro-polymerization process due to the lower availability of reactive sites on oligomeric chains and/or free monomers in solution due to steric hindrance and chemical interactions with TGF-β1. As a result of this contribution, the currents recorded during the backward potential scan were larger than those at the same potential during the forward potential scan, due to the inhibition of the polymerization process leading to the observed oxidation current loops. Moreover, this aspect suggests the incorporation of TGF-β1 in the polymeric matrices which is favoured by electrostatic interactions. In order to improve the device sensitivity, an ultrathin polymer film was formed on the electrode surface using just five cycles of CV, although the polymer thickness could be easily increased by working on specific parameters during the electro-polymerization such as the scan rate, the number of cycles and the monomer concentration. In particular, before choosing the optimum value of 5 scans, the number of cycles during electropolymerization was considered as a variable influencing the polymer thickness, keeping the scan rate and the monomer concentration constant. In this sense, electropolymerization was also performed by 3 scans and 10 scans of CV. Electropolymerization by 3 scans led to the formation of an uneven and unstable polymer film, while the use of 10 scans gave no performance results in terms of template removal, probably because the thickness of the polymer film increased too much. This is the reason why, after testing 3 and 10 scans during electropolymerization, a value of 5 scans has been selected as optimum, since it gives the best performance in MIP fabrication.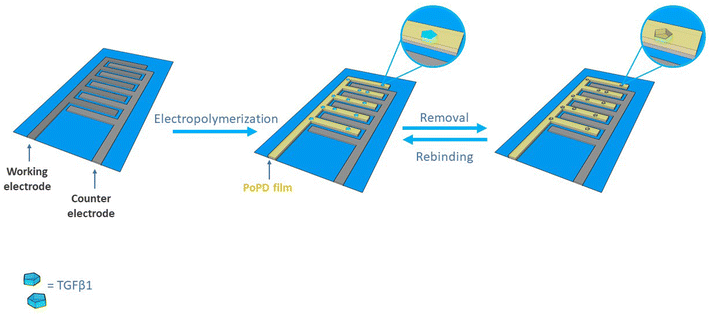 | ||
| Scheme 1 Schematic representation of TGF-β1 MIP receptor fabrication using in situ electrochemical polymerization on single-use platinum microelectrodes patterned on glass substrates. | ||
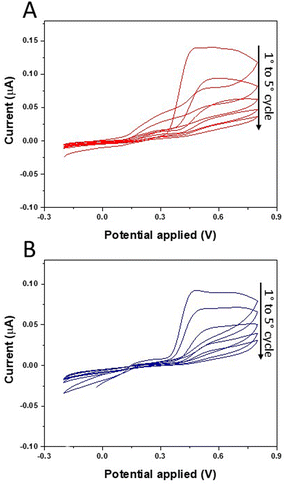 | ||
| Fig. 1 TGF-β1 MIP receptor formation using in situ electrochemical polymerization. Cyclic voltammetric deposition of 0.1 mg mL−1o-PD in the presence of 1 μg mL−1 TGF-β1 (A) and in its absence (B). | ||
The polymer morphology has been studied by STEM in Z-contrast mode on a TEM grid after PoPD coating: high-magnification STEM images (Fig. 2) of the polymer-surface highlight an average thickness of about 10 nm and a uniform PoPD deposition.
The obtained MIP receptor films were then treated with different solutions in order to explore the best conditions for template removal. Solvent extraction is an important aspect in the preparation of MIP, affecting the resulting binding properties. The solvent should suppress the interaction between the polymer and the template and allow fast diffusion of the template from the polymer film. Among the different bathing solutions reported for template extraction,13 two were tested in this work, namely a solution of acetic acid 5% and an alkaline medium based on ethanol–water (EtOH/H2O 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) solution containing 0.25 M NaOH. Imprinted electrodes before and after the template removal by solutions were investigated by the DPV technique. The solution of acetic acid 5% was found to be a poor solvent for PoPD, leading to an ineffective release of the template (Fig. S2†). In contrast, the alkaline medium tested for 15 min at 50 °C proved to be more effective in quantitatively removing the template11 (Fig. 3). Specifically, the alkaline medium suppresses the interaction between the positively charged polymer and the TGF-β1 adsorbed on it by electrostatic interactions30 and also prevents the degradation of the PoPD film and the denaturation of template molecules at extreme pH.13 As a matter of fact, the DPV results of imprinted electrodes before and after template removal carried out in the presence of 10 mM K3[Fe(CN)6]/K4[Fe(CN)6], (1
1 v/v) solution containing 0.25 M NaOH. Imprinted electrodes before and after the template removal by solutions were investigated by the DPV technique. The solution of acetic acid 5% was found to be a poor solvent for PoPD, leading to an ineffective release of the template (Fig. S2†). In contrast, the alkaline medium tested for 15 min at 50 °C proved to be more effective in quantitatively removing the template11 (Fig. 3). Specifically, the alkaline medium suppresses the interaction between the positively charged polymer and the TGF-β1 adsorbed on it by electrostatic interactions30 and also prevents the degradation of the PoPD film and the denaturation of template molecules at extreme pH.13 As a matter of fact, the DPV results of imprinted electrodes before and after template removal carried out in the presence of 10 mM K3[Fe(CN)6]/K4[Fe(CN)6], (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at room temperature show that the washing step with NaOH in EtOH/H2O increases the peak current since after the template removal the empty imprinted sites increase the surface area and the diffusion on the electrode surface, thus favoring the electron transfer between the solution and the electrode. The obtained data indicate that the use of NaOH in EtOH/H2O as washing solution leads to an effective release of the template molecule, and thus it has been selected for the fabrication of the MIP sensor.
1) at room temperature show that the washing step with NaOH in EtOH/H2O increases the peak current since after the template removal the empty imprinted sites increase the surface area and the diffusion on the electrode surface, thus favoring the electron transfer between the solution and the electrode. The obtained data indicate that the use of NaOH in EtOH/H2O as washing solution leads to an effective release of the template molecule, and thus it has been selected for the fabrication of the MIP sensor.
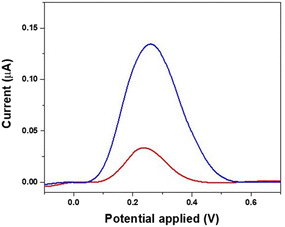 | ||
| Fig. 3 DPV characterization of the MIP before washing (red line) and after template removal (blue line) with NaOH in EtOH/H2O solution. | ||
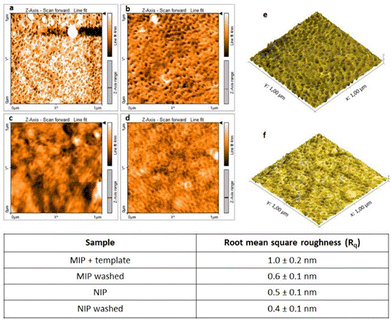
An atomic force microscope (AFM) was used to investigate the surface topography of the polymeric layers. The AFM images of imprinted and non-imprinted polymers, before and after the template removal, are shown in Fig. 4.
The AFM images reveal a marked difference in the roughness. As shown in Fig. 4, the NIP film was relatively more uniform and flatter than the MIP one with a calculated RMS value of 0.5 nm; on the other hand the MIP showed a rougher surface, with a calculated RMS value of 1 nm, which may be due to the presence of TGF-β1 template molecules inside the polymeric film which changes the polymerization process and thus the film structure. After the washing step to remove the template the roughness values decrease in both cases (MIP and NIP) without significant differences from each other. Even if the RMS values are very similar, the morphologies of MIP and NIP samples show a clear difference in topographic features: the NIP sample shows a uniform surface while the MIP one is characterized by the presence of several nanoholes.
Rebinding tests and affinity of the TGF-β1 MIP receptor
The evaluation of MIP binding properties and the characterization of the MIP-based biosensors were performed by incubating (1 h) the samples with spiked solutions with different concentrations of TGF-β1. Unspecifically adsorbed molecules were removed from the electrode surface by washing in PBS solution. The NIP receptor was used as a negative control. The device responses were characterized by electrochemical measurements in the presence of 10 mM K3[Fe(CN)6]/K4[Fe(CN)6] (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
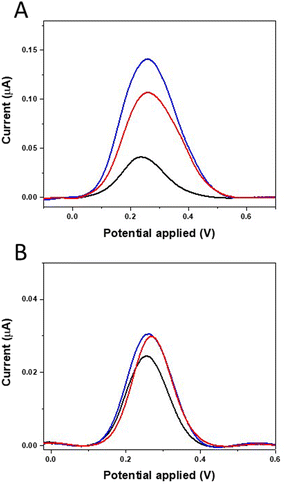
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at room temperature. The electrochemical characterization of MIP-modified electrodes was carried out by the DPV technique in the potential range −0.2–0.8 V at a scan rate of 100 mV s−1, and the results are reported in Fig. 5A. After the removal of TGF-β1 with NaOH in EtOH/H2O, an increase of the redox peaks (blue line) was observed as a result of the presence of empty cavities in the MIP which can favor the diffusion of the redox probe at the electrode surface. The rebinding test with TGF-β1 20 ng mL−1 (red line) then showed a reduction in the peak current, since some of the cavities were occupied by TGF-β1 molecules impeding the electron transfer between the electrode and the redox probe. After rebinding, the reduction of the redox peak suggests an efficient steric hindrance among TGF-β1 and the biomimetic sites. DPV measurements recorded on a control electrode modified with the non-imprinted polymer (NIP) revealed instead the absence of significant differences between the peak current values obtained during the three steps of NIP construction (Fig. 5B). These data indicate the absence of unspecific interactions and further support the specificity of the achieved interaction among the TGF-β1 target and the realized MIP film together with high sensitivity of the MIP-based sensor.
1) at room temperature. The electrochemical characterization of MIP-modified electrodes was carried out by the DPV technique in the potential range −0.2–0.8 V at a scan rate of 100 mV s−1, and the results are reported in Fig. 5A. After the removal of TGF-β1 with NaOH in EtOH/H2O, an increase of the redox peaks (blue line) was observed as a result of the presence of empty cavities in the MIP which can favor the diffusion of the redox probe at the electrode surface. The rebinding test with TGF-β1 20 ng mL−1 (red line) then showed a reduction in the peak current, since some of the cavities were occupied by TGF-β1 molecules impeding the electron transfer between the electrode and the redox probe. After rebinding, the reduction of the redox peak suggests an efficient steric hindrance among TGF-β1 and the biomimetic sites. DPV measurements recorded on a control electrode modified with the non-imprinted polymer (NIP) revealed instead the absence of significant differences between the peak current values obtained during the three steps of NIP construction (Fig. 5B). These data indicate the absence of unspecific interactions and further support the specificity of the achieved interaction among the TGF-β1 target and the realized MIP film together with high sensitivity of the MIP-based sensor.
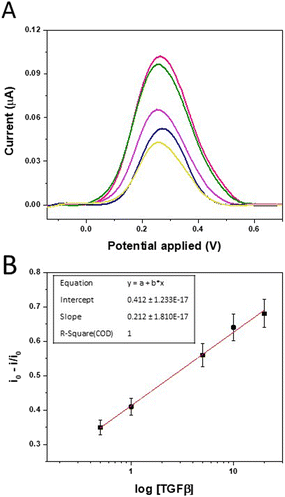
To further investigate the MIP-TGF-β1 binding and evaluate the sensing performance, the biosensors were incubated with binding buffer solution containing increasing concentrations of TGF-β1, ranging from 0.5 to 20 ng mL−1, for 1 hour; then, a washing step was performed. Every electrode was regenerated by the washing step and used for rebinding tests for a maximum of three concentrations for calibration, since beyond this value the degradation of the polymer film could affect the measurement, resulting in a variability lower than 6%. Each measurement was performed three times and the relative error was estimated as 6% of the mean value. DPV was used to monitor the ferro/ferrocyanide probe response as affected by TGF-β1 binding on the MIP-receptor. Data reported in Fig. 6A show a decrease of redox peak currents of the ferro/ferrocyanide couple with increasing TGF-β1 concentration, since more and more imprinted sites are rebound and the binding of the protein with MIP blocks the electron transfer on the electrode surface. The calibration plot, constructed by plotting the MIP current intensities (i) normalized with respect to the washed MIP current intensity (i0) as a function of the logarithm of TGF-β1 concentration, is reported in Fig. 6B. The MIP-modified electrode displayed a linear response in the tested concentration range from 0.5 to 20 ng mL−1 with a detection limit of 0.09 ng mL−1 calculated as 3.3s/slope (where s = 0.00597 and the slope = 0.2129), a sensitivity of 0.2129 and R2 = 1.
The imprinting factor, defined as the ratio between MIP and NIP currents recorded and calculated for a concentration of 20 ng mL−1, showed a high value equal to 32.38 demonstrating a strong increase of the interaction between TGF-β1 and the imprinted polymer compared to the non-imprinted one.
In order to demonstrate the selectivity of the electrochemical biosensor towards the target molecule, the developed MIP was incubated with a buffer solution containing interleukine-10 at a concentration of 20 ng mL−1. IL-10 was chosen as the interfering molecule since it has a chemical structure similar to TGFβ-1. The electrochemical characterization of MIP-modified electrodes was carried out by the DPV technique and the results are reported in Fig. 7. The obtained data showed a considerable difference between the TGF-β1 signal and the peak current of interleukine-10. The rebinding test with TGF-β1 20 ng mL−1 (red line) gave a peak current value similar to that obtained after electro-polymerization of o-PD (black line). The data obtained after the binding with interleukine-10 revealed instead that there are no significant differences between the peak current values obtained during the washing step of MIP construction, indicating the absence of specific interactions between interleukine-10 and the biomimetic sites. These results indicate that the significant difference in the interaction between the proteins and the binding cavities in the polymer further supports the high affinity and specificity of the synthetic receptor for the template molecule realized in this work.
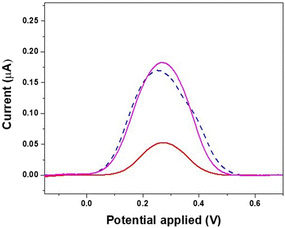 | ||
| Fig. 7 Electrochemical evaluation of template rebinding with interleukine-10 (purple line) with respect to the washing step (dashed blue line) and rebinding with TGF-β1 (red line). | ||
Determination of TGF-β1 in saliva samples
To verify the ability of our MIP based sensor to work in a real diagnostic context we tested it in a complex matrix such as artificial saliva to mimic a real sample. Artificial saliva NeutraSal®, when dissolved in water, forms an electrolyte solution resembling human saliva. The use of artificial saliva instead of human saliva is justified by the fact that human saliva is a very varying matrix and its composition can vary not only from patient to patient, but also during the day. Thus, using artificial saliva allows to mimic if not the variety of biological parameters, at least the physico-chemical features of human saliva. The calibration plot obtained for TGF-β1 in buffer solution was compared with the measurements recorded with spiked saliva samples (Fig. S3†). Also in this case, a proportional decrease in the current signal with TGF-β1 concentration in real samples was observed due to the specific binding of the analyte in the MIP cavities (Fig. S4†). These data demonstrate a very poor interaction of the polymeric interface with the several components (proteins, salts, sugars, etc.) of the complex matrix and its high selectivity towards target molecules also despite higher viscosity, since no further dilution or treatment of saliva was necessary. The efficacy of the proposed MIP sensor was estimated by performing a recovery test for TGF-β1 in artificial saliva and the results are reported in Table 1. The mean recovery values for the TGF-β1 spiked sample are close to real values for all tested concentrations equal to (95 ± 2), (102 ± 3) and (96 ± 3) for TGF-β1 1 ng mL−1, 5 ng mL−1 and 20 ng mL−1, confirming the selectivity of the proposed MIP sensors and their potential use for determining TGF-β1 in real saliva samples.| Samples | Added (ng mL−1) | Found (ng mL−1) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| TGF-β1 | 1 | 0.095 | 95 ± 2 | 23 |
| TGF-β1 | 5 | 5.100 | 102 ± 3 | 34 |
| TGF-β1 | 20 | 19.200 | 96 ± 3 | 39 |
Conclusions
In this paper, highly selective artificial receptors specific to TGF-β1 were realized by exploiting the remarkable potential of molecularly imprinted polymers in biorecognition. They were synthesized via electrochemical polymerization on platinum interdigitated microelectrodes patterned on glass substrates, in the presence of TGF-β1 as a template, followed by target removal.The polymer surface/nanofilm was characterized by AFM, STEM and electrochemical measurements in order to monitor all the steps of MIP fabrication, namely synthesis, template removal and subsequent target analyte binding. The MIP based sensor demonstrated high selectivity and high sensitivity with a linear response to the TGF-β1 concentration in buffer solution and a LOD of 0.09 ng ml−1 (corresponding to 3.6 pM) that is largely below the pathological levels of TGF-β1 in saliva. As a proof of concept, the realized MIP was tested with spiked artificial saliva, mimicking real conditions under which the assay could be used for diagnostic screening. The features of the MIP and its ability to bind the selected analyte were not affected by the different fluids used, thus demonstrating the readiness of the developed platform to be used in real settings.
In Table 2, the main characteristics of our sensing platform have been compared with those of other biosensors reported in the literature against TGF-β1.
| Reference | LOD | Sample | Detection methods | Capture probe | Main features |
|---|---|---|---|---|---|
| 32 | 10 pg mL−1 | Urine | Amperometry | Antibodies | Sandwich immunoassay with biotin-anti-TGF conjugated with peroxidase-labelled streptavidin polymer |
| 33 | 0.95 pg mL−1 | Human saliva | Amperometry | Antibodies | Sandwich immunoassay with V-Phe SWCNT(HRP)-anti-TGF conjugates as carrier tags |
| 34 | 20.1 pM−1 | Cell culture medium | Fluorescence | Antibodies | Microfluidic device integrated with the fluorescent microbead-based assay |
| 35 | 1000 pg mL−1 | Hepatic stellate cells | Voltammetry | Aptamers | Aptamers labelled with redox reporters |
| 36 | 570 pg mL−1 | Clinical serum | Impedance | Antibodies | Completely label free |
| Present work | (3.6 pM) | Artificial saliva | Voltammetry | MIP | Completely label free |
At the moment, few methods are available for the determination of TGF-β1: in particular, commercial ELISA colorimetric kits based on sandwich-immunoassay are usually employed for TGF-β1 quantification in the range from hundreds to thousands of pg mL−1. This benchtop approach is not suitable for quick and near-to-bed diagnosis and requires trained staff to perform all the analytical steps. To overcome the limits of traditional techniques some other groups developed sensing platforms for TGF-β1 determination. Similar electrochemical devices exploiting the use of antibodies or aptamers as recognition elements have been proposed but the performances in terms of LOD and simplicity of the test of our MIP-based sensor demonstrated competitive results since it does not require any labelling step. Sanchez-Tirado and co-workers developed analytical methods with very low LOD and tested them on saliva samples. It is important to underline that to achieve results, a standard approach requires a sandwich-based assay and the use of enzyme and electroactive molecules to amplify the signal. It represents an additional cost and requires additional steps to perform the analysis. Conversely, our device is very fast and simple to be used since the analysis can be done in a single step; for these reasons it is an ideal candidate as a tool for POC diagnostics.
The use of MIP instead of biological probes offers several advantages in terms of cost and physical and chemical stability (long shelf-life and no need for refrigerated storage), two aspects that have a paramount importance in the development of devices to be used outside a research laboratory and in near-to-bed diagnosis and self-diagnostics.
The novelty of our MIP-based approach relies on the fact that the developed miniaturized sensor is sensitive and selective and could be successfully applied in liquid biopsy at the POC; in fact, it can detect molecules at very low concentrations as required in biofluids and it demonstrated no unspecific interaction with interfering molecules and complex biological matrices such as saliva.
Author contributions
G. S. and E. P. contributed equally to this work, came up with the research concept and cowrote the manuscript. G. S. synthesized materials and conducted electrochemical analysis. M. A. S. and L. V. characterized the morphology of MIP and NIP by AFM analysis. M. E. investigated the MIP thickness using STEM analysis. M. S. C. and E. P. co-supervised this project. M. S. C., F. F., A. T. and G. G. edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the following funding programmes: Progetto PON ARS01_00906 “TITAN Nanotecnologie per l'immunoterapia dei tumori”, finanziato dal FESR nell'ambito del PON “Ricerca e Innovazione” 2014-2020 Azione II-OS (1.b); PRIN 2017 Project – “Prostate cancer: disentangling the relationships with tumor microenvironment to better model and target tumor progression” (grant number: Prot. 20174PLLYN); and “Tecnopolo per la medicina di precisione” (TecnoMed Puglia) Regione Puglia: DGR n.2117 del 21/11/2018, CUP: B84I180 0 0540 0 02”.References
- K. Ramajayam, S. Ganesan, P. Ramesh, M. Beena, T. Kokulnathan and A. Palaniappan, Biomimetics, 2023, 8, 245 CrossRef CAS PubMed.
- M. J. Whitcombe, I. Chianella, L. Larcombe, S. A. Piletsky, J. Noble, R. Porter and A. Horgan, Chem. Soc. Rev., 2011, 40, 1547–1571 RSC.
- N. Karimian, M. Vagin, M. H. A. Zavar, M. Chamsaz, A. P. F. Turner and A. Tiwari, Electrochem. Commun., 2013, 36, 92–95 CrossRef CAS.
- G. K. Ali and K. M. Omer, Talanta, 2022, 236, 122878 CrossRef CAS PubMed.
- X. Dong, C. Zhang, X. Du and Z. Zhang, Nanomaterials, 2022, 12, 1913 CrossRef CAS PubMed.
- L. Wang, M. Pagett and W. Zhang, Sens. Actuators Rep., 2023, 5, 100153 CrossRef.
- Y. L. Mustafa, A. Keirouz and H. S. Leese, J. Mater. Chem. B, 2022, 10, 7418–7449 RSC.
- F. M. Suzaei, A. D. Batista, B. Mizaikoff, S. Rahimi, S. M. Daryanavard and M. Abdel-Rehim, Microchim. Acta, 2022, 189, 255 CrossRef CAS PubMed.
- K. Zhang, H. Wu and Y. Zhang, Int. J. Electrochem. Sci., 2022, 17(11), 221185 CrossRef.
- Z. Cheng, E. Wang and X. Yang, Biosens. Bioelectron., 2001, 16, 179–185 CrossRef CAS PubMed.
- H. Peng, Y. Zhang, J. Zhang, Q. Xie, L. Nie and S. Yao, Analyst, 2001, 126, 189–194 RSC.
- L. Feng, Y. Liu, Y. Tan and J. Hu, Biosens. Bioelectron., 2004, 19, 1513–1519 CrossRef CAS PubMed.
- N. Karimian, A. P. F. Turner and A. Tiwari, Biosens. Bioelectron., 2014, 59, 160–165 CrossRef CAS PubMed.
- V. Ratautaite, S. N. Topkaya, L. Mikoliunaite, M. Ozsoz, Y. Oztekin, A. Ramanaviciene and A. Ramanavicius, Electroanalysis, 2013, 25, 1169–1177 CrossRef CAS.
- C. Neuzillet, A. Tijeras-Raballand, R. Cohen, J. Cros, S. Faivre, E. Raymond and A. de Gramont, Pharmacol. Ther., 2015, 147, 22–31 CrossRef CAS PubMed.
- D. R. Prinicpe, J. A. Doll, J. Bauer, B. Jung, H. G. Munshi, L. Bartholin, B. Pasche, C. Lee and P. J. Grippo, J. Natl. Cancer Inst., 2014, 106, 2 Search PubMed.
- A. Vander Ark, J. Cao and X. Li, Cell. Signalling, 2018, 52, 112–120 CrossRef CAS PubMed.
- J. Massague, Cell, 2008, 134, 215–230 CrossRef CAS PubMed.
- Y. E. Zhang, Cell Res., 2009, 19, 128–139 CrossRef CAS PubMed.
- M. Tian and W. P. Schiemann, Future Oncol., 2009, 5, 259–271 CrossRef CAS PubMed.
- Y. Drabsch and P. ten Dijke, Cancer Metastasis Rev., 2012, 31, 553–568 CrossRef CAS PubMed.
- M. Chen, W. Wang, P. Lin, K. Lee and W. Chen, Clin. Sci., 2012, 122, 459–472 CrossRef CAS PubMed.
- J. Sabarathinam, S. Dharman and J. Selvaraj, J. Pharm. Res. Int., 2020, 32, 36–45 CrossRef.
- M. Polz-Dacewicz, M. Strycharz-Dudziak, J. Dworzański, A. Stec and J. Kocot, Infect. Agents Cancer, 2016, 11, 45 CrossRef PubMed.
- A. Patel, S. Patel, P. Patel and V. Tanavde, Front. Oncol., 2022, 12, 828434 CrossRef CAS PubMed.
- F. Ferrara, S. Zoupanou, E. Primiceri, Z. Ali and M. Chiriacò, Biosens. Bioelectron., 2022, 196, 113698 CrossRef CAS PubMed.
- S. Zoupanou, A. Volpe, E. Primiceri, C. Gaudiuso, A. Ancona, F. Ferrara and M. S. Chiriacò, Micromachines, 2021, 12, 885 CrossRef PubMed.
- V. Garzarelli, F. Ferrara, E. Primiceri and M. Chiriacò, MethodsX, 2022, 9, 101759 CrossRef CAS PubMed.
- V. Garzarelli, M. Chiriacò, M. Cereda, I. Autuori and F. Ferrara, Clin. Chim. Acta, 2022, 536, 104–111 CrossRef CAS PubMed.
- M. Yamamoto, Y. Tabata, L. Hong, S. Miyamoto, N. Hashimoto and Y. Ikada, J. Controlled Release, 2000, 64, 133–142 CrossRef CAS PubMed.
- I. Losito, F. Palmisano and P. Zambonin, Anal. Chem., 2003, 75, 4988–4995 CrossRef CAS.
- E. Sánchez-Tirado, G. Martínez-García, A. González-Cortés, P. Yáñez-Sedeño and J. M. Pingarrón, Biosens. Bioelectron., 2017, 88, 9–14 CrossRef PubMed.
- E. Sánchez-Tirado, L. M. Arellano, A. González-Cortés, P. Yáñez-Sedeño, F. Langa and J. M. Pingarrón, Biosens. Bioelectron., 2017, 98, 240–247 CrossRef PubMed.
- K. Son, P. Gheibi, G. Stybayeva, A. Rahimian and A. Revzin, Microsyst. Nanoeng., 2017, 3, 17025 CrossRef CAS PubMed.
- Z. Matharu, D. Patel, Y. Gao, A. Haque, Q. Zhou and A. Revzin, Anal. Chem., 2014, 86, 8865–8872 CrossRef CAS PubMed.
- Y. Yao, J. Bao, Y. Lu, D. Zhang, S. Luo, X. Cheng, Q. Zhang, S. Li and Q. Liu, Sens. Actuators, B, 2016, 222, 127–132 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00958k |
| This journal is © The Royal Society of Chemistry 2023 |