Open Access Article
Open Access ArticleEnhanced fluorescence detection of miRNA by means of Bloch surface wave-based biochips†
Agostino
Occhicone
 *ab,
Francesco
Michelotti
*ab,
Francesco
Michelotti
 ab,
Paola
Rosa
a,
Daniele
Chiappetta
ab,
Tommaso
Pileri
a,
Paola
Del Porto
c,
Norbert
Danz
d,
Peter
Munzert
d,
Giuseppe
Pignataro
e and
Alberto
Sinibaldi
ab
ab,
Paola
Rosa
a,
Daniele
Chiappetta
ab,
Tommaso
Pileri
a,
Paola
Del Porto
c,
Norbert
Danz
d,
Peter
Munzert
d,
Giuseppe
Pignataro
e and
Alberto
Sinibaldi
ab
aSAPIENZA Università di Roma, Department of Basic and Applied Sciences for Engineering, Via A. Scarpa, 16, 00161 Roma, Italy. E-mail: agostino.occhicone@uniroma1.it
bCenter for Life Nano and Neuro Science, Istituto Italiano di Tecnologia (IIT), Viale Regina Elena 291, 00161 Rome, Italy
cDepartment of Biology and Biotechnology “C. Darwin”, Sapienza University of Rome, 00185 Rome, Italy
dFraunhofer Institute for Applied Optics and Precision Engineering, A.-Einstein-Str. 7, 07745 Jena, Germany
eDivision Pharmacology, Department of Neuroscience, School of Medicine, “Federico II” University of Naples, Italy
First published on 14th July 2023
Abstract
We report on the use of biochips based on one-dimensional photonic crystals sustaining Bloch surface waves to specifically detect target miRNA that is characteristic of hemorrhagic stroke (miR-16-5p) at low concentration in a buffer solution. The biochips were functionalized with streptavidin and ssDNA oligonucleotides to enable miRNA detection. To discriminate the target miRNA from a non-specific control (miR-101a-3p), we made use of an optical platform developed to work both in label-free and fluorescence detection modes. We demonstrate that the limit of detection provided when operating in the fluorescence mode allows us to specifically detect the target miRNA down to 1 ng mL−1 (140 pM), which matches the recommendations for diagnostic miRNA assays, 5 ng mL−1. The low costs open the way towards the application of these disposable optical biochips based on 1DPC sustaining Bloch surface waves as a promising tool for early disease detection in a liquid biopsy format.
1. Introduction
Microribonucleic acids (miRNAs) have emerged as a potential new class of biomarkers for non-invasive early diagnosis of a variety of diseases, including cancer, viral pathogenesis, and stroke.1–3 Concerning stroke, miRNAs are involved in several risk factors including hypertension, atherosclerosis, atrial fibrillation, diabetes, and dyslipidemia.4,5 Animal studies demonstrated the differential expression of specific miRNAs in the brain and blood following either an ischemic or a haemorrhagic insult and confirmed their potential use as diagnostic and prognostic markers.6During the last few decades, several research studies have focused on the development of point-of-care biosensors for the detection of biochemical interactions exploiting electrical, optical and electrochemical transducing systems.7 Optical biosensors have gained increasing interest since they provide clear advantages over traditional analytical techniques in terms of real-time, label-free and fluorescence detection of biological molecules in a highly sensitive, specific and cost-effective manner.8 Recent progress in miRNA detection based on surface-enhanced Raman scattering and surface plasmon polariton coupled fluorescence has been reviewed by X. Lu et al.9 The accurate and specific detection of miRNAs is a great challenge, as the short-chain microRNAs usually lack chemical stability. By combining nanomaterial assembling strategies with a wide variety of nucleic acid amplification methods, trace levels of miRNAs in biofluids can be sensitively detected.10 However, simple, cost-effective, and robust methodologies that satisfy the deployment of point-of-care testing are still needed for analysing clinical samples.
In the following, we demonstrate experimentally that disposable optical biochips based on one-dimensional photonic crystals (1DPC) sustaining Bloch surface waves (BSW) can efficiently and specifically detect miRNAs associated with haemorrhagic stroke. For the purpose of the present study, two models of rat miRNAs from Rattus norvegicus were selected, rno-miR-16-5p and rno-miR-101a-3p, which are specifically expressed in either ischemic or haemorrhagic stroke at different concentration levels depending on either the type of stroke or time delay after the insult.11
BSWs are surface waves that exist at the interface between a truncated periodic dielectric stack (1DPC) and an external dielectric medium.12,13 Similar to surface plasmon resonance (SPR) biosensors,14 BSW biosensors pertain to the class of evanescent-field-based photonic biosensors, which can conveniently probe liquid analytes in contact with their surface.15 The two main characteristics of the BSW evanescent field are that it decays exponentially along an axis perpendicular to the sensing surface, with a decay length on the order of 100 nm, and that it is resonantly enhanced at the surface of the 1DPC. Such a localization enhances the light–matter coupling and provides both sensitivity towards changes of the complex refractive index16 and enhancement of the emission of fluorescent labels17 in proximity to the interface. Moreover, the use of dielectric materials for the fabrication of the 1DPC sustaining BSW, compared to metals used in surface plasmon enhanced fluorescence schemes, prevents undesirable quenching phenomena during fluorescence emission.18,19 BSWs have been proposed for a variety of sensing applications, including standard protein detection protocols,20 cancer protein biomarker detection in complex biological media,21 labelled miRNA hybridization in buffer solutions19 and gas sensing.22–24
Here, we show that BSW coupled fluorescence operation can lead to a limit of detection (LoD) down to the ng mL−1 level for miRNAs dissolved in a buffer solution. Due to the 1DPC design and the optical readout scheme we adopted, such a resolution is better than that obtained in previous work with a different BSW based biosensing scheme for miRNA detection.19
2. Methods
2.1. Disposable 1DPC biochips
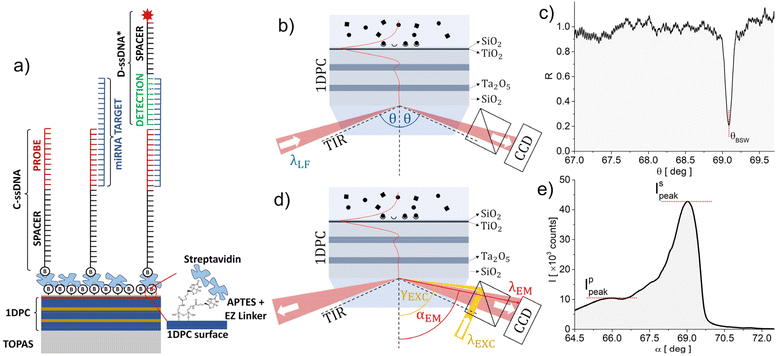
The biochips were obtained by depositing a 1DPC by plasma ion-assisted electron beam evaporation25 on an optical quality injection-moulded TOPAS26 substrate with a refractive index ns = 1.526 at a wavelength λLF = 670 nm for label-free operation. The substrate cross section is prismatic and allows coupling an external laser beam with the BSW under total internal reflection (TIR) conditions27 as shown in Fig. 1. An injection-moulded hard polymer flow cell with a patterned elastomer contact layer was coupled to the biochip to define a microchannel (width: 0.8 mm and height: 0.1 mm) and fluid connectors.28The 1DPC was designed29 to sustain a BSW in the visible region when operating in an aqueous external medium (na = 1.328 at λLF) and features 2.5 periods of SiO2 (low refractive index) and Ta2O5 (high refractive index) layers, with the thickness dL = 275 nm and dH = 120 nm, respectively. Two additional TiO2 and SiO2 layers, each dt = 20 nm thick, top the 1DPC. These top layers are used to position the BSW dispersion in the middle of the 1DPC photonic bandgap, to tune the absorption losses and to provide a SiO2 termination for subsequent silane-based chemical surface functionalization.21 The complex refractive indices at λLF of SiO2, Ta2O5, and TiO2 are nL = 1.450 + j5 × 10−6, nH = 2.095 + j5 × 10−5 and nT = 2.294 + j1.8 × 10−3, respectively. They were measured by either reflection/transmission spectroscopy on single supported thin films or ellipsometry on test 1DPC. For such a design, the BSW is associated with an exponentially decaying evanescent tail of the field inside the external medium with a penetration depth Lpen = 120 nm.
During the assays, the biochips were kept at a constant temperature (30 °C) by means of a Peltier element to reduce the signal variations originating from thermo-optical and thermo-mechanical effects.
2.2. Optical read-out system
The optical read-out system can operate in two alternative ways, by measuring either the refractive index change (label-free, LF) or the intensity of the emission of given labels (fluorescence, FLUO). In both cases the signals originate from biomolecules binding at the biochip's surface in the thin layer probed using the BSW evanescent tail in the liquid analyte. The two operation principles are sketched in Fig. 1.In the LF mode (Fig. 1b), an s-polarized laser beam at λLF = 671 nm is focused through the coupling prism in a wide range of angles θ above TIR. The BSW excitation at the biochip surface causes dissipation of the incident intensity and the appearance of a resonant dip at θBSW in the angular spectrum of the reflected beam, as shown in Fig. 1c. The minimum angular reflectance position depends on the dispersion of the BSW at λLF. In a LF assay, tracking the θBSW shift due to binding of biomolecules at the surface allows for real-time monitoring of the binding kinetics.
In the FLUO mode (Fig. 1d), the biochip is excited via a dichroic beam splitter in an epifluorescence configuration at an angle γEXC above TIR and λEXC = 637 nm by an s-polarized and slightly focused (ΔγEXC = 0.64°) laser beam. Alexa Fluor 647 was selected as a fluorescent label, since λEXC and λLF match its absorption (λABS = 647 nm) and emission (λEM = 671 nm) peak wavelengths, respectively.30 The angle γEXC is tuned to excite the BSW at λEXC with a resonantly enhanced intensity at the 1DPC surface, giving rise to an increased excitation rate. The fluorescence emission pattern is dominated by the large s and p polarized BSW density of states.31 Due to the dispersion of these leaky surface wave states,17 each spectral component at λFLUO > λEXC is emitted through the substrate at a different angle α(λFLUO) < γEXC as shown in Fig. 1e.
The layout of the optical system that implements the functions sketched in Fig. 1 is described in detail elsewhere.28 The system performs a cylindrical focusing of the laser beams to illuminate a line along the microfluidic channel. A 6 mm wide part of this region is imaged onto one dimension of the detector with a lateral resolution below 60 μm. A Fourier image of the light's angular distribution is generated along the other dimensions of the camera to obtain spatially and angularly resolved data. Since λFLUO is around λLF, it is possible to make use of the same collection optics, which guarantees an angular field of view of 2.7° and 8°, for the LF and FLUO cases, respectively. A couple of electronically controlled shutters allow switching between the LF and FLUO modes and performing either LF or FLUO assays or any temporal combination. A detailed description of the read-out system is given in section S.1 of the ESI.†
2.3. Bioassay format
The miRNA detection protocol is sketched in Fig. 1a and makes use of two synthetic single-stranded ssDNA: an anti-miRNA (C-ssDNA) and a detection probe (D-ssDNA*). The C-ssDNA (MW = 8592.7 Da) is composed of a probe sequence (12 bases) bound through a spacer sequence (15 thymine bases) to a biotin, which is used to anchor the C-ssDNA to a streptavidin layer previously conjugated to the biochip surface. The C-ssDNA probe sequence is complementary to a portion of the target miRNA. The D-ssDNA* is composed of a probe sequence (10 bases), which is complementary to the second portion of the target miRNA, conjugated through a spacer (11 thymine bases) to an Alexa Fluor 647 dye label (NHS Ester), to enable the fluorescence detection of miRNAs. The ssDNA probes and the miRNAs were purchased from Integrated DNA Technologies (IDT).Two different rat miRNAs were considered for the 1DPC biochip detection assays: rno-miR-16-5p (5′-GCG-GUU-AUA-AAU-GCA-CGA-CGA-U-3′), the target miRNA related to ischemic stroke (MW = 7065.3 Da, 22 RNA bases), and rno-miR-101a-3p (5′-AAG-UCA-AUA-GUC-UCA-UGA CAU-3′), the non-target miRNA used as a negative control (MW = 6705.1 Da, 21 RNA bases).
The ssDNA and miRNAs used in the assay were resuspended in IDTE 1× TE solution (10 mM Tris, 0.1 mM EDTA), pH 8, from Integrated DNA Technologies.
2.4. Chemical functionalization of the biochips
The chemical functionalization selected to immobilize the C-ssDNA relies on the deposition of a self-assembled organosilane layer onto the sensitive surface of the biochips, by using as a silane coupling reagent (3-aminopropyl)triethoxysilane (APTES, C9H23NO3Si, MW = 221.37 g mol−1, 99%, Merck).32The 1DPC coated portion of the biochips was cleaned with a piranha solution for 2 min, rinsed thoroughly with deionized water (DI-H2O) and dried under a stream of air. The clean biochips were immersed in a 2% APTES solution in an ethanol/water (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 v/v) mixture at ambient temperature (TA) for 1 h. Ethanol, rather than other solvents such as toluene, was used to avoid degradation of the TOPAS substrates. The biochips were removed from the APTES bath, sonicated in ethanol, rinsed with pure ethanol and dried under a stream of air. Finally, the biochips were soft baked at 50 °C for 1 h, to stop the polymerization of the siloxane layer and to stabilize the APTES film.
5 v/v) mixture at ambient temperature (TA) for 1 h. Ethanol, rather than other solvents such as toluene, was used to avoid degradation of the TOPAS substrates. The biochips were removed from the APTES bath, sonicated in ethanol, rinsed with pure ethanol and dried under a stream of air. Finally, the biochips were soft baked at 50 °C for 1 h, to stop the polymerization of the siloxane layer and to stabilize the APTES film.
The primary amines (NH2) provided by the APTES film were used to graft an EZ-Link-Sulfo-NHS-LC-Biotin (EZ-linker, C20H29O9N4S2Na, MW = 556.59 g mol−1, No-Weigh™ Format, Thermo Fisher Scientific) and obtain a densely packed biotin film. The EZ-linker from a single-use 1 mg vial was dissolved immediately before use in sterile phosphate-buffered saline (PBS 1×) to a concentration of 1 mg mL−1 and its immobilization was performed either on the whole sensitive surface of the biochip (simplex assay), by dropping a volume of ∼120 μL of the solution with a pipette, or on a series of 5 different zones aligned along the illumination/measurement strip, by means of a nano-plotter (multiplex assay). The nano-plotter is a piezoelectric actuated microarrayer system that allows for the deployment of small sized droplets (∼600 pL) with a high level of reproducibility (xy-axes repetition accuracy ±10 μm). The 5 circular zones spanned over a 0.6 mm diameter, owing to the hydrophilicity of the biochip surface after APTES functionalization. The biochips were then kept in a covered Petri dish for 1 h, after pipetting at TA and after plotting at 4 °C to avoid evaporation of the nano-drops. After that, the biochips were rinsed with sterile PBS 1× and dried under an air stream. In the end, the biochip surface was passivated with a bovine serum albumin solution (BSA, 1 mg mL−1 in PBS 1×, Merck) overnight at 4 °C, rinsed with PBS 1×, and dried under an air stream.
In a practical application, the functionalization would be completed by immobilizing by means of the EZ-linker the streptavidin and then the biotinylated C-ssDNA, so as to produce a ready-to-use biochip. However, since we wanted to investigate and optimize the immobilization conditions, the biochip was coupled to its microfluidic cover already at this stage and such successive steps were carried out by directly injecting the solutions in the complete biochip at the beginning of the assays, while monitoring the LF signal with the chip mounted on the optical read-out system.
3. Results and discussion
All bioassays carried out with 1DPC biochips reported here started with a regeneration process, in which the injection of a solution of glycine (20 mM) and HCl (added to reach pH 2.5) was carried out to remove the excess BSA from the 1DPC surface, followed by washing with PBS 1×. The volume used for all the washing steps was the same, i.e., 400 μL at a flow rate of 2.266 μL s−1.Then the bioconjugation of streptavidin (from Thermo Fisher Scientific) was carried out by injecting 180 μL of a 20 μg mL−1 solution of streptavidin in PBS 1× at a flow rate of 1.37 μL s−1, followed by washing with the PBS 1× and the TE buffers. The streptavidin interaction time was 10 min, as it was for all other subsequent bioconjugation steps.
3.1. Simplex assays
Fig. 2 shows a typical LF signal recorded during an assay (biochip S15 listed in Table 1). The θBSW resonance angle is tracked as a function of time, while interactions at the surface take place. The signal is the average over 10 adjacent 200 μm wide spots along the illumination strip.| Chip no. | 1st assay | 2nd assay | |
|---|---|---|---|
| Target miR-16-5p | Non-target miR-101a-3p | Target miR-16-5p | |
| [ng mL−1] | |||
| 14 chips were used in preliminary tests and partial assays and are not listed in the table. | |||
| S6 | 1000 | ||
| S10 | 100 | ||
| S11 | 100 | ||
| S14 | 1 | 100 | |
| S15 | 5 | 20 | |
| S18 | 1 | 1000 | |
| S20 | 1 | 5 | |
| S21 | 1 | 5 | |
| S22 | 1 | 5 | |
| S23 | 0 | 1000 | |
| S24 | 0 | 1000 | |
During a first phase the preparation of the biochip for the assays is completed. During the glycine/HCl regeneration (light grey band), the steep changes of θBSW were due to the change of the refractive index of the solutions in contact with the biochip surface; after washing, a negative residual shift was observed, indicating that non-specifically bound molecules were removed from the surface. The slight drift observed after regeneration in the running buffer (white band) is due to temperature and pump pressure drifts. Such drifts can be ruled out by operating in a differential configuration (see ESI, Fig. S.3†). During the streptavidin incubation (green band), a clear binding kinetics signal was observed, witnessing the efficient capture at the surface. The residual signal after washing with PBS 1× was due to the streptavidin capture. The subsequent change in the θBSW was attributed to the replacement of PBS 1× with the TE running buffer. The preparation proceeded with the injection and incubation of the C-ssDNA (1 μg mL−1 in TE, yellow band), leading to a biochip that was ready for a miRNA detection assay.
With reference to Fig. 2, we performed two consecutive assays with the same biochip. In each assay, the target miRNA (first at 5 ng mL−1 and then at 20 ng mL−1 in TE) (light violet bands) and the D-ssDNA* (1 μg mL−1 in TE, pink bands) are injected in a sequence. After each incubation step, washing of the biochip was carried out by injecting the TE buffer. The LF shifts recorded upon immobilization/hybridization of the ssDNA oligonucleotides are much smaller compared to the streptavidin protein case, showing that the LF resolution does not allow us to conclude anything concerning the miRNA detection, at such low concentrations. To overcome such a limitation, given the better resolution of the FLUO mode,21 we collected the fluorescence intensities in the FLUO mode at fixed times, before (dark grey bands) and after (red bands) the injection and incubation of the D-ssDNA*, in order to evaluate a differential signal.
Despite its limited resolution, the LF signal is a precious tool for the development of the assay protocols, since it allows monitoring, in real time and in the very same region probed by the FLUO signal, the biochip preparation process and assessing its quality. Taking advantage of the LF signal, it was, for example, possible to assess the efficiency and the repeatability of streptavidin bioconjugation. The LF sensograms reported in Fig. 3 are related to 18 different biochips functionalized with APTES and an EZ-linker and recorded upon injection of the same streptavidin solution (180 μL at 20 μg mL−1 in PBS 1×). The sensograms show very good repeatability with a 3.5% relative standard deviation and an average residual signal ΔθBSW = 116 mdeg. From ΔθBSW we can evaluate the mass surface coverage of streptavidin as:33
 | (1) |
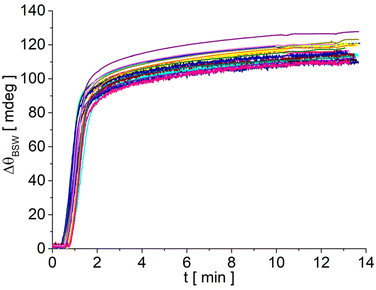 | ||
| Fig. 3 LF sensograms recorded during streptavidin injection for 18 different biochips. The curves were shifted by setting ΔθBSW to zero in the running buffer after the regeneration step. | ||
Similar studies were carried out to optimize the concentration of the C-ssDNA and D-ssDNA* solutions, within the limit of the LF resolution in assays in which the concentrations are larger than those used for the assay shown in Fig. 2 (see ESI, Fig. S.3†).
The reason for performing two consecutive miRNA assays as shown in Fig. 2, either target or non-target, is to increase the number of experimental data points and to demonstrate multiple uses of the biochips. In any case, the second concentration was always larger than the first.
With this aim in mind, after collecting the FLUO signal emitted by the labels at the end of the first assay, the biochips were photobleached in order to obtain a low background fluorescence signal before the successive assay. Photobleaching of the Alexa Fluor 647 dye under resonant CW excitation and field enhancement at γBSW is a fast process.28 Illumination at λEXC and γBSW for 3 min was sufficient to extinguish the fluorescence signal (see Fig. S.2a in the ESI†).
The list of the assays carried out with several different biochips is provided in Table 1.
Fig. 4 shows the background subtracted angular fluorescence emission recorded at the end of the first assay for six experiments carried out as shown in Fig. 2 with different biochips and for three different target miRNA concentrations (0 ng mL−1 (blank), 1 ng mL−1, and 5 ng mL−1).
 | ||
| Fig. 4 Background subtracted angularly resolved fluorescence emission spectra recorded in the FLUO mode at the end of target miRNA detection assays carried out with different biochips. | ||
The fluorescence emission from the 10 adjacent spots was acquired with a CCD camera at resonant excitation γEXC. Care was taken that each spot was excited resonantly. After subtracting the background fluorescence signal and normalizing by the excitation laser intensity, which has a Gaussian intensity distribution along the illumination strip, the spot signals were averaged to give the curves shown with their standard deviation. The curves are characterized by s (right) and p (left) polarized emission bands. Clearly the curves recorded at 1 ng mL−1 can be distinguished from the blank and from that obtained at 5 ng mL−1. The four different biochips at 1 ng mL−1 show a dispersion of the signal, which can be due to chip-to-chip inhomogeneity, derived from both biochip fabrication and chemical functionalization.
The results obtained in the FLUO mode at the end of the first assay for each biochip quoted in Table 1 were analysed in detail in order to estimate the LoD and to evaluate the chip-by-chip repeatability. Fig. 5 shows a bar chart of the relative intensities of the σ peak amplitudes shown in Fig. 4 and of those obtained in similar assays (not shown).
 | ||
| Fig. 5 Mean peak amplitudes Is of the s bands; the error bars are the standard deviation of the mean over 10 adjacent spots along the sensitive region of each biochip. The non-target and target miRNAs are miRNA-101a-3p and miRNA-16-5p, respectively. The red and black horizontal lines mark the means (solid) of either the negative or the positive samples and the standard deviation interval of the distributions (dotted), respectively. Inset: isothermal adsorption plots obtained with data produced by the 1st assays (dot) and 2nd assays (squares). The dashed curves are guides for the eyes according to eqn (3). | ||
We showed elsewhere34 that the amplitude of the s peak is proportional to the total fluorescence intensity given by the integral of the curves shown in Fig. 4, if anisotropic photobleaching can be neglected and the relative amplitude of the s and p bands remains constant.17,28 Using such a parameter therefore allows for a quantitative analysis, especially when part of the angular emission spectrum falls outside the angular field of view of the optical read-out system, as in the case shown in Fig. 4. In every FLUO measurement, the biochip was excited under the same resonant conditions and illumination time (5.0 ± 0.1 s), showing that photobleaching affects all fluorescence intensity levels by the same reduction factor. The uncertainty in the illumination time (0.1 s) corresponds to uncertainty in the peak intensity level of 90 counts, which is well below the standard error provided by averaging the intensity measured in each spot.
Fig. 5 confirms the scattering of the results obtained for the miRNA solutions at 1 ng mL−1, which, however, are always well distinguishable from the levels observed for the blank and for the non-target miRNA solutions at much larger concentration. The peak intensities of the positive (miR-16-5p at 1 ng mL−1) and negative (miR-101a-3p at 100 ng mL−1 and 1000 ng mL−1, and blanks) samples are distributed with mean (kcounts), standard deviation (kcounts) and coefficient of variation values (Ī,σ,σ/Ī) equal to (23.6, 6.8, 0.29) and (5.2, 3.2, 0.62), respectively. Interestingly, the intensities of the negative samples are smaller than the those of blanks, probably due to their non-specific binding and partial blocking against the D-ssDNA*.
Since the difference ĪPOS − ĪNEG overcomes by 1.84 times σPOS + σNEG, the two distributions can be distinguished with 99% probability. Therefore, the technique is capable (99%) of detecting the target miRNA in buffer down to a directly measured LoDFLUO = 1 ng mL−1 = 140 pM, below the threshold of 5 ng mL−1 (700 pM) desired for the clinical application of assays detecting circulating miRNAs related to ischemic stroke.35 The LoDFLUO found is smaller than that found with biochips based on a different grating assisted BSW coupled fluorescence for similar miRNA targets in buffered solutions.19 In that case the assays were limited to single hybridization of dye-labelled miRNA to a C-ssDNA probe and the LoD was not assessed towards blank or non-target solutions.19 Therefore, if similar LoD can be achieved when conducting measurements in biological media, the technique demonstrates potential for clinical applications.
We can estimate a potential LoDESTFLUO in buffer, by assuming that a concentration is detected if the average intensity overcomes ĪNEG by 3σNEG:
 | (2) |
If the LoDESTFLUO is assessed by using as a negative only the results obtained with the non-target miRNA (three data points in Fig. 5), which is closer to the real operation conditions, then, we obtain LoDESTFLUO = 0.16 ng mL−1 = 22 pM. However, since we did not assay at such LoDESTFLUO, we cannot claim such a resolution and just quote the measured value at LoDFLUO = 1 ng mL−1 in buffer.
The mean intensities shown in Fig. 5 can be plotted (dots) as a function of the target concentration together with those (squares) measured in the second assay carried out with each biochip and analysed in a similar way, as shown in the inset of Fig. 5. The dashed lines are a guide to the eye following the modified Langmuir isotherm model:36
 | (3) |
Finally, Fig. 2 shows that, when starting an assay with a biochip prepared up to the C-ssDNA immobilization step, an miRNA detection assay with the present platform may last less than 30 min, demonstrating the competitiveness of the technique with respect to the gold-standards such as the ELISA37 or qPCR38 techniques, which typically take up to 2.5–6 hours.
3.2. Multiplex assays
The biochips that were patterned with the nano-plotter, once mounted on the read-out system, were used in assays that started with the same glycine/HCl cleaning and streptavidin immobilization steps used for the simplex assays. Fig. 6a and b show the angular images collected by the CCD camera in the LF mode for an exemplary assay, before and after streptavidin immobilization, respectively. Each of the 90 rows corresponds to a 67 μm spot along the 6 mm long illumination strip. The real-time LF sensograms recorded in both the SIG and CTRL zones are reported in Fig. S.3 of the ESI.† The blue region marks the angular position of the BSW resonant dip in every spot. The clear modulation of θBSW allows the verification of the effectiveness of the selective functionalization of the surface in the 5 signal (SIG) zones and of the BSA blocking in the intermediate control zones (CTRL).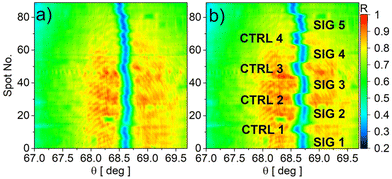 | ||
| Fig. 6 Reflectance map in the LF mode recorded before (a) and after (b) the streptavidin immobilization for a biochip that was patterned by means of the nano-plotter. | ||
After streptavidin immobilization, the assay proceeded according to the same protocol used for the simplex assays. In Table 2 we list the biochips that were used, with the concentrations of the ssDNA and miRNA solutions. The aim of the assays, besides demonstrating the possibility of different multiplex assays on the same biochip, was to test different conditions and optimize the protocols. Therefore, different combinations of the concentrations of the solutions were used.
| Chip no. | C-ssDNA | rno-miR-16-5p | D-ssDNA* |
|---|---|---|---|
| [μg mL−1] | |||
| M1 | 10 | 1 | 1 |
| M2 | 50 | 1 | 1 |
| M3 | 100 | 0.1 | 0.1 |
As in that case, the LoDLF value did not permit verification in a label-free manner whether the C-ssDNA, miRNA and D-ssDNA* were captured at the surface. However, the introduction of the fluorescent label via hybridization with D-ssDNA* provides such a resolution. Fig. 7 shows the background corrected CCD images collected after D-ssDNA* hybridization. The images were also normalized along the vertical direction to take into account the Gaussian shape of the fluorescence excitation laser. Also in this case, each of the 90 rows corresponds to a 67 μm spot along the fluorescent strip. The 5 different signal zones can be clearly distinguished, with a fluorescence intensity that depends on the specific biochip. Such a result demonstrates that the technique provides the lateral resolution that would be needed to perform multiplex assays with a single biochip if different C-ssDNAs were immobilized in different zones.
 | ||
| Fig. 7 Angular fluorescence emission maps recorded for the biochips M1 (a), M2 (b) and M3 (c). The s and p polarized emission bands appear as two vertical segmented bands. | ||
The quantification of the fluorescence intensities was achieved by integrating the angular fluorescence emission spectra shown in Fig. 7 over the CCD angular range of collection (8°) and averaging inside each signal zone. The results of the analysis are shown in Fig. 8. They show that the response of the 5 signal zones (intra-biochip) is relatively uniform, with an average coefficient of variation CV = 0.28, which is in the range of that found above for assays carried out at 1 ng mL−1 with five different biochips (inter-biochip). Such a result suggests that the variability of the biochip response is due to the assay biochemical steps rather than a dispersion of the physical properties of the 1DPC transductors upon fabrication.
A direct comparison of biochips M1 and M2 (red and green) leads to the expected conclusion that the emitted fluorescence is a function of the immobilized quantity of C-ssDNA (while maintaining the same concentration of the other injected oligonucleotides). This aspect is critically highlighted by a further comparison of the biochips M2 and M3. In this case, by doubling the concentration of the C-ssDNA, an even larger amount of fluorescence is collected by using one tenth of the target and D-ssDNA* concentrations (Table 2).
4. Conclusions
In conclusion, the study reports the successful development of consumable biochips for miRNA detection using a 1D photonic crystal platform. The biochips were prepared by a regeneration process followed by bioconjugation of streptavidin and immobilization of probe ssDNA oligonucleotides. The biochips were used in test assays with target and non-target miRNA in buffer solutions at different concentrations, using both reflectance and fluorescence spectroscopy. The lower resolution of the reflectance spectroscopy was useful for monitoring the biochip preparation process and optimizing bioconjugation and ssDNA immobilization. The fluorescence spectroscopy provided better sensitivity for miRNA detection.The results obtained with biochips during their first use directly demonstrated that the target miRNA can be detected in buffer at a concentration as low as 1 ng mL−1 (140 pM) in less than 30 minutes.
The achieved resolution is definitively adequate for the analysis of serum samples, since 5 ng mL−1 is defined as the threshold between basal and abnormal miRNA expression levels related to ischemic/haemorrhagic stroke. The fundamental difference is the substantial reduction of the assay duration with respect to gold-standard techniques, such as ELISA or qPCR, which may take from 2.5 to 6 hours.
The study demonstrates the potential of 1D photonic crystal platforms for miRNA detection with high sensitivity and specificity. Further optimization and validation with clinical samples could lead to the development of a diagnostic tool for miRNA-based diseases. Such future work will have to address the challenges due to fouling that may affect the LoD and the assessment of the relevant concentration range in actual blood samples. Moreover, using the same read-out system, there is a possibility of multiplexed and real-time investigations utilizing a spotting tool system, therefore enabling the simultaneous detection of different miRNAs.
Author contributions
The manuscript was written through contributions of all authors. Conceptualization, A.S., F.M. and G.P.; methodology, A.O., A.S., T.P., P.D.P., and D.C.; software, F.M., N.D. and A.O.; validation, A.O., P.R. and A.S.; formal analysis, A.O. and P.R.; investigation, A.O., P.R., T.P., D.C., P.M. and A.S.; resources, D.C., P.D.P., P.M. and A.S.; data curation, A.O., P.R. and T.P.; writing—original draft preparation, A.O., A.S. and F.M.; writing—review and editing, A.O., A.S. P.D.P., N.D. and F.M.; supervision, A.S. and F.M.; project administration, A.S. and F.M.; and funding acquisition, F.M. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. S. acknowledges funding from PNRR-Rome Technopole Flagship Project 7 (B83C22002820006), F. M. from PNRR-PNC-D34Health (B53C22006120001), and A. O. from Regione Lazio through the projects BIOLIGHT (B86J20001690002). A. S. and F. M. acknowledge funding from the Italian MUR Ministry through the project NEON (ARS01_00769).The authors wish to thank Paola Di Matteo for her assistance with the chemical functionalization of the biochips, Frank Sonntag for providing the nano-plotter used in the experiments and Marco Magi for technical assistance.
References
- S. M. Hammond, Adv. Drug Delivery Rev., 2015, 87, 3–14 CrossRef CAS PubMed.
- P. S. Mitchell, R. K. Parkin, E. M. Kroh, B. R. Fritz, S. K. Wyman, E. L. Pogosova-Agadjanyan, A. Peterson, J. Noteboom, K. C. O′briant, A. Allen, D. W. Lin, N. Urban, C. W. Drescher, B. S. Knudsen, D. L. Stirewalt, R. Gentleman, R. L. Vessella, P. S. Nelson, D. B. Martin and M. Tewari, Proc. Natl. Acad. Sci. USA, 2008, 105, 10513–10518 CrossRef CAS PubMed.
- Y. Wu, Q. Li, R. Zhang, X. Dai, W. Chen and D. Xing, Clin. Chim. Acta, 2021, 516, 46–54 CrossRef CAS PubMed.
- World Stroke Organization, https://www.world-stroke.org, (accessed 8 February 2023).
- M. Katan and A. Luft, Semin. Neurol., 2018, 38, 208–211 CrossRef PubMed.
- G. Koutsis, G. Siasos and K. Spengos, Curr. Top. Med. Chem., 2013, 13, 1573–1588 CrossRef CAS PubMed.
- S. Shrivastava, T. Q. Trung and N.-E. Lee, Chem. Soc. Rev., 2020, 49, 1812–1866 RSC.
- H. Altug, S. H. Oh, S. A. Maier and J. Homola, Nat. Nanotechnol., 2022, 17, 5–16 CrossRef CAS PubMed.
- X. Lu, C. Yao, L. Sun and Z. Li, Biosens. Bioelectron., 2022, 203 Search PubMed.
- F. Yin, H. Liu, Q. Li, X. Gao, Y. Yin and D. Liu, Anal. Chem., 2016, 88, 4600–4604 CrossRef CAS PubMed.
- P. Cepparulo, O. Cuomo, A. Vinciguerra, M. Torelli, L. Annunziato and G. Pignataro, Front. Neurol., 2021, 12, 736474 CrossRef PubMed.
- P. Yeh, A. Yariv and C.-S. Hong, J. Opt. Soc. Am., 1977, 67, 423–438 CrossRef.
- I. Tamm, Z. Phys., 1932, 849–850 CrossRef CAS.
- J. Homola, Chem. Rev., 2008, 108, 462–493 CrossRef CAS PubMed.
- A. Sinibaldi, N. Danz, E. Descrovi, P. Munzert, U. Schulz, F. Sonntag, L. Dominici and F. Michelotti, Sens. Actuators, B, 2012, 174, 292–298 CrossRef CAS.
- M. Shinn and W. M. Robertson, Sens. Actuators, B, 2005, 105, 360–364 CrossRef CAS.
- F. Michelotti and E. Sepe, J. Phys. Chem. C, 2019, 123, 21167–21175 CrossRef CAS.
- K. Toma, E. Descrovi, M. Toma, M. Ballarini, P. Mandracci, F. Giorgis, A. Mateescu, U. Jonas, W. Knoll and J. Dostálek, Biosens. Bioelectron., 2013, 43, 108–114 CrossRef CAS PubMed.
- F. Frascella, S. Ricciardi, L. Pasquardini, C. Potrich, A. Angelini, A. Chiadò, C. Pederzolli, N. De Leo, P. Rivolo, C. F. Pirri and E. Descrovi, Analyst, 2015, 140, 5459–5463 RSC.
- V. N. Konopsky and E. V. Alieva, Anal. Chem., 2007, 79, 4729–4735 CrossRef CAS PubMed.
- A. Sinibaldi, C. Sampaoli, N. Danz, P. Munzert, L. Sibilio, F. Sonntag, A. Occhicone, E. Falvo, E. Tremante, P. Giacomini and F. Michelotti, Biosens. Bioelectron., 2017, 92, 125–130 CrossRef CAS PubMed.
- F. Michelotti, B. Sciacca, L. Dominici, M. Quaglio, E. Descrovi, F. Giorgis and F. Geobaldo, Phys. Chem. Chem. Phys., 2010, 12, 502–506 RSC.
- V. N. Konopsky, D. V. Basmanov, E. V. Alieva, S. K. Sekatskii and G. Dietler, Appl. Phys. Lett., 2012, 100, 083108 CrossRef.
- Y. Chen, Z. Kong, W. Sun, J. Liang, J. Xing, S. Lin, S. Zhu, H. Zhang, Z. Shen and J. Lu, Opt. Express, 2022, 30, 34510 CrossRef PubMed.
- P. Munzert, N. Danz, A. Sinibaldi and F. Michelotti, Surf. Coat. Technol., 2017, 314, 79–84 CrossRef CAS.
- TOPAS Advanced Polymers GmbH, https://www.topas.com, (accessed 10 February 2023).
- E. Kretschmann and H. Raether, Z. Naturforsch., A: Astrophys., Phys. Phys. Chem., 1968, 23, 2135–2136 CrossRef CAS.
- E. Sepe, A. Sinibaldi, N. Danz, P. Munzert and F. Michelotti, J. Phys. Chem. C, 2019, 123, 21176–21184 CrossRef CAS.
- F. Michelotti, R. Rizzo, A. Sinibaldi, P. Munzert, C. Wächter and N. Danz, Opt. Lett., 2017, 42, 2798 CrossRef CAS PubMed.
- Alexa Fluor™ 647 NHS Ester, https://www.thermofisher.com/order/catalog/product/A20006, (accessed 10 February 2023).
- S. D. Choudhury, R. Badugu and J. R. Lakowicz, Acc. Chem. Res., 2015, 48, 2171–2180 CrossRef PubMed.
- N. S. K. Gunda, M. Singh, L. Norman, K. Kaur and S. K. Mitra, Appl. Surf. Sci., 2014, 305, 522–530 CrossRef CAS.
- J. Vörös, Biophys. J., 2004, 87, 553–561 CrossRef PubMed.
- A. Occhicone, P. Del Porto, N. Danz, P. Munzert, A. Sinibaldi and F. Michelotti, Crystals, 2021, 11, 1517 CrossRef CAS.
- A. Mompeón, L. Ortega-Paz, X. Vidal-Gómez, T. J. Costa, D. Pérez-Cremades, S. Garcia-Blas, S. Brugaletta, J. Sanchis, M. Sabate, S. Novella, A. P. Dantas and C. Hermenegildo, Sci. Rep., 2020, 10, 5373 CrossRef PubMed.
- P. Atkins, J. de Paula and J. Keeler, Atkins’ Physical Chemistry, Oxford University Press, Oxford, 12th edn, 2023 Search PubMed.
- miREIA – miRNA enzyme immunoassay, https://www.biovendor.com/mirna, (accessed 27 February 2023).
- TaqMan Advanced miRNA Assays, https://www.thermofisher.com/it/en/home/life-science/pcr/real-time-pcr/real-time-pcr-assays/mirna-ncrna-taqman-assays/taqman-advanced-mirna-assays.html, (accessed 27 February 2023).
Footnote |
| † Electronic supplementary information (ESI) available: Detailed description of the read-out system (S1), of the photobleaching phenomena (S2) and of the real time differential operation of the multiplex biochips (S3). See DOI: https://doi.org/10.1039/d3an00804e |
| This journal is © The Royal Society of Chemistry 2023 |