Open Access Article
Open Access ArticleLubricin (PRG-4) anti-fouling coating for surface-enhanced Raman spectroscopy biosensing: towards a hierarchical separation system for analysis of biofluids†
Mingyu
Han
 *acg,
Saimon M.
Silva
*acg,
Saimon M.
Silva
 bcd,
Matthew J.
Russo
a,
Pauline E.
Desroches
a,
Weiwei
Lei
bcd,
Matthew J.
Russo
a,
Pauline E.
Desroches
a,
Weiwei
Lei
 a,
Anita F.
Quigley
ce,
Robert M. I.
Kapsa
ce,
Simon E.
Moulton
bcd,
Paul R.
Stoddart
a,
Anita F.
Quigley
ce,
Robert M. I.
Kapsa
ce,
Simon E.
Moulton
bcd,
Paul R.
Stoddart
 *f and
George W.
Greene
*abc
*f and
George W.
Greene
*abc
aInstitute for Frontier Materials, Deakin University, Waurn Ponds, Victoria 3216, Australia. E-mail: wren.greene@deakin.edu.au; han383@csiro.au; Tel: +613924 68278
bARC Centre of Excellence for Electromaterials Science, School of Science, Computing and Engineering Technologies, Swinburne University of Technology, Hawthorn, Victoria 3122, Australia
cThe Aikenhead Centre for Medical Discovery, St Vincent's Hospital Melbourne, Fitzroy, Victoria 3065, Australia
dIverson Health Innovation Research Institute, Swinburne University of Technology, Hawthorn, Victoria 3122, Australia
eSchool of Electrical and Biomedical Engineering, RMIT University, Melbourne, Victoria 3001, Australia
fSchool of Science, Computing and Engineering Technologies, Swinburne University of Technology, Hawthorn, Victoria 3122, Australia. E-mail: pstoddart@swin.edu.au; Tel: +61392145839
gCommonwealth Scientific and Industrial Research Organization (CSIRO), Agriculture and Food, 671 Sneydes Road, Werribee, Victoria 3030, Australia
First published on 7th November 2023
Abstract
Surface-enhanced Raman Spectroscopy (SERS) is a powerful optical sensing technique that amplifies the signal generated by Raman scattering by many orders of magnitude. Although the extreme sensitivity of SERS enables an extremely low limit of detection, even down to single molecule levels, it is also a primary limitation of the technique due to its tendency to equally amplify ‘noise’ generated by non-specifically adsorbed molecules at (or near) SERS-active interfaces. Eliminating interference noise is thus critically important to SERS biosensing and typically involves onerous extraction/purification/washing procedures and/or heavy dilution of biofluid samples. Consequently, direct analysis within biofluid samples or in vivo environments is practically impossible. In this study, an anti-fouling coating of recombinant human Lubricin (LUB) was self-assembled onto AuNP-modified glass slides via a simple drop-casting method. A series of Raman spectra were collected using rhodamine 6G (R6G) as a model analyte, which was spiked into NaCl solution or unprocessed whole blood. Likewise, we demonstrate the same sensing system for the quantitative detection of L-cysteine spiked in undiluted milk. It was demonstrated for the first time that LUB coating can mitigate the deleterious effect of fouling in a SERS sensor without compromising the detection of a target analyte, even in a highly fouling, complex medium like whole blood or milk. This feat is achieved through a molecular sieving property of LUB that separates small analytes from large fouling species directly at the sensing interface resulting in SERS spectra with low background (i.e., noise) levels and excellent analyte spectral fidelity. These findings indicate the great potential for using LUB coatings together with an analyte-selective layer to form a hierarchical separation system for SERS sensing of relevant analytes directly in complex biological media, aquaculture, food matrix or environmental samples.
Introduction
The analysis of biofluids such as blood, saliva, urine, tears, and milk is a critical activity in a wide range of fields, including medical diagnostics, forensics, and food production. Given the versatility and sensitivity of surface-enhanced Raman scattering (SERS),1 several researchers have tried to exploit this technique for identifying different states of body fluid (e.g. diseased versus healthy) or the detection of specific components in body fluids.2,3–6 Since the most significant and valuable biochemical information is contained in the clinical blood sample, blood analysis using the SERS technique is attracting increasing attention. However, SERS analysis of clinical blood samples is subject to many of the same challenges as other analytical techniques, given that the samples are complex, unstable mixtures of many potentially interfering components with a wide range of absorptivity. Due to the complex composition of blood, it makes a major difference if the measurement is performed on whole blood, plasma, or serum, and if the sample is filtered or dried. Even for plasma or serum samples, serum proteins have been found to limit SERS enhancement by preventing nanoparticle aggregation when colloidal nanoparticles (typically gold or silver) are used as the substrate.7 In addition, the potential selective adsorption of some proteins on the SERS substrate can change intensities in the spectra even at very low concentrations.8 Therefore, to obtain more reliable and quantitative Raman enhancements, it is usually necessary to involve intensive pre-treatments or chemically modify the SERS substrate surface to filter the fouling molecules (e.g. proteins, fats) from biofluid samples.Although a few studies have attempted to analyze unprocessed samples,9,10 the majority of reports use a variety of separation technologies such as nitrocellulose membrane and HPLC instrument,11,12 or protective coatings (e.g. polyethylene glycol and silica)2 to detect specific analytes that might not otherwise be resolved in the presence of other dominant or interfering species due to the substantial non-specific adsorption of proteins or cells in clinical blood samples. Given these challenges, a number of strategies reported in the literature have focused on how to remove interfering components before analysis or employed selective recognition elements such as peptide-based receptors and antibodies on the surface to detect target analytes that interact weakly with the SERS substrate.13 SERS nanoparticles with epidermal growth factor peptide as a targeting ligand have been used to detect circulating tumor cells in unprocessed peripheral blood.14
Polymers, especially zwitterionic polymers such as poly(glycidyl methacrylate-co-sulfobetaine methacrylate),15L-cysteine,16–18 and poly(carboxybetaine),19 are often used as an anti-fouling layer to combat the non-specific adsorption of proteins from blood samples. However, the zwitterionic polymer-modified SERS substrate still can only be used to analyze the serum,16,17 or blood plasma,19 instead of unprocessed whole blood. Furthermore, in previous works that reported analysis of serum or blood plasma using SERS, the SERS substrate had to be washed with Milli-Q water or buffer solution to minimize the fouling effect of serum or plasma prior to conducting Raman measurements.16–18
Despite these and a few other examples, in comparison to more conventional electrochemical biosensors and plasmonic techniques,20,21 the use of anti-fouling surfaces in SERS to reduce the confounding effects of cells and larger biomolecules appears to have been relatively neglected. In order to advance these promising applications of SERS in biofluid analysis, particularly for the rapid, point-of-care detection of small molecules such as metabolites, drugs and hormones in whole samples, it is clear that improved methods of separation are required to manage interference from unwanted biomolecules.
Here we investigate the use of lubricin (LUB) as an anti-fouling surface for the SERS analysis of unprocessed whole blood samples spiked with rhodamine 6G (R6G) as a model analyte, and an undiluted milk sample spiked with cysteine as a clinical analyte. This comprises a first step towards a novel hierarchical, in situ approach to sample separation for SERS analysis. LUB is a large glycoprotein that is found in articular joints and self-assembles on a range of different substrates.22–24 It has recently been shown to form anti-adhesive,25–30 size-selective coatings that inhibit biofouling of electrode surfaces with minimal loss in electrochemical activity.31,32 It is therefore reasonable to expect that the anti-fouling and size-selective properties of LUB coating can be transferred to the SERS sensing platform for the purpose of resisting unwanted adsorption. Compared to other polymers used as anti-fouling and protecting layers, there is no need for LUB coating to employ complex surface chemistry to modify the surface, which can change the surface properties of SERS substrates.
To showcase the effectiveness of the self-assembled lubricin (LUB) coating as an anti-fouling surface for SERS analysis of a ‘real’ biological analyte in highly complex fluid media, L-cysteine was quantitatively detected in undiluted milk. Cysteine is a thiol-functionalized amino acid that plays an important role in the human body as one of the twenty essential amino acids.33 As the only natural amino acid with a sulfhydryl group, which participates in protein synthesis and protein folding,34,35 and the metabolism of phospholipids in human organs such as the liver as one of the major intermediate products,36,37 cysteine has been widely used as a biomarker in bioanalysis and food processing industries.38 Cysteine has also been added to dairy foods such as milk as a high nutritional supplement to increase antioxidant capacity,39 and improve various biomarkers of endothelial dysfunction and cardiovascular disease.40,41 Prior to adding cysteine as additional nutrition content in milk, it is important to understand the original concentration of cysteine. It has been reported that the concentration of free cysteine in bovine mammary secretions beginning 3 to 5 days after last milking is 0.66 μmol L−1 before drying off and 6.66 μmol L−1 after drying off.42 Also, the difference of cysteine concentration and methionine to cysteine ratio in goat, cow, and human milk has been confirmed.43 It is therefore significant to selectively and sensitively detect the additional cysteine levels in food matrices like milk. Several analytical methods such as electrochemical determination,44,45 colourimetric determination,46 Fourier transform infrared spectroscopy,47 and fluorescence probes have been applied,48–51 however, to the best of our knowledge, there is no reported SERS-based sensing technique for the direct detection of cysteine in a milk sample, especially detecting the cysteine in the undiluted milk sample.
The ability to perform SERS analysis directly within complex biological fluids such as blood and milk with effectively no sample preparation (i.e. dilution, extraction, rinsing, etc.) has the potential to open up a new world of future applications for SERS including in vivo continuous drug monitoring and in-line screening of food and beverage feedstocks for harmful toxins or pathogens.
Materials and methods
Materials
Rhodamine (R6G, 99%) and NaCl were purchased from Chem-Supply Pty Ltd, H2SO4 and H2O2 were purchased from ASIS Scientific, aminopropyltriethoxysilane (APTES, ≥98%), sodium tetrachloroaurate(III) dehydrate (AuCl4Na2·H2O), L-cysteine, and sodium citrate tribasic dihydrate (Na3C6H5O7·2H2O) was purchased from Sigma-Aldrich, and the unprocessed whole blood samples used in this work were commercially available products (anticoagulated with K2 EDTA) acquired from Innovative Research (Novi, USA) from a healthy human donor. Undiluted, ‘full cream’ milk was purchased from a local supermarket (Woolworths, Melbourne, Australia). The Lubricin (LUB) used in this work was a full sequence recombinant human LUB, supplied by Lubris BioPharma (Framingham, MA USA). The original LUB was a 2.3 mg mL−1 solution in PBS 150 mM NaCl (10 mM sodium phosphate, 137 mM sodium chloride, and 2.7 mM potassium chloride at pH 7.4, at 25 °C). An additional 0.1% polysorbate 20 stabilizing agent was added as a surfactant and potentially adhesion to the substrate surface. The purity of the LUB was >99.5%, assessed by Lubris Biopharma. 2.5 mL of LUB solution was dialyzed against a 500 mL polysorbate free solution of PBS using a Slide-ALyzer 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecular weight cut-off dialysis cassette. The dialysis was performed over a period of 24 h with the dialyzing solution exchanged for a fresh solution at approximately 4 and 8 h. After the dialysis, the LUB solution was divided into smaller volumes and flash-frozen using liquid nitrogen for storage until use.
000 molecular weight cut-off dialysis cassette. The dialysis was performed over a period of 24 h with the dialyzing solution exchanged for a fresh solution at approximately 4 and 8 h. After the dialysis, the LUB solution was divided into smaller volumes and flash-frozen using liquid nitrogen for storage until use.
Substrates and samples fabrication
The Au nanoparticles were synthesized as described by Vega et al.52 In detail, 100 mL of 1 mM AuCl4Na2·H2O solution was brought to the boiling point, and then 15 mL of 1 wt% of sodium citrate was rapidly added to the AuCl4Na2·H2O solution while stirring. The solution was kept at 4 °C once cooled down. The AuNPs coated substrate was prepared based on the previous work, reported by Hou et al. with a slight modification.53 Briefly, the glass slide (SiO2) was firstly rinsed through a large amount of Milli-Q water, then immersed in freshly prepared piranha solution consisting of 98% H2SO4 and 30% H2O2 with a volume ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 1 h at 80 °C to remove any organic contaminations on the surface and obtain a completely smooth and hydroxylation layer. The modified glass slide was then rinsed thoroughly with Milli-Q water and dried in a stream of nitrogen gas. In this study, the organosilane layer containing a homogeneous amine group surface was fabricated by the vapour deposited method. In detail, the piranha solution cleaned glass slide was put into a vacuum desiccator with 1 mL of pure APTES in the desiccator overnight. After rinsing with Milli-Q water and curing at 110 °C for 15 min, the APTES layer was deposited onto the cleaned surface. Finally, the modified glass slide was immersed in the gold colloid solution at room temperature for 20 min to obtain the AuNPs surface. For the LUB-SERS, 50 μl of LUB solution (100 μg mL−1) was directly dropped on the cleaned and AuNP-coated surface and left for 20 min to allow LUB self-assembled onto the surface, followed by Milli-Q water rinse to remove the free LUB molecules. 20 min has proven to be long enough for LUB to form a stable layer on a wide range of surfaces in previous works.22,24,27,31,54
1 for 1 h at 80 °C to remove any organic contaminations on the surface and obtain a completely smooth and hydroxylation layer. The modified glass slide was then rinsed thoroughly with Milli-Q water and dried in a stream of nitrogen gas. In this study, the organosilane layer containing a homogeneous amine group surface was fabricated by the vapour deposited method. In detail, the piranha solution cleaned glass slide was put into a vacuum desiccator with 1 mL of pure APTES in the desiccator overnight. After rinsing with Milli-Q water and curing at 110 °C for 15 min, the APTES layer was deposited onto the cleaned surface. Finally, the modified glass slide was immersed in the gold colloid solution at room temperature for 20 min to obtain the AuNPs surface. For the LUB-SERS, 50 μl of LUB solution (100 μg mL−1) was directly dropped on the cleaned and AuNP-coated surface and left for 20 min to allow LUB self-assembled onto the surface, followed by Milli-Q water rinse to remove the free LUB molecules. 20 min has proven to be long enough for LUB to form a stable layer on a wide range of surfaces in previous works.22,24,27,31,54
In the study of R6G detection in unprocessed whole blood, two types of analytes were prepared which were R6G (10−4 M) spiked in 150 mM NaCl and R6G spiked in unprocessed whole blood. R6G (10−4 M) was prepared by dissolving R6G in 150 mM NaCl solution, while the R6G-blood mixture was prepared by directly dissolving R6G with a concentration of 10−4 M in unprocessed whole blood. For the calibration measurements, the R6G-blood mixture was again prepared, but with a series of different concentrations including 10−4 M, 10−5 M, 10−6, 10−7 and 10−8 M, respectively.
In the study of cysteine detection in undiluted milk, two types of milk samples were prepared which were cysteine (10−4 M) spiked in 150 mM NaCl and cysteine spiked in undiluted milk. Cysteine (10−4 M) was prepared by dissolving cysteine in 150 mM NaCl solution, while the cysteine-milk mixture was prepared by directly dissolving cysteine with a concentration of 10−4 M in undiluted milk. For the calibration measurements, the cysteine-milk mixture was again prepared, but with a series of different concentrations including 10−2 M, 10−3 M, 10−4, 10−5 and 10−6 M, respectively.
SERS characterization
The Renishaw inVia Raman spectrometer coupled to a research-grade Leica microscope was employed in this study, and specifically, the helium–neon laser (633 nm) was used for excitation. The suitable grating and filter combinations were installed, and calibration was conducted before each set of measurements. For the Raman measurements, the sample was inverted on the microscope stage to allow the laser to be aligned, positioned and focussed on the sample through the backside if the slide, which has been shown to increase the SERS enhancement.55 The parameters for all sets of measurements were the same, including a 10 s or 30 s integration time, 25 mW or 5 mW laser power on the sample and the spectral range was from 500 to 1750 cm−1. The Raman spectra were collected by a 50× objective lens from Leica. Fluorescence in the collected spectra was removed by a background subtraction algorithm by Wire 5.4, followed by a process using MATLAB R2018b software package.56To assess the fouling effect of the unprocessed whole blood on the Raman spectra of R6G, and the undiluted milk on the Raman spectra of cysteine, 20 μl of each of R6G spiked in 150 mM NaCl or blood, and cysteine spiked in 150 mM NaCl or undiluted milk was deposited on the bare SERS substrate, respectively, for 20 min to reach equilibrium. The size-selective transport property of the LUB layer was investigated by depositing the same series of 20 μl samples on LUB-SERS. The calibration measurements of R6G spiked blood, or cysteine in milk were conducted by depositing 20 μl of each of R6G spiked in blood, or cysteine in milk with different concentrations on LUB-SERS, respectively.
Results and discussion
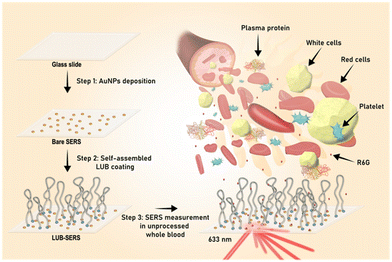
For these experiments, the simplest of SERS sensors was used consisting of citrate stabilized Au nanoparticles (AuNPs, 4 nm avg. radius) physically adsorbed at the surface of a commercial, borosilicate glass microscope slide (see Fig. 1). Over the top of this AuNP layer, a self-assembled LUB brush layer was applied by drop-casting a 100 μg ml−1 recombinant LUB solution in phosphate buffered saline. It has been conclusively demonstrated in the literature that LUB self-assembly results in a telechelic polymer brush coating (see Fig. 1) that is a product of the unique LUB structure where the molecules adhere to surfaces via adhesive domains on either end of a 200 nm long, anti-adhesive, and heavily glycosylated ‘mucin’ section that forms the extended ‘loop’.22,24,57 As detailed in previous reports, the ∼100 nm thick LUB brush structure is very diffuse; consisting of >95% water.24 This diffuse nature of the LUB brush gives rise to size-selective transport properties which block large fouling molecules (e.g. proteins) while permitting small molecules (e.g. drugs, peptides, ions) to diffuse across the LUB brush layer to access the underlying interface.31 Most importantly, the adsorbing end-domains have an average grafting distance on SiO2 surfaces of ∼9 nm and cover less than 15% of the surface area24 meaning that the majority remains unmodified and unaffected by the assembled LUB brush.24,25,27,31,58 This low surface coverage of the LUB brush means that it has minimal impact on the ability of diffusing small molecules to interact with buried active interfaces; in this case, the SERS-active AuNPs.The impact of self-assembled LUB layer on bare SERS
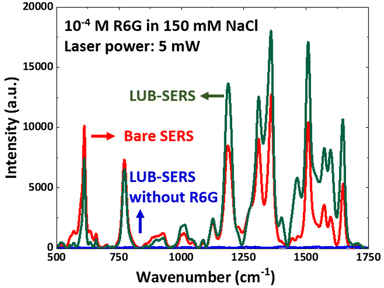
The Raman scattering will be enhanced for any Raman-active species within just a few nm of the AuNPs. Therefore, it was essential to determine the level, if any, that the assembled LUB brush may contribute to the ‘background’ of the Raman signal. Fig. 2 shows the Raman spectrum obtained using a 633 nm helium–neon laser in a clean 150 mM NaCl solution (i.e., saline) from the LUB-modified SERS (LUB-SERS) sensor. Somewhat unexpectedly, as seen in Fig. 2, no discernable SERS peaks were observed arising from the surface-adsorbed LUB molecules. The lack of a background signal indicates that the LUB end-domains, which are rich in amine, carboxyl, and hydroxyl functional groups known to be good Raman scatterers, do not adhere to or interact strongly with the surface bound AuNPs. Instead, the assembly must occur, preferentially, within the interstitial space between the nanoparticles, which suggests that there must be a strong (electrostatic) repulsion between the LUB end-domains and the AuNPs that lead the adsorbing end-domains to maximize their distance from the nanoparticles.To assess the ability of small molecules to traverse the anti-fouling LUB brush layer to access the buried AuNPs, the 150 mM NaCl solution was replaced with a solution of 10−4 M R6G in 150 mM NaCl and a SERS spectrum was again collected using a 633 nm laser. The SERS spectrum collected for R6G using the LUB-SERS sensor was then compared with the R6G spectrum collected using a bare-SERS sensor (i.e., no LUB coating) in the same solution under the same measurement conditions. As seen in Fig. 2, the spectra collected using the bare SERS and LUB-SERS substrates both show strong Raman scattering with little noise and peak positions consistent with R6G as reported in previous studies.59–61 A clear change in the peak intensities and the appearance of a few new peaks (e.g. at 1480 cm−1) was observed in the R6G spectra collected using the LUB-SERS sensor compared with the bare sensor, possibly due to the LUB brush influencing the orientation of the R6G molecules as they diffuse through the LUB brush structure to bind with AuNPs surface and consistent with previous reports on molecular orientational effects on SERS spectra.59,62 In general, it appears that the orientational influence of the LUB brush on R6G molecules leads to a general enhancement in the Raman peak magnitude for wavenumbers above about 1100 cm−1. Despite these apparent orientational effects of the LUB layer, it is clear from the overall intensity of the R6G spectra from the LUB-SERS sensor relative to that of the bare sensor that the presence of the LUB brush does not significantly inhibit the ability of R6G molecules to access and interact with the AuNPs or reduce the number of interacting R6G molecules.
SERS measurement of R6G spiked in unprocessed whole blood using a self-assembled LUB-modified SERS platform
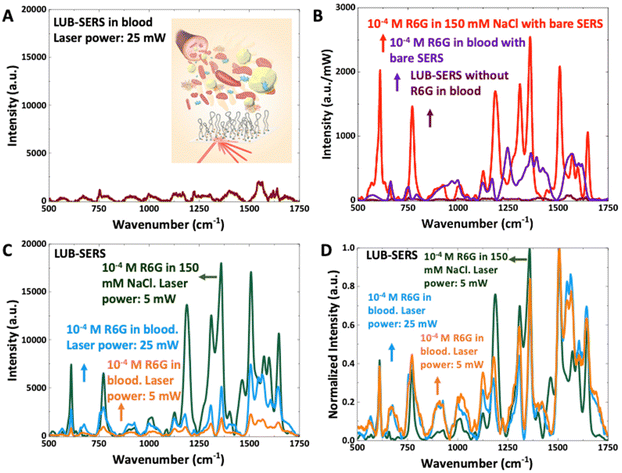
The LUB-SERS sensors were next challenged against undiluted, whole human blood (stabilized by EDTA). Fig. 3A shows the SERS spectrum collected from the undiluted blood using the LUB-SERS sensor. Unlike previously with 150 mM NaCl (see. Fig. 2), a low intensity background spectrum is observed at a laser intensity of 25 mW. This background spectrum was observed to be uniform in terms of the relative peak intensities across different regions of the sensor surface. The background spectrum itself was likewise found to be similar to previously reported blood spectra7–10,63 and found to most closely conform with the normal Raman spectrum for liquid whole blood.10 This background thus arises because the thickness of the confocal plane (calculated to be 500 nm in this system) is significantly thicker than the ∼100 nm thickness of the LUB brush. Therefore, the measurement is sampling a non-trivial volume of unseparated blood lying outside of the LUB brush; however, because the LUB prevents macromolecular components of the blood from accessing the embedded AuNPs, the intensity of the background remains low.The low intensity and relatively uniform background observed in undiluted whole blood with the LUB-SERS sensor is in sharp contrast with that observed using a bare-SERS sensor in whole blood after normalizing the spectral intensities of LUB-SERS only in blood, 10−4 M R6G in 150 mM NaCl or blood with bare-SERS against laser power (see Fig. 3B and S1A in ESI†). The background spectrum observed in blood using the bare SERS sensor in the absence of protection by the anti-fouling LUB brush is significantly higher in intensity and, highly non-uniform (compare Fig. 3B, S1A, and S1B†), changing (by approximately 10×) both in the intensity, number, and position of observed peaks at different positions on the sensor surface. This non-uniformity and high intensity of the background is the product of severe and uncontrolled fouling of the sensor surface by blood cells, proteins, and both large and small molecule constituents of the blood. This fouling is so severe, that when the same measurement is repeated (with a new bare-SERS sensor) using blood ‘spiked’ with R6G to a concentration of 10−4 M, no spectral peaks can be measured at any position on the bare sensor surface that can be attributed to or correlated with R6G. The lack of correlation can be seen by comparing the R6G spectra collected in 150 mM NaCl (see Fig. 2) with those collected in blood shown in Fig. 3B and Fig. S1A,† using bare-SERS.
To assess the ability of the LUB brush to prevent sensor surface fouling by blood and resolve the SERS spectral signal of R6G, the R6G spiked blood was pipetted onto a LUB-SERS sensor surface and the SERS spectrum was collected using laser powers of 5 mW and 25 mW (see Fig. 3C). The higher laser power was used to provide similar spectral intensities to aid visual comparison purposes and there was no indication of thermal damage observed. Although the overall intensity of the SERS spectrum is greatly reduced compared with previous spectra collected for R6G in clean 150 mM NaCl, a consequence of the turbidity and high light scattering of the whole blood and/or the reduction in ‘free’ R6G concentration due to binding with blood proteins and cell surfaces,64–67 the SERS spectrum of R6G is clearly resolved, with few (if any) additional peaks due to fouling. Importantly, the same R6G spectrum, although varying slightly in relative peak intensity as discussed in the previous report,68 was observed at all sampling positions on the LUB-SERS sensor surface. The strong correlation between the R6G spectra collected in spiked whole blood and in clean 150 mM NaCl, in which the concentration of R6G was 10−4 M, is seen in the high similarities of the intensity normalized spectra (see Fig. 3D). Increasing the laser power to 25 mW to compensate for the intensity loss due to blood scattering/turbidity enhances the R6G intensity and shows that the low intensity background observed for blood (see Fig. 3A) does not significantly distort the peak intensities or introduce significant numbers of extraneous peaks. Again, comparing the intensity of normalized R6G spectra collected in clean 150 mM NaCl and in undiluted whole blood confirms the LUB anti-fouling layer's ability to separate the R6G molecules from the complex blood matrix enabling its SERS detection.
Calibration of R6G spiked in unprocessed whole blood with LUB-SERS
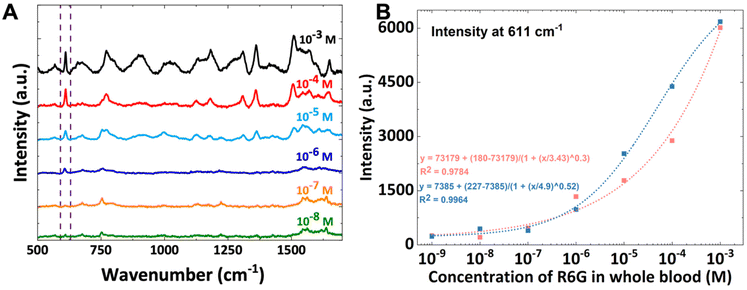
Finally, in order to ascertain the proportionality of the R6G peak intensities to the concentration of R6G and assess the obtainable limit of R6G detection in undiluted whole blood, a calibration curve was generated using a serial dilution (with additional whole blood) until no further peaks originating from the R6G could be detected. Fig. 4A shows representative spectra collected over the serial dilution of R6G spiked in blood from an initial concentration of 10−3 M to 10−8 M after subtracting the background SERS spectrum collected with the LUB-SERS sensor in unspiked blood (i.e. no R6G). Although it is clear, particularly at the very low R6G concentrations, that the background subtraction fails to remove all of the background signal, a characteristic peak corresponding to only the R6G scattering was identified at 611 cm−1 and used to quantify the R6G concentration in blood. The relationship between the 611 cm−1 peak intensity as a function of R6G concentration is shown in Fig. 4B and results in a nearly linear correlation over approximately 4 orders of magnitude before deviating significantly below 10−7 M (i.e., 100 nM) as the intensity of the R6G spectrum falls below that of the (normal Raman) blood background spectrum. While the 611 cm−1 peak provided the highest sensitivity to R6G concentration, the relationship between the intensity of other spectral peaks to R6G concentration in blood gave similar, although slightly lower quality, correlations (see Fig. S2B–D†). From the obtained calibration curve in Fig. 4B, the lowest detectable concentration of R6G in the undiluted whole blood is thus 1 × 10−7 M (100 nM) which is within a concentration regime associated with many promising, blood circulating biomarkers. To the best of our knowledge, the detection of such a low concentration of any molecular component or analyte within whole, undiluted, and unprocessed blood has never been achieved in any other SERS-based system. We also note that the dye R6G and related compounds of the Rhodamine family have been shown to bind to cell surfaces69 and blood proteins such as albumin.64,67,70,71 The LUB-SERS sensor is sensitive to only the ‘free’ (i.e., unbound) population of R6G molecules since any protein or cell-bound R6G would be unable to transit the anti-fouling LUB layer to interact with the SERS-active AuNPs.72 For this reason, it is likely that the actual concentration of R6G available for detection and quantification (i.e. the free fraction) is much lower than the lowest detectable concentration determined from the calibration curve which does not discriminate between the bound and unbound states.Clinical analyte analysis in complex food matrix with LUB-SERS
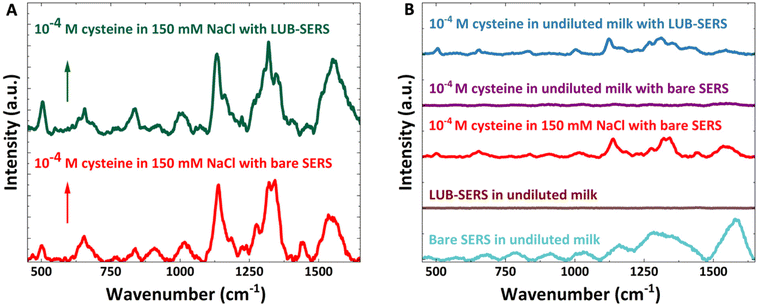
To demonstrate the effectiveness of the same LUB-SERS optical sensor for detecting a clinical analyte in a highly fouling environment, cysteine was chosen as the target analyte and undiluted milk was used as the fouling medium. Similar to the detection of R6G in whole blood with the LUB-SERS sensor, the assessment of cysteine spiked in 150 mM NaCl was first carried out using bare SERS followed by the LUB-SERS sensor. Fig. 5A shows the SERS spectra of cysteine in 150 mM NaCl collected with bare SERS and LUB-SERS sensors. As seen in Fig. 5A, the SERS spectra collected using the bare SERS and LUB-SERS substrates both showed strong Raman scattering with little noise and peak positions consistent with cysteine as reported in previous studies.73–75 Briefly, the band at about 504 cm−1 is from the stretching vibration of S–S bond of cysteine, indicating that there were gauche–gauche–gauche disulfide bridges formed between cysteine molecules.73,76 The peak at about 656 cm−1 is from C–S stretching, 838 cm−1 is from HCS bend, 1130 cm−1 is from C–C stretching, while the bands at about 1279 cm−1, 1319 cm−1, and 1345 cm−1 are assigned to CCH bend, HNH bend, and CH2 twist, respectively.73,74 Similar to the differences between R6G SERS spectra collected by SERS and LUB-SERS, the SERS spectra of cysteine in 150 mM NaCl collected by bare SERS and LUB-SERS also showed clear differences in peak intensities. It has been reported that there are different microstructures when cysteine molecules interact with gold particles due to the three rotational conformers of cysteine.73,74 It is also possible that the orientational influence of the LUB brush on cysteine contributes to the difference in peak intensities for cysteine spectra collected with bare SERS or LUB-SERS. Regardless, it is again clear that the microenvironment of the LUB brush has an impact on the relative intensities of the SERS peaks, resulting in the enhancement and suppression of different peaks in the spectrum. However, similar to the effect of the LUB brush on the Raman spectrum of R6G, it is clear that the LUB brush does not interfere with nor inhibit the binding of cysteine molecules to the AuNPs.To accurately detect the cysteine spiked in undiluted milk, the SERS spectrum of undiluted milk only was measured using bare SERS and LUB-SERS, as shown in Fig. 5B. There is no detectable SERS spectrum of undiluted milk at the laser wavelength and power used on either the bare SERS or the LUB-SERS. The SERS spectrum of cysteine spiked in undiluted milk with bare SERS was then collected (Fig. 5B) to investigate the fouling impact of undiluted milk on the detection of cysteine with bare SERS. As clearly seen in Fig. 5B, without the anti-fouling property of the LUB brush, no cysteine spectrum is measured in milk with the bare SERS even at high concentrations (i.e. 10−4 M). In the case of the milk, the surface fouling by proteins, hydrocolloids, and other molecular components interferes with and effectively blocks the cystine from approaching and interacting with the AuNPs and thus from being detected.
In contrast, the anti-fouling and size-selective properties of LUB-SERS enables a strong cystine spectrum to be collected in the undiluted milk containing the spiked cysteine at a concentration of 10−4 M, as also seen in Fig. 5B. Similar to the fouling prevention ability observed when detecting R6G spiked in unprocessed whole blood, an almost identical cysteine spectrum was collected at all positions scanned from the LUB-SERS in milk as was collected from the LUB-SERS in non-fouling NaCl solution (see Fig. 5A).
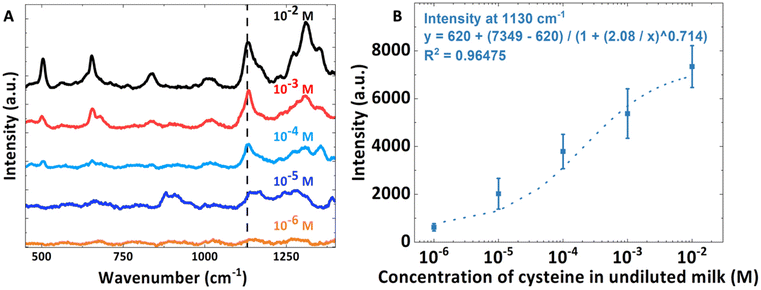
Finally, a series of SERS spectra of cysteine spiked in undiluted milk were collected using a serial dilution to ascertain the sensitivity of the LUB-SERS sensor. The resulting calibration curve spanning four orders of magnitude of concentration from 10−2 M to 10−6 M is shown in Fig. 6A together with representative SERS spectra collected over the different concentrations. The characteristic cysteine scattering peak at 1130 cm−1 was used to quantify the cysteine concentration in undiluted milk. The relationship between peak intensity of the 1130 cm−1 band and the concentration of cysteine in undiluted milk was plottedin Fig. 6B, together with a polynomial fit. From the obtained calibration curve in Fig. 6B, the lowest detectable concentration of cysteine in the undiluted milk is 10−5 M (10 μM).
Conclusions
In conclusion, we demonstrate a proof-of-concept, fouling-resistant SERS sensor platform enabled by the anti-adhesive and size-selective transport properties of self-assembled LUB brushes. The ability of the LUB brush to prevent the fouling of SERS substrates while selectively separating small molecule analytes from complex media thus enables SERS analysis even in the most fouling of biological fluids, i.e., unprocessed, undiluted, whole blood, or undiluted milk. The demonstrated success of detecting analytes in highly fouling environments and the mitigation of unwanted surface fouling effects strongly supports the application of this and other LUB-embodied sensor architectures to other fouling bodily fluids, including saliva, urine, or tear fluids for the detection and quantification of analytes such as drugs, toxins, or other clinically relevant biomarkers. It is important to highlight that the self-adsorption of analytes onto the AuNPs does not require any additional nanoparticle surface modification. Since LUB has the capacity to self-assemble onto virtually any solid interface, the methodology and mechanism described in this report should be easily transferable to any SERS active interface including 2D-nanomaterials such as graphene-based 2D-nanomaterials, boron nitride, MXene, and MoS2.22,23,25,28,77,78,78,78 Future work will focus on the development of selective surfaces using analyte recognition elements such as aptamer-coated AuNP surfaces or molecularly imprinted polymer-coated surfaces.79 The sensitivity could be further enhanced by using larger particles or more enhancing geometries (e.g. noble nanoparticles modified 2D-materials). Regarding the target analytes, native components of the blood or other biofluids will be analyzed by combining selective affinity-based coatings with the non-selective lubricin anti-fouling coating as a hierarchical separation system in a layered structure.Author contributions
The manuscript was written through the contributions of all authors. All authors have given approval for the final version of the manuscript.Ethical statement
All experiments were performed in accordance with the Guidelines “National Statement on Ethical Conduct in Human Research 2007 (updated 2018)” and approved by the ethics committee at Swinburne University of Technology (20203067-4757). Informed consents were obtained from human participants of this study. Blood samples used in this study were from a blood biobank, where samples were collected retrospectively. Our experiments didn't involve any form of contact with the blood donor. The blood donor had to sign a consentient form when donating blood to the company that we bought the blood from. The consentient form signed by the donor was captured in the ethics protocol documents.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
We acknowledge the financial support from the Australian Research Council in the Discovery Project (DP180102287). The authors would also like to thank Lubris Biopharma for supporting this research through the gift of their recombinant lubricin materials.References
- A. Balcytis, Y. Nishijima, S. Krishnamoorthy, A. Kuchmizhak, P. R. Stoddart, R. Petruskevicius and S. Juodkazis, From Fundamental toward Applied SERS: Shared Principles and Divergent Approaches, Adv. Opt. Mater., 2018, 6(16) DOI:10.1002/adom.201800292.
- L. E. Jamieson, S. M. Asiala, K. Gracie, K. Faulds and D. Graham, Bioanalytical Measurements Enabled by Surface-Enhanced Raman Scattering (SERS) Probes, in Annual Review of Analytical Chemistry, ed. R. G. Cooks and J. E. Pemberton, Annual Review of Analytical Chemistry, 2017, vol. 10, pp. 415–437 Search PubMed.
- C. Zong, M. X. Xu, L. J. Xu, T. Wei, X. Ma, X. S. Zheng, R. Hu and B. Ren, Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges, Chem. Rev., 2018, 118(10), 4946–4980, DOI:10.1021/acs.chemrev.7b00668 . Review.
- C. Muehlethaler, M. Leona and J. R. Lombardi, Review of Surface Enhanced Raman Scattering Applications in Forensic Science, Anal. Chem., 2016, 88(1), 152–169, DOI:10.1021/acs.analchem.5b04131.
- B. Yu, M. Ge, P. Li, Q. Xie and L. Yang, Development of surface-enhanced Raman spectroscopy application for determination of illicit drugs: Towards a practical sensor, Talanta, 2019, 191, 1–10, DOI:10.1016/j.talanta.2018.08.032.
- C. Zong, M. Xu, L.-J. Xu, T. Wei, X. Ma, X.-S. Zheng, R. Hu and B. Ren, Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges, Chem. Rev., 2018, 118(10), 4946–4980, DOI:10.1021/acs.chemrev.7b00668.
- A. Bonifacio, S. Dalla Marta, R. Spizzo, S. Cervo, A. Steffan, A. Colombatti and V. Sergo, Surface-enhanced Raman spectroscopy of blood plasma and serum using Ag and Au nanoparticles: a systematic study, Anal. Bioanal. Chem., 2014, 406(9–10), 2355–2365, DOI:10.1007/s00216-014-7622-1.
- S. Boyd, M. F. Bertino, D. X. Ye, L. S. White and S. J. Seashols, Highly Sensitive Detection of Blood by Surface Enhanced Raman Scattering, J. Forensic Sci., 2013, 58(3), 753–756, DOI:10.1111/1556-4029.12120.
- M. Casella, A. Lucotti, M. Tommasini, M. Bedoni, E. Forvi, F. Gramatica and G. Zerbi, Raman and SERS recognition of beta-carotene and haemoglobin fingerprints in human whole blood, Spectrochim. Acta, Part A, 2011, 79(5), 915–919, DOI:10.1016/j.saa.2011.03.048.
- W. R. Premasiri, J. C. Lee and L. D. Ziegler, Surface-Enhanced Raman Scattering of Whole Human Blood, Blood Plasma, and Red Blood Cells: Cellular Processes and Bioanalytical Sensing, J. Phys. Chem. B, 2012, 116(31), 9376–9386, DOI:10.1021/jp304932g.
- H. Torul, H. Ciftci, D. Cetin, Z. Suludere, I. H. Boyaci and U. Tamer, Paper membrane-based SERS platform for the determination of glucose in blood samples, Anal. Bioanal. Chem., 2015, 407(27), 8243–8251, DOI:10.1007/s00216-015-8966-x.
- G. Trachta, B. Schwarze, B. Sagmuller, G. Brehm and S. Schneider, Combination of high-performance liquid chromatography and SERS detection applied to the analysis of drugs in human blood and urine, J. Mol. Struct., 2004, 693(1–3), 175–185, DOI:10.1016/j.molstruc.2004.02.034.
- M. Li, S. K. Cushing, J. Zhang, S. Suri, R. Evans, W. P. Petros, L. F. Gibson, D. Ma, Y. Liu and N. Wu, Three-Dimensional Hierarchical Plasmonic Nano-Architecture Enhanced Surface-Enhanced Raman Scattering Immunosensor for Cancer Biomarker Detection in Blood Plasma, ACS Nano, 2013, 7(6), 4967–4976, DOI:10.1021/nn4018284.
- X. Wang, X. M. Qian, J. J. Beitler, Z. G. Chen, F. R. Khuri, M. M. Lewis, H. J. C. Shin, S. M. Nie and D. M. Shin, Detection of Circulating Tumor Cells in Human Peripheral Blood Using Surface-Enhanced Raman Scattering Nanoparticles, Cancer Res., 2011, 71(5), 1526–1532, DOI:10.1158/0008-5472.Can-10-3069.
- K. Sivashanmugan, P.-C. Liu, K.-W. Tsai, Y.-N. Chou, C.-H. Lin, Y. Chang and T.-C. Wen, An anti-fouling nanoplasmonic SERS substrate for trapping and releasing a cationic fluorescent tag from human blood solution, Nanoscale, 2017, 9(8), 2865–2874, 10.1039/c6nr08077d.
- S. S. Panikar, N. Banu, J. Haramati, G. Y. Gutierrez-Silerio, B. E. Bastidas-Ramirez, M. C. Tellez-Bañuelos, T. A. Camacho-Villegas, S. d. Toro-Arreola and E. De la Rosa, Anti-fouling SERS-based immunosensor for point-of-care detection of the B7–H6 tumor biomarker in cervical cancer patient serum, Anal. Chim. Acta, 2020, 1138, 110–122, DOI:10.1016/j.aca.2020.09.019.
- S. S. Panikar, N. Banu, E.-R. Escobar, G.-R. García, J. Cervantes-Martínez, T.-C. Villegas, P. Salas and E. De la Rosa, Stealth modified bottom up SERS substrates for label-free therapeutic drug monitoring of doxorubicin in blood serum, Talanta, 2020, 218, 121138, DOI:10.1016/j.talanta.2020.121138.
- S. S. Panikar, G. Ramírez-García, S. Sidhik, T. Lopez-Luke, C. Rodriguez-Gonzalez, I. H. Ciapara, P. S. Castillo, T. Camacho-Villegas and E. De la Rosa, Ultrasensitive SERS Substrate for Label-Free Therapeutic-Drug Monitoring of Paclitaxel and Cyclophosphamide in Blood Serum, Anal. Chem., 2019, 91(3), 2100–2111, DOI:10.1021/acs.analchem.8b04523.
- F. Sun, H.-C. Hung, A. Sinclair, P. Zhang, T. Bai, D. D. Galvan, P. Jain, B. Li, S. Jiang and Q. Yu, Hierarchical zwitterionic modification of a SERS substrate enables real-time drug monitoring in blood plasma, Nat. Commun., 2016, 7(1), 13437, DOI:10.1038/ncomms13437.
- P. H. Lin and B. R. Li, Antifouling strategies in advanced electrochemical sensors and biosensors, Analyst, 2020, 145(4), 1110–1120, 10.1039/c9an02017a.
- E. Mauriz, Low-Fouling Substrates for Plasmonic Sensing of Circulating Biomarkers in Biological Fluids, Biosensors, 2020, 10(6) DOI:10.3390/bios10060063.
- M. Han, S. M. Silva, W. Lei, A. Quigley, R. M. I. Kapsa, S. E. Moulton and G. W. Greene, Adhesion and Self-Assembly of Lubricin (PRG4) Brush Layers on Different Substrate Surfaces, Langmuir, 2019, 35(48), 15834–15848, DOI:10.1021/acs.langmuir.9b01809.
- M. Han, J. D. Berry, S. M. Silva, M. L. P. Vidallon, W. Lei, A. F. Quigley, R. M. I. Kapsa, S. E. Moulton, R. Tabor and G. W. Greene, Self-Assembly of Lubricin (PRG-4) Brushes on Graphene Oxide Affords Stable 2D-Nanosheets in Concentrated Electrolytes and Complex Fluids, ACS Appl. Nano Mater., 2020, 3(11), 11527–11542, DOI:10.1021/acsanm.0c02621.
- G. W. Greene, R. Thapa, S. A. Holt, X. Wang, C. J. Garvey and R. F. Tabor, Structure and Property Changes in Self-Assembled Lubricin Layers Induced by Calcium Ion Interactions, Langmuir, 2017, 33(10), 2559–2570, DOI:10.1021/acs.langmuir.6b03992.
- H. Ye, M. Han, R. Huang, T. A. Schmidt, W. Qi, Z. He, L. L. Martin, G. D. Jay, R. Su and G. W. Greene, Interactions between Lubricin and Hyaluronic Acid Synergistically Enhance Antiadhesive Properties, ACS Appl. Mater. Interfaces, 2019, 11(20), 18090–18102, DOI:10.1021/acsami.9b01493.
- P. E. Desroches, S. M. Silva, S. W. Gietman, A. F. Quigley, R. M. I. Kapsa, S. E. Moulton and G. W. Greene, Lubricin (PRG4) Antiadhesive Coatings Mitigate Electrochemical Impedance Instabilities in Polypyrrole Bionic Electrodes Exposed to Fouling Fluids, ACS Appl. Bio Mater., 2020, 3(11), 8032–8039, DOI:10.1021/acsabm.0c01109.
- G. W. Greene, L. L. Martin, R. F. Tabor, A. Michalczyk, L. M. Ackland and R. Horn, Lubricin: A versatile, biological anti-adhesive with properties comparable to polyethylene glycol, Biomaterials, 2015, 53(C), 127–136, DOI:10.1016/j.biomaterials.2015.02.086.
- M. J. Russo, M. Y. Han, A. F. Quigley, R. M. I. Kapsa, S. E. Moulton, E. Doeven, R. Guijt, S. M. Silva and G. W. Greene, Lubricin (PRG4) reduces fouling susceptibility and improves sensitivity of carbon-based electrodes, Electrochim. Acta, 2020, 333, 135574, DOI:10.1016/j.electacta.2019.135574.
- L. Ali, C. Jin and N. G. Karlsson, ‘Glycoproteomics of Lubricin-Implication of Important Biological Glyco- and Peptide-Epitopes in Synovial Fluid’, in Rheumatoid Arthritis – Etiology, Consequences and Co-Morbidities, InTech, 2012, DOI:10.5772/25657.
- C. S. Manasa, S. M. Silva, P. E. Desroches, J. Dennaoui, M. J. Russo, M. Han, A. F. Quigley, G. W. Greene, R. M. I. Kapsa and S. E. Moulton, Lubricin as a tool for controlling adhesion in vivo and ex vivo, Biointerphases, 2021, 16(2), 020802, DOI:10.1116/6.0000779 (acccessed 2021/04/18).
- G. W. Greene, V. Ortiz, C. Pozo-Gonzalo, S. E. Moulton, X. Wang, L. L. Martin, A. Michalczky and P. C. Howlett, Lubricin Antiadhesive Coatings Exhibit Size-Selective Transport Properties that Inhibit Biofouling of Electrode Surfaces with Minimal Loss in Electrochemical Activity, Adv. Mater. Interfaces, 2018, 5(7), 1701296–1701210, DOI:10.1002/admi.201701296.
- S. M. Silva, D. P. Langley, L. R. Cossins, A. N. Samudra, A. F. Quigley, R. M. I. Kapsa, R. W. Tothill, G. W. Greene and S. E. Moulton, Rapid Point-of-Care Electrochemical Sensor for the Detection of Cancer Tn Antigen Carbohydrate in Whole Unprocessed Blood, ACS Sens., 2022, 7(11), 3379–3388, DOI:10.1021/acssensors.2c01460.
- S. Aluri and S. P. de Visser, The Mechanism of Cysteine Oxygenation by Cysteine Dioxygenase Enzymes, J. Am. Chem. Soc., 2007, 129(48), 14846–14847, DOI:10.1021/ja0758178.
- G. I. Giles, K. M. Tasker and C. Jacob, Hypothesis: the role of reactive sulfur species in oxidative stress, Free Radicals Biol. Med., 2001, 31(10), 1279–1283, DOI:10.1016/S0891-5849(01)00710-9.
- C. E. Paulsen and K. S. Carroll, Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery, Chem. Rev., 2013, 113(7), 4633–4679, DOI:10.1021/cr300163e.
- M. H. Stipanuk, J. E. Dominy, J.-I. Lee and R. M. Coloso, Mammalian Cysteine Metabolism: New Insights into Regulation of Cysteine Metabolism12, J. Nutr., 2006, 136(6), 1652S–1659S, DOI:10.1093/jn/136.6.1652S.
- J. E. Dominy Jr., L. L. Hirschberger, R. M. Coloso and M. H. Stipanuk, Regulation of cysteine dioxygenase degradation is mediated by intracellular cysteine levels and the ubiquitin–26 S proteasome system in the living rat, Biochem. J., 2006, 394(1), 267–273, DOI:10.1042/BJ20051510 (acccessed 9/2/2023).
- M. Gagaoua, A. L. Dib, N. Lakhdara, M. Lamri, C. Botineştean and J. M. Lorenzo, Artificial meat tenderization using plant cysteine proteases, Curr. Opin. Food Sci., 2021, 38, 177–188, DOI:10.1016/j.cofs.2020.12.002.
- I. T. Khan, M. Nadeem, M. Imran, R. Ullah, M. Ajmal and M. H. Jaspal, Antioxidant properties of Milk and dairy products: a comprehensive review of the current knowledge, Lipids Health Dis., 2019, 18(1), 41, DOI:10.1186/s12944-019-0969-8.
- T. Chitapanarux, P. Tienboon, S. Pojchamarnwiputh and D. Leelarungrayub, Open-labeled pilot study of cysteine-rich whey protein isolate supplementation for nonalcoholic steatohepatitis patients, J. Gastroenterol. Hepatol., 2009, 24(6), 1045–1050, DOI:10.1111/j.1440-1746.2009.05865.x (acccessed 2023/09/01).
- P. Kanikarla-Marie and S. K. Jain, L-Cysteine, supplementation reduces high-glucose and ketone-induced adhesion of monocytes to endothelial cells by inhibiting ROS, Mol. Cell. Biochem., 2014, 391(1), 251–256, DOI:10.1007/s11010-014-2009-3.
- R. W. Brown and M. N. Mickelson, Lactoperoxidase, thiocyanate, and free cystine in bovine mammary secretions in early dry period and at the start of lactation and their effect on Streptococcus agalactiae growth, Am. J. Vet. Res., 1979, 40(2), 250–255 CAS . PubMed.
- C. G. Prosser, Compositional and functional characteristics of goat milk and relevance as a base for infant formula, J. Food Sci., 2021, 86(2), 257–265, DOI:10.1111/1750-3841.15574 (acccessed 2023/09/01).
- D. R. Kumar, M. L. Baynosa and J.-J. Shim, Cu2+-1,10-phenanthroline-5,6-dione@electrochemically reduced graphene oxide modified electrode for the electrocatalytic determination of L-cysteine, Sens. Actuators, B, 2019, 293, 107–114, DOI:10.1016/j.snb.2019.04.122.
- X. Zhai, S. Li, X. Chen, Y. Hua and H. Wang, Coating silver metal-organic frameworks onto nitrogen-doped porous carbons for the electrochemical sensing of cysteine, Microchim. Acta, 2020, 187(9), 493, DOI:10.1007/s00604-020-04469-3.
- S. Sahu, S. Sharma, T. Kant, K. Shrivas and K. K. Ghosh, Colorimetric determination of L-cysteine in milk samples with surface functionalized silver nanoparticles, Spectrochim. Acta, Part A, 2021, 246, 118961, DOI:10.1016/j.saa.2020.118961.
- B. R. Khalkho, M. K. Deb, R. Kurrey, B. Sahu, A. Saha, T. K. Patle, R. Chauhan and K. Shrivas, Citrate functionalized gold nanoparticles assisted micro extraction of L-cysteine in milk and water samples using Fourier transform infrared spectroscopy, Spectrochim. Acta, Part A, 2022, 267, 120523, DOI:10.1016/j.saa.2021.120523.
- G. S. Yeom, I.-h. Song, S. J. Park, A. Kuwar and S. B. Nimse, Development and application of a fluorescence turn-on probe for the nanomolar cysteine detection in serum and milk samples, J. Photochem. Photobiol., A, 2022, 431, 114074, DOI:10.1016/j.jphotochem.2022.114074.
- J. Wang, H. Wang, Y. Hao, S. Yang, H. Tian, B. Sun and Y. Liu, A novel reaction-based fluorescent probe for the detection of cysteine in milk and water samples, Food Chem., 2018, 262, 67–71, DOI:10.1016/j.foodchem.2018.04.084.
- B. Zhang, L. Chen, M. Zhang, C. Deng and X. Yang, A gold-silver bimetallic nanocluster-based fluorescent probe for cysteine detection in milk and apple, Spectrochim. Acta, Part A, 2022, 278, 121345, DOI:10.1016/j.saa.2022.121345.
- S. Gong, A. Qin, Y. Zhang, M. Li, X. Chen, Y. Liang, X. Xu, Z. Wang and S. Wang, A new ratiometric AIE fluorescent probe for detecting cysteine in food samples and imaging in the biological system, Food Chem., 2023, 400, 134108, DOI:10.1016/j.foodchem.2022.134108.
- M. M. Vega, A. Bonifacio, V. Lughi, S. Marsi, S. Carrato and V. Sergo, Long-term stability of surfactant-free gold nanostars, J. Nanopart. Res., 2014, 16(11) DOI:10.1007/s11051-014-2729-z.
- T. Hou, L. L. Martin, R. G. Horn and G. W. Greene, Use of optical interferometry to measure gold nanoparticle adsorption on silica, Colloids Surf., A, 2016, 506, 383–392, DOI:10.1016/j.colsurfa.2016.07.022.
- S. Das, X. Banquy, B. Zappone, G. W. Greene, G. D. Jay and J. N. Israelachvili, Synergistic Interactions between Grafted Hyaluronic Acid and Lubricin Provide Enhanced Wear Protection and Lubrication, Biomacromolecules, 2013, 14(5), 1669–1677, DOI:10.1021/bm400327a.
- S. Jayawardhana, L. Rosa, S. Juodkazis and P. R. Stoddart, Additional Enhancement of Electric Field in Surface-Enhanced Raman Scattering due to Fresnel Mechanism, Sci. Rep., 2013, 3(1), 2335, DOI:10.1038/srep02335.
- P. J. Cadusch, M. M. Hlaing, S. A. Wade, S. L. McArthur and P. R. Stoddart, Improved methods for fluorescence background subtraction from Raman spectra, J. Raman Spectrosc., 2013, 44(11), 1587–1595, DOI:10.1002/jrs.4371 (acccessed 2023/09/24).
- B. Zappone, M. Ruths, G. W. Greene, G. D. Jay and J. N. Israelachvili, Adsorption, lubrication, and wear of lubricin on model surfaces: Polymer brush-like behavior of a glycoprotein, Biophys. J., 2007, 92(5), 1693–1708 CrossRef CAS PubMed.
- S. M. Silva, A. F. Quigley, R. M. I. Kapsa, G. W. Greene and S. E. Moulton, Lubricin on Platinum Electrodes: A Low–Impedance Protein–Resistant Surface Towards Biomedical Implantation, ChemElectroChem, 2019, 6(6), 1939–1943, DOI:10.1002/celc.201900237.
- H. K. Turley, Z. Hu, L. Jensen and J. P. Camden, Surface-Enhanced Resonance Hyper-Raman Scattering Elucidates the Molecular Orientation of Rhodamine 6G on Silver Colloids, J. Phys. Chem. Lett., 2017, 8(8), 1819–1823, DOI:10.1021/acs.jpclett.7b00498.
- G. Kostovski, D. J. White, A. Mitchell, M. W. Austin and P. R. Stoddart, Nanoimprinted optical fibres: Biotemplated nanostructures for SERS sensing, Biosens. Bioelectron., 2009, 24(5), 1531–1535, DOI:10.1016/j.bios.2008.10.016.
- R. Goul, S. Das, Q. Liu, M. Xin, R. Lu, R. Hui and J. Z. Wu, Quantitative analysis of surface enhanced Raman spectroscopy of Rhodamine 6G using a composite graphene and plasmonic Au nanoparticle substrate, Carbon, 2017, 111(C), 386–392, DOI:10.1016/j.carbon.2016.10.019.
- M. Moskovits, D. P. DiLella and K. J. Maynard, Surface Raman spectroscopy of a number of cyclic aromatic molecules adsorbed on silver: selection rules and molecular reorientation, Langmuir, 1988, 4(1), 67–76, DOI:10.1021/la00079a012.
- P. Hu, X. S. Zheng, C. Zong, M. H. Li, L. Y. Zhang, W. Li and B. Ren, Drop-coating deposition and surface-enhanced Raman spectroscopies (DCDRS and SERS) provide complementary information of whole human tears, J. Raman Spectrosc., 2014, 45(7), 565–573, DOI:10.1002/jrs.4499.
- V. K. Verma, K. Tapadia, T. Maharana and A. Sharma, Convenient and ultra-sensitive fluorescence detection of bovine serum albumin by using Rhodamine-6G modified gold nanoparticles in biological samples, Luminescence, 2018, 33(8), 1408–1414, DOI:10.1002/bio.3563 (acccessed 2021/07/27).
- D. K. Das, T. Mondal, A. K. Mandal and K. Bhattacharyya, Binding of Organic Dyes with Human Serum Albumin: A Single-Molecule Study, Chem. – Asian J., 2011, 6(11), 3097–3103, DOI:10.1002/asia.201100272.
- P. Mandal, M. Bardhan and T. Ganguly, A detailed spectroscopic study on the interaction of Rhodamine 6G with human hemoglobin, J. Photochem. Photobiol., B, 2010, 99(2), 78–86, DOI:10.1016/j.jphotobiol.2010.02.009.
- N. Manjubaashini, M. P. Kesavan, J. Rajesh and T. Daniel Thangadurai, Multispectroscopic and bioimaging approach for the interaction of rhodamine 6G capped gold nanoparticles with bovine serum albumin, J. Photochem. Photobiol., B, 2018, 183, 374–384, DOI:10.1016/j.jphotobiol.2018.05.005.
- R. Buividas, N. Dzingelevičius, R. Kubiliūtė, P. R. Stoddart, V. Khanh Truong, E. P. Ivanova and S. Juodkazis, Statistically quantified measurement of an Alzheimer's marker by surface-enhanced Raman scattering, J. Biophotonics, 2015, 8(7), 567–574, DOI:10.1002/jbio.201400017 , PubMed.
- S. K. Das, P. Ghosh, I. Ghosh and A. K. Guha, Adsorption of rhodamine B on Rhizopus oryzae: Role of functional groups and cell wall components, Colloids Surf., B, 2008, 65(1), 30–34, DOI:10.1016/j.colsurfb.2008.02.020.
- H. Rao, W. Qi, R. Su, Z. He and X. Peng, Mechanistic and conformational studies on the interaction of human serum albumin with rhodamine B by NMR, spectroscopic and molecular modeling methods, J. Mol. Liq., 2020, 316, 113889, DOI:10.1016/j.molliq.2020.113889.
- H.-H. Cai, X. Zhong, P.-H. Yang, W. Wei, J. Chen and J. Cai, Probing site-selective binding of rhodamine B to bovine serum albumin, Colloids Surf., A, 2010, 372(1), 35–40, DOI:10.1016/j.colsurfa.2010.09.017.
- M. J. Russo, A. F. Quigley, R. M. I. Kapsa, S. E. Moulton, R. Guijt, S. M. Silva and G. W. Greene, A Simple Electrochemical Swab Assay for the Rapid Quantification of Clonazepam in Unprocessed Saliva Enabled by Lubricin Antifouling Coatings, ChemElectroChem, 2020, 7(13), 2851–2858, DOI:10.1002/celc.202000393 , (acccessed 2021/07/27).
- G. Yao and Q. Huang, DFT and SERS Study of L-Cysteine Adsorption on the Surface of Gold Nanoparticles, J. Phys. Chem. C, 2018, 122(27), 15241–15251, DOI:10.1021/acs.jpcc.8b00949.
- C. Jing and Y. Fang, Experimental (SERS) and theoretical (DFT) studies on the adsorption behaviors of L-cysteine on gold/silver nanoparticles, Chem. Phys., 2007, 332(1), 27–32, DOI:10.1016/j.chemphys.2006.11.019.
- J. Pei, Y. Sun, X. Yu, Z. Tian, S. Zhang, S. Wei, Y. Zhao and R. Boukherroub, Au NPs decorated holey g-C3N4 as a dual-mode sensing platform of SERS and SALDI-MS for selective discrimination of L-cysteine, J. Colloid Interface Sci., 2022, 626, 608–618, DOI:10.1016/j.jcis.2022.06.176.
- N. N. Brandt, A. Y. Chikishev, A. V. Kargovsky, M. M. Nazarov, O. D. Parashchuk, D. A. Sapozhnikov, I. N. Smirnova, A. P. Shkurinov and N. V. Sumbatyan, Terahertz time-domain and Raman spectroscopy of the sulfur-containing peptide dimers: Low-frequency markers of disulfide bridges, Vib. Spectrosc., 2008, 47(1), 53–58, DOI:10.1016/j.vibspec.2008.01.014.
- M. J. Russo, M. Han, P. E. Desroches, C. S. Manasa, J. Dennaoui, A. F. Quigley, R. M. I. Kapsa, S. E. Moulton, R. M. Guijt and G. W. Greene, et al., Antifouling Strategies for Electrochemical Biosensing: Mechanisms and Performance toward Point of Care Based Diagnostic Applications, ACS Sens., 2021, 6(4), 1482–1507, DOI:10.1021/acssensors.1c00390.
- M. J. Russo, M. Han, N. G. Menon, A. F. Quigley, R. M. I. Kapsa, S. E. Moulton, R. M. Guijt, S. M. Silva, T. A. Schmidt and G. W. Greene, Novel Boundary Lubrication Mechanisms from Molecular Pillows of Lubricin Brush-Coated Graphene Oxide Nanosheets, Langmuir, 2022, 38(18), 5351–5360, DOI:10.1021/acs.langmuir.1c02970.
- G. Seniutinas, G. Gervinskas, R. Verma, B. D. Gupta, F. Lapierre, P. R. Stoddart, F. Clark, S. L. McArthur and S. Juodkazis, Versatile SERS sensing based on black silicon, Opt. Express, 2015, 23(5), 6763–6772, DOI:10.1364/oe.23.006763 , From NLM.
Footnote |
| † Electronic supplementary information (ESI) available: Raman spectra of bare-SERS in R6G spiked in 150 mM NaCl, bare or modified SERS in R6G spiked in blood, the relationship between the intensity of the peaks at 1174 cm−1, 1310 cm−1 and 1361 cm−1versus the concentration of R6G in unprocessed whole blood over the whole tested concentration range and the Method section. See DOI: https://doi.org/10.1039/d3an00910f |
| This journal is © The Royal Society of Chemistry 2024 |