Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Towards non-invasive monitoring of non-melanoma skin cancer using spatially offset Raman spectroscopy†
Martha Z.
Vardaki
 *ab,
Eleftherios
Pavlou
a,
Nikolaos
Simantiris
c,
Evangeli
Lampri
d,
Konstantinos
Seretis
*ab,
Eleftherios
Pavlou
a,
Nikolaos
Simantiris
c,
Evangeli
Lampri
d,
Konstantinos
Seretis
 e and
Nikolaos
Kourkoumelis
e and
Nikolaos
Kourkoumelis
 a
a
aDepartment of Medical Physics, School of Health Sciences, University of Ioannina, 45110 Ioannina, Greece
bInstitute of Chemical Biology, National Hellenic Research Foundation, 48 Vassileos Constantinou Avenue, Athens, 11635, Greece
cIonian University, Department of Informatics, Corfu, 49132, Greece
dDepartment of Pathology, Faculty of Medicine, University of Ioannina, 45110 Ioannina, Greece
eDepartment of Plastic Surgery, School of Health Sciences, University of Ioannina, 45110 Ioannina, Greece
First published on 4th August 2023
Abstract
BCC (basal cell carcinoma) and SCC (squamous cell carcinoma) account for the vast majority of cases of non-melanoma skin cancer (NMSC). The gold standard for the diagnosis remains biopsy, which, however, is an invasive and time-consuming procedure. In this study, we employed spatially offset Raman spectroscopy (SORS), a non-invasive approach, allowing the assessment of deeper skin tissue levels and collection of Raman photons with a bias towards the different layers of epidermis, where the non-melanoma cancers are initially formed and expand. Ex vivo Raman measurements were acquired from 22 skin biopsies using conventional back-scattering and a defocused modality (with and without a spatial offset). The spectral data were assessed against corresponding histopathological data to determine potential prognostic factors for lesion detection. The results revealed a positive correlation of protein and lipid content with the SCC and BCC types, respectively. By further correlating with patient data, multiple factor analysis (MFA) demonstrated a strong clustering of variables based on sex and age in all modalities. Specifically for the defocused modality (zero and 2 mm offset), further clustering occurred based on pathology. This study demonstrates the utility of the SORS technology in NMSC diagnosis prior to histopathological examination on the same tissue.
Introduction
The incidence of non-melanoma skin cancers (NMSCs), which are the most common human malignancies, is rising globally.1 The prevalent forms of NMSC are the basal cell carcinoma (BCC), followed by the squamous cell carcinoma (SCC). However, few studies have evaluated the incidence of BCCs and SCCs separately, because, in the majority of European countries, distinct forms of NMSC are not separated in their national cancer registry data.2 Although their incidence rates and aetiologies differ significantly, their overall occurrence rate is increasing. This is probably related to increased exposure to the sun and artificial tanning devices, as well as to more efficient registration methods. Due to insufficient skin pigmentation, Caucasian populations have a significantly higher risk of developing skin cancer, which is exacerbated by excessive UV exposure.Even though NMSCs have low mortality rate, they usually appear on the head and neck, posing a substantial impact on the quality of life while introducing a sizeable financial burden on health care systems.3,4 The danger of metastasis linked with NMSC is low, especially for BCC. In contrast, SCC has a relatively low but recognised rate of metastasis (0.3%–3.7%), accounting for 20% of skin cancer-related deaths.5 NMSC lesions propagate in all directions affecting the integrity of the adjacent skin tissue especially after a postponed or inadequate therapy. Moreover, a known history of NMSC increases by ten-fold the likelihood of a second NMSC.6 Although an invasive and time-consuming procedure, histopathological evaluation remains the gold standard approach for assessing the pathology of surgically removed skin tissues. The study of the excised tissue is performed ex vivo with light microscopy by qualified physicians, who provide a biochemical tumour assessment. Raman spectroscopy has been suggested as a complementary histopathological tool in terms of time management as well as workload issues.7 The biochemical profile of the deep layers of the lesion can also provide lesion vascular characteristics that can be used to diagnose the type of BCC.8 If a technology could reliably and non-invasively assess a specimen and possibly provide biochemical information from the tissue layers, where the NMSCs initially form and expand, that would assist surgeons to diagnose the malignancy with greater accuracy.
Reflectance confocal microscopy (RCM), optical coherence tomography (OCT), and ultrasonography have emerged as alternative non-invasive techniques to assess skin lesions. However, limited spatial resolution, penetration depth and contrast hinder their diagnostic capability. In fact, the penetration depth of RCM and OCT is restricted to the first millimetre, due to intense photon scattering in the skin and the mismatch of the refractive index of the inhomogeneous tissue.9 Similarly, ultrasound contrast is often restricted due to the inability to distinguish hypoechoic skin lesions from normal hypoechoic skin components.9
Raman spectroscopy is a form of molecular spectroscopy, which relies on the inelastic scattering of photons, exchanging energy via vibrational modes of molecules. Raman-active molecules, featuring anisotropic polarizability, provide spectroscopic signals with significant chemical and compositional information. In turn, Raman spectra contain chemical bond-specific bands and provide a “molecular fingerprint” of the material under study. Raman spectroscopy can detect subtle biochemical changes in biological samples since it does not interfere with water, it is non-invasive, and requires minimal sample preparation. The methodology has been utilised as a reliable approach for the in situ detection of malignancies at the tissue level and the investigation of the subcellular molecular composition of tissues.10 In addition, it has been used in ex vivo studies of various skin pathologies11 and in in vivo skin measurements for real-time skin cancer screening,12 as well as in the detection of the stages of tumour growth, and in the assessment of the effectiveness of radiation therapy.13 Depending on the type of skin cancer, the origin of the cancer can be found in different depths of the tissue. Yet, the majority of recent Raman studies have focused on confocal measurements, which allow for improved resolution and, consequently, malignancy discrimination at shallow sub-surface depths.14
Deep Raman spectroscopy, and especially spatially offset Raman spectroscopy (SORS), is a relatively new technique that provides, non-invasively, the biochemical profile of the skin by collecting Raman photons with a bias toward deeper skin layers where cancer initially develops. In SORS measurements, the excitation and collection signals on the surface of the material are decoupled and their distance (i.e., offset) corresponds to the depth of Raman photon origin,15 which can reach up to a few centimeters,16 revealing the chemical composition of sub-surface components that are covered by superficial, turbid layers. Although SORS has been employed in a number of biomedical applications, such as transfusion blood17–19 and breast cancer,20,21 studies on skin are still limited to wound assessment on rodents22 and salmon skin assessment.23
To our knowledge, our group is the only one to have employed a spatially offset mode to investigate skin-deep layer profiles of healthy human skin,24 and with this study we aim to extend the application to skin cancer biopsies. Alternative approaches have been suggested to achieve deeper penetration depths of up to 100 μm,25,26 or even higher, with OCT-combined systems.27 These methods include defocusing the excitation beam, utilizing a depth-sensitive microscope approach, and coupling it with spatially separated collection fibers.28 However, these methods have limitations in terms of depth signal collection, which restricts flexibility and increases measurement acquisition time and power usage. In contrast to breast cancer studies, in which SORS has been effectively deployed to retrieve a signal of calcifications in tissue depth,21 it is currently uncertain but promising, as demonstrated by preliminary studies, that the deep Raman modality can assist in skin cancer diagnostics.
In this study, we employed conventional and spatially offset Raman spectroscopy to explore the diagnostic potential of the technique on a number of biopsies as part of the standard clinical protocol. We then compared the different modalities to identify the one that leads to the most efficient discrimination between BCC and SCC. We further employed multi factor analysis to demonstrate that despite sex and age variability in our samples, biochemical information acquired from deeper layers through SORS measurements results in a more accurate (compared to superficial conventional measurements) pathology-based classification between the non-melanoma cancer subtypes. Given the biochemical information revealed by deep Raman spectroscopy for the differences in the composition of BCC and SCC tissue, this study demonstrates that Raman spectroscopy could support the development of a screening tool for future non-invasive skin cancer applications.
Materials and methods
Human tissue samples
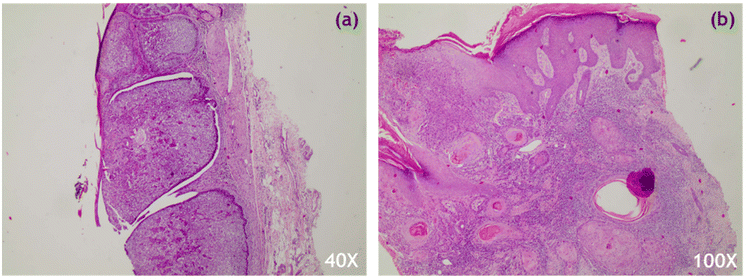
22 skin tumour biopsy samples removed from 20 patients were provided by the Pathology Department of the University Hospital of Ioannina, following standard clinical protocols. The samples were rinsed with saline to remove excess blood and then positioned on a stainless-steel substrate at the top of aluminum foil in a Petri dish for measurement acquisition. To ensure that the biopsy lesion was properly illuminated, circumferential and lesion-specific measurements were taken. Following Raman measurements, the biopsy samples were sectioned, hematoxylin and eosin (H&E) stained (Fig. 1) and evaluated by an expert histopathologist. Only Raman spectra from cases verified as basosquamous, nodular or micronodular BCC, and SCC of high or medium grade were subjected to further analysis (Table SI†). Samples removed from the hairline or with an extremely uneven surface were excluded from the analysis. The results were compared to corresponding histopathological data to determine potential prognostic factors for lesion detection.Raman setup
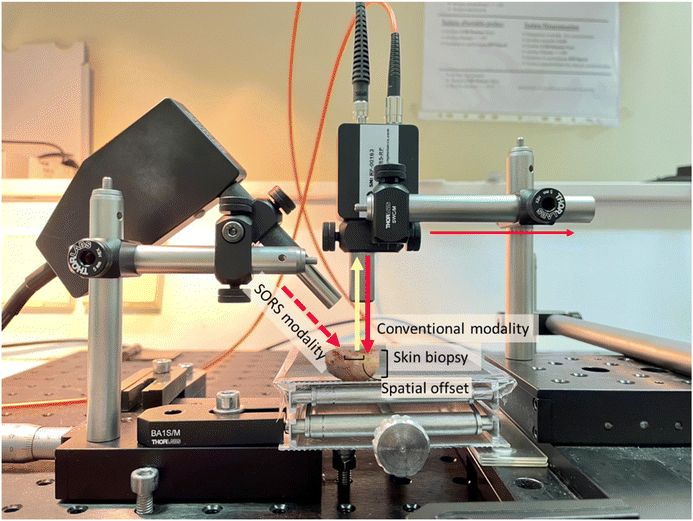
Ex vivo Raman measurements were acquired from the lesion areas of 22 skin tissue biopsies using the conventional mode (back-scattering) and the SORS mode of a custom-built Raman system, previously optimized on skin tissue phantoms with geometry and optical properties similar to skin (Fig. 2).24 A 785 nm laser (Mini-Benchtop Stabilized Laser, Coherent, Santa Clara, CA, USA) was focused on the sample through a multimode fiber (M43L01, Thorlabs), which was in turn coupled to a standard probe with a 4 cm long and 11 mm working distance probe tip (Wasatch Photonics, Morrisville, NC, USA). A second fiber (M29L01, Thorlabs) was coupled to the same probe to collect the Raman scattered photons in a back-scattering conventional mode. The collection fiber was coupled to an f/1.3 Raman spectrometer (WP 785 ER Raman Spectrometer, Wasatch Photonics). The laser spot size employed in conventional modality was 170 μm. For the SORS measurements, a second excitation probe (BAC102, B&W TEK) was instead coupled to the laser delivering a beam at a 45° angle with respect to the sample (Fig. 2). The incident beam on the sample surface was elliptical, with the shorter radius being 2 mm and the longer 3 mm, essentially consisting of a defocused excitation beam with much lower confocality compared to the conventional mode. The excitation probe employed for the SORS measurements was mounted on an individual XY translational stage (Thorlabs, Inc., Newton, NJ, USA) to adjust the spatial offset.Raman measurements
Each Raman spectrum was collected using 5 accumulations of 5–10 s depending on the CCD saturation levels. Dark noise was recorded prior to each measurement and automatically removed by the software (Enlighten Spectroscopy software, Wasatch Photonics). The system was calibrated using Raman bands of an aspirin tablet (acetylsalicylic acid). All samples were measured with 50%–100% laser power (150–300 mW) on the sample. In the SORS mode, the spatial offset employed was 0 and 2 mm using a diffuse laser beam. A visual examination of the tissue revealed no heat degradation due to the laser beam as (i) the sample was kept hydrated between measurements with saline and (ii) the large volume of each tissue sample (compared to tissue sections) allowed for efficient heat dissipation. 62 Raman spectra were collected from 20 biopsies using the conventional back-scattering modality. 66 Raman spectra were collected from 21 and 22 biopsies using the zero and 2 mm offset modality, respectively.Data analysis
All data were exported through the Enlighten Spectroscopy software (Wasatch Photonics) and preprocessed using Python 3.7. Preprocessing of the obtained spectra involved the removal of cosmic rays and spikes using a custom-made algorithm built around the signal.find_peaks method of Scipy (1.7.3), Savitzky–Golay smoothing using a 3rd degree polynomial in a 15-point window, background removal using the SNIP algorithm29 (15 iterations), and unit vector normalization after the spectra were cropped in the 400–1800 cm−1 fingerprint region. For principal component analysis (PCA), the preprocessed data were used without further processing.For the statistical analysis of the data, multiple factor analysis (MFA)30,31 was performed in order to evaluate the statistical relationships and similarities between heterogeneous variables from the same set of patients. The analysis was carried out on Matlab (version 2021a). MFA is an extension of the common principal component analysis (PCA) that is able to handle a mixture of different types of variables (quantitative and qualitative) collected on the same observations. In this study, the quantitative data used were the Raman spectra (400–3600 cm−1 spectral range) from the skin samples of the patients, and the qualitative data were the cancer type, sex, and age of the patients. For the quantitative data, PCA was performed to obtain the first eigenvalue of the dataset. Then, the dataset was normalized/weighted by dividing all of its elements by the square root of the first eigenvalue. For the qualitative data, an indicator variable was associated with each category (e.g. 1 for male, 0 for female), followed by a multiple correspondence analysis (MCA). Age groups were defined by less or more than 77 years old. Similar to the quantitative data, the qualitative data were weighted by dividing all elements by the square root of the first eigenvalue obtained from the MCA. The separate weighting of the quantitative and qualitative data allows for the merging of the data into a weighted dataset, in which a global PCA was performed to obtain a set of principal factor scores for the observations (patients) and principal factor loadings for the variables (Raman, cancer type, sex, and age). Furthermore, we performed an eigenanalysis of the spectral data correlation matrix, which showed that only the first four eigenvectors exhibited eigenvalues greater than 1. These three eigenvalues express more than 99% of the variability in the spectral dataset. Nevertheless, the factors from the eigenvector matrix are orthogonal and uncorrelated.32 Thus, we performed oblique rotations, specifically Promax rotations,33 in order to allow factors to be correlated and simplify the interpretation of the findings.
Ethical approval
All experiments were performed in compliance with the guidelines of the Declaration of Helsinki and approved by the Institutional Ethical Review Committee of the University Hospital of Ioannina, which confirmed the proposed protocol and the anonymity of the donors (reference number: 8(17)/14.04.2021).Results and discussion
PCA analysis in the fingerprint region
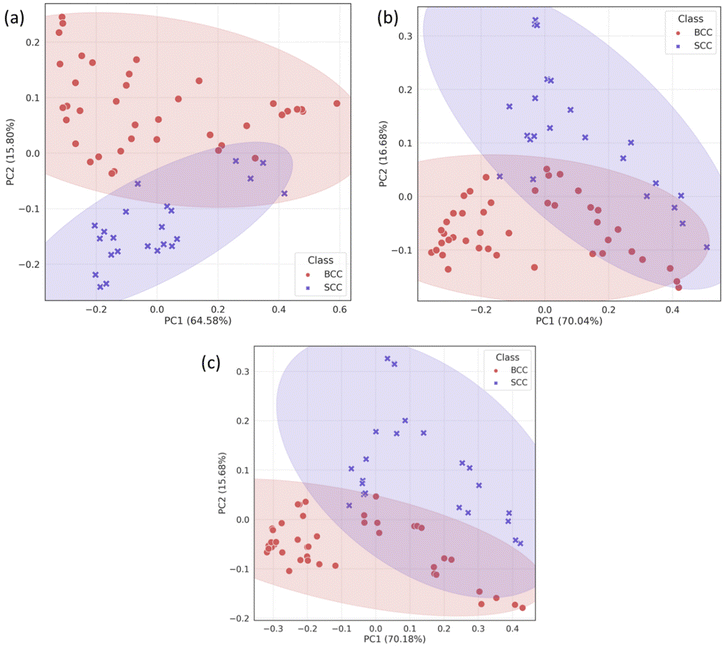
Due to the lack of strong visual differences between BCC and SCC Raman spectra,34 we employed PCA to uncover them. Principal components were evaluated on the basis of successful discrimination between BCC and SCC. Pathology discrimination (BCC vs. SCC) was explored in terms of the spatial offset. Two of the principal components were chosen for each mode (conventional, zero and 2 mm offset), based on the best discrimination achieved and their scores were plotted for each spectrum in Fig. 3.In the PCA scatter plot of the biopsy samples (Fig. 3), BCC and SCC are discriminated mainly across principal component 1 (PC1) and 2 (PC2), the loadings of which explain a total of 80%–85% of the spectral variance, depending on the measurement mode (Fig. S1†). The separation of the pathological groups is more evident in the defocused beam (0 and 2 mm), and occurs primarily over PC2. The calculation of the Euclidean distance (0.213 in the conventional vs. 0.248 and 0.261 in zero and 2 mm offset, respectively) confirmed this observation. Furthermore, loadings corresponding to PC1 and PC2 are almost identical for the 0 and 2 mm offsets (Fig. 4), implying the same physical basis for the separation in both modes. For both PC1 and PC2 in the 0 and 2 mm modes, the corresponding loadings are positively associated with bands assigned to SCC (Fig. 4). Those bands include 937, 1003, 1125, 1339 and 1640 cm−1 and are moving in the opposite direction from 507, 631, 772, 870, 1076, 1189, 1268, 1300, 1435, 1531, 1650 and 1743 cm−1, which are assigned to BCC typical bands negatively associated with the loading for PC2.
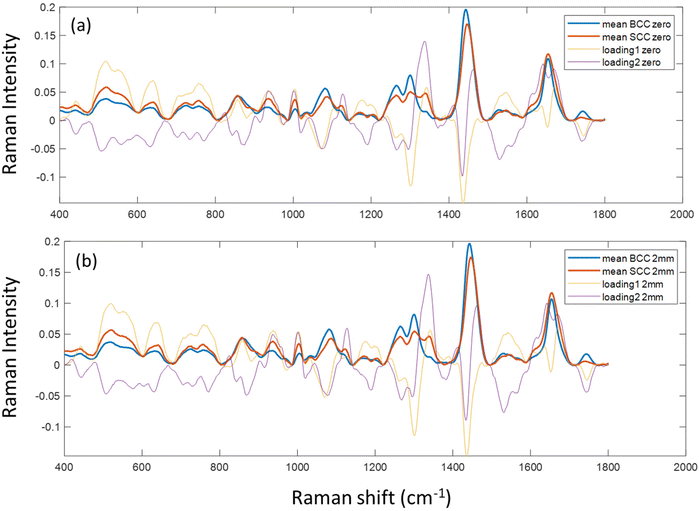 | ||
| Fig. 4 PC1 and 2 loadings of biopsy samples measured with (a) zero and (b) 2 mm SORS, subplotted with the mean Raman spectra of BCC and SCC tissues measured at each mode. | ||
To confirm the physical basis of the PCA separation, mean BCC and SCC spectra were subplotted with loadings 1 and 2 for the 0 (Fig. 4a) and 2 mm (Fig. 4b) offsets. As shown in the PCA scatter plots, certain spectral features separating the two subtypes include bands assigned to phenylalanine (1003 and 1339 cm−1), skeletal (937 cm−1) and amide I (1640 cm−1) collagen vibrations, which are more prominent in the SCC type. Spectral bands moving the opposite way (negative) in PC2 loading (Fig. 4) are assigned to tryptophan (772 cm−1), lipids (1076 cm−1, 1300 cm−1 and 1650 cm−1) and proteins (amide III at 1268 cm−1 and hydroxyproline at 870 cm−1) and are prominent in the BCC group, as previously reported in the literature.26,35,36
Specifically for the spectral bands at 1339 cm−1 and 1076 cm−1, they have been previously assigned to nucleic acids, bases (adenine) and backbone as well as lipids and proteins.12,35 However, as the strongest nucleic acid contribution (785 cm−1) is not discernible in our data and other bands assigned to nucleic acids (i.e. 1531 cm−1) are relatively stable in the processed spectra, we assume that 1076 cm−1 is assigned to lipids (v(C–C), v(C–O)) and 1339 cm−1 to either lipids or phenylalanine. The latter is in line with the trend of the characteristic phenyl ring band at 1003 cm−1, which is also more prominent in the SCC spectra as confirmed by Silveira et al. previously.34 This band can be attributed to phenylalanine, but also to keratin as the phenyl ring structure is abundant in the keratin molecule.37 Furthermore, SCC has been shown to positively correlate with phenyl ring content due to the presence of hyperkeratosis,38 which is in agreement with our findings.
We also noticed a stronger contribution of the 1743 cm−1 band in the BCC spectra. Although this may be due to lipid contribution from the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching mode of the ester groups in the glycerol heads of TAG in the adipose tissue underneath the dermis,24,39 it may also imply an increased amount of melanin due to more pigmented skin in the BCC patient group.36
O stretching mode of the ester groups in the glycerol heads of TAG in the adipose tissue underneath the dermis,24,39 it may also imply an increased amount of melanin due to more pigmented skin in the BCC patient group.36
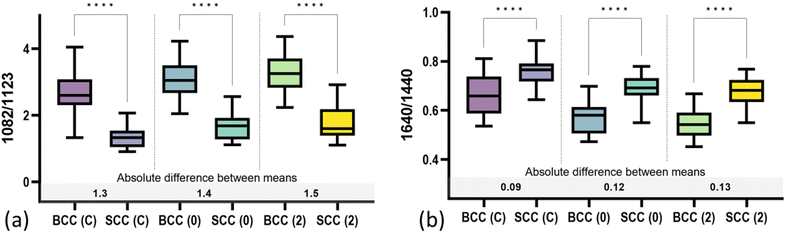
By further studying the lipid contributions in PC2 loadings (Fig. 4), we noticed that spectral features of lipids are included in both the BCC and SCC groups. More specifically, PC2 loadings present negative peaks (characteristic of the BCC group) at 1080 cm−1, 1260–1310 cm−1 and 1659 cm−1 assigned to unsaturated lipids and mainly triolein from adipocytes, as well as positive peaks (characteristic of the SCC group) at 1128 cm−1 and 1283 cm−1 assigned to saturated fatty acids of ceramides from the epidermis and phospholipids sphingomyelin and phosphatidylcholine from the cell membrane.34 Estimation of the ratio of characteristic bands from unsaturated (1080 cm−1) and saturated (1128 cm−1) lipids revealed statistically significant differences between the 2 pathology groups in all measurement modes (Fig. 5a).
A negative correlation between BCC and ceramide has already been reported by Tunnell et al.38 A comparison of the two groups has demonstrated a higher abundance of ceramide in SCC tissue and lower in triolein, although the results were not as statistically strong.
In terms of collagen content, PCA (Fig. 4) indicates an increased contribution of bands at 937 cm−1 and 1640 cm−1, which are assigned to the proline and hydroxyproline backbone and the amide I vibrations of collagen, respectively,40 in the SCC spectra compared with the BCC spectra. The difference is shown to be statistically significant when collagen (1640 cm−1) is normalized to protein content (1339 cm−1) (Fig. 5b). Although the abundance of collagen in non-melanoma cancers is not well-studied yet, the negative correlation of collagen with BCC has been confirmed by a small number of studies supporting the low abundance of connective tissue and therefore collagen in BCC sites.41,42
Multivariate factor analysis (MFA)
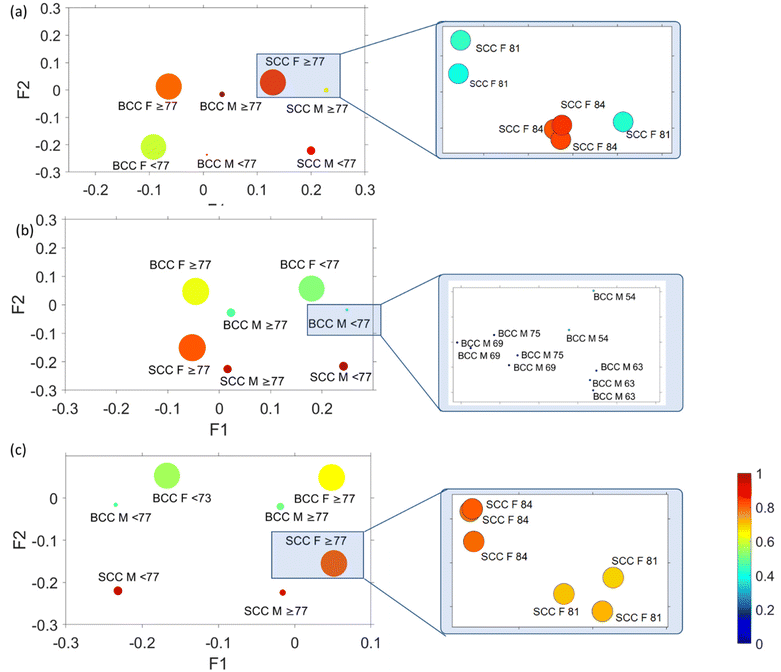
In order to correlate spectral results with age and sex sample parameters, we employed MFA to assess each mode separately. MFA is an extension of PCA and is able to analyse quantitative (Raman data) and qualitative variables (sex, age, and cancer type) on the same patients by projecting them into a high-dimensional space (Fig. 6).In Fig. 6, factor loadings are plotted together for each of the modes, where the size of the circles represents factor 3 and circle colour factor 4. MFA and plotting of factor loading values provide an assessment of the spectral variability between the different pathologies of all patients. In all cases (conventional, 0 mm and 2 mm), we noticed a very strong clustering of variables according to sex (female or male) and age (older or younger than 77 years old). The factor loading values for each distinct group are so similar that an extreme magnification is required to observe the individual variables as they highly overlap. Comparing the data with the corresponding histopathological imaging, we noticed that there is no clustering due to the pathology observed in the MFA scatter plot of the conventional mode (Fig. 6a). In contrast, the variables in the conventional mode seem to rather separate along factor loading 2 according to age. In spatial offset modes (Fig. 6b and c), apart from the extremely tight age and sex clustering, the MFA scatter plots also exhibit loose variable separation along factor 2 loading based on tissue pathology (BCC or SCC). As no separation occurs across factor 3 and factor 4, their scores are not analysed further.
Focusing on the two spatial offset cases (Fig. 6b and c), where clustering of different pathology-groups is observed, we noticed that data separation occurred across factor 2. Factor 2 loadings are plotted in Fig. 7, indicating that the main discriminating features in both cases are 875, 1076, 1268, 1300, 1435, 1745, 2850 and 2892 cm−1 bands, which correlate positively with the BCC subtype and 937, 1003, 1339, 1640, 1671 and 2948 cm−1 bands (characteristic of SCC).
 | ||
| Fig. 7 Factor score 2 spectra obtained from MFA plotted against wavenumber in the case of (a) zero and (b) 2 mm offset modes. | ||
We noticed that the spectral bands in the fingerprint region in factor loading 2, which are responsible for the separation in MFA analysis, are identical to those of PC2 loading, causing BCC–SCC separation in PCA (Fig. 3 and 4). In terms of the high wavenumber region, MFA indicated contributions of lipids and proteins in both pathology groups, which in turn implies the involvement of lipids and proteins in the pathogenesis of both BCC and SCC.
In order to explore the contribution of each factor loading to different measurement modalities, we calculated the averaged factor loading values for all patients (Fig. 8). The factor loading values indicate the strength of the relationship (Pearson correlation) between each modality and the respective factor. Although the factor loadings between different modalities are not directly comparable due to different sampling volumes and measurements, we observe that factor loading 1 (Fig. S2†) is the one that correlates the most with all modalities (although as noted earlier is not responsible for the separation) and decreases slightly with depth. Factor loading 2 (plotted in Fig. 7), which is responsible for pathology discrimination, increases in the 2 mm mode compared to the zero mode.
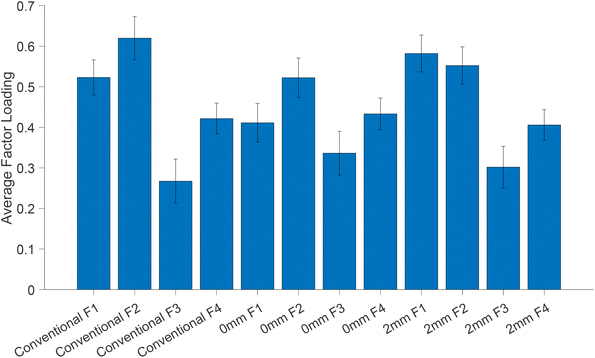 | ||
| Fig. 8 Average factor loadings (normalised and scaled) from MFA. Error bars indicate the standard deviation of the mean. | ||
Conclusions
In this study, we explored the application of SORS technology in non-melanoma cancer diagnosis (BCC and SCC). Our findings highlight spectral separation based on collagen, phenyl ring and lipid vibrations between non-melanoma subtypes. More specifically, we show that bands assigned to protein content (phenylalanine and proline/hydroxyproline in collagen) are more abundant in the SCC type, whereas lipid vibrations are more abundant in the BCC type. A statistically significant difference in unsaturated (triolein) vs. saturated (ceramide) lipid spectral ratio has also been observed in our measurements. The results are in agreement with those of previous studies38 whereas the lack of ceramide in the skin is thought to result in the mutation of basal cells in the epidermis and initiation of BCC growth.43 It should be noted that components such as lipids and water vary significantly between individuals, as well as between anatomical sites of the same individual, and could therefore influence our conclusions.While the utilization of the confocal modality results in a more precise sampling volume and, subsequently, a better discrimination, we propose that a larger (i.e., deeper) measuring volume could potentially mitigate interpatient variability by increased Raman sampling of skin layers. Additionally, this approach could enable the acquisition of biochemical information from deeper skin layers. Similar studies have utilized a defocused beam to look into a 100 μm depth of skin malignancies, demonstrating that classification accuracy decreases with depth,25 and have eventually chosen an extremely shallow depth measurement in subsequent studies.26
Here, by employing multivariate factor analysis, we conclude that despite a strong impact of patient sex and age, differences in biochemical compositions between BCC and SCC as they are reflected on Raman spectra are significant enough to lead to a pathology-based separation. Our preliminary data demonstrate that Raman spectroscopy in a defocused mode (either 0 or 2 mm) is able to overcome the interpatient variability and therefore constitutes a promising approach for non-melanoma screenings in clinics. We believe that the results of our study will put forward the SORS approach, which will pave the way for an automated and non-invasive skin cancer clinical screening prior to histopathological imaging.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research is co-financed by Greece and the European Union (European Social Fund-ESF) through the Operational Programme “Human Resources Development, Education and Lifelong Learning” in the context of the projects: “Strengthening Human Resources Research Potential via Doctorate Research” (MIS-5000432) and “Reinforcement of Postdoctoral Researchers-2nd Cycle” (MIS-5033021), both implemented by the State Scholarships Foundation (IKY).References
- A. Lomas, J. Leonardi-Bee and F. Bath-Hextall, Br. J. Dermatol., 2012, 166, 1069–1080 CrossRef CAS PubMed.
- M. Ciazynska, G. Kaminska-Winciorek, D. Lange, B. Lewandowski, A. Reich, M. Slawinska, M. Pabianek, K. Szczepaniak, A. Hankiewicz, M. Ulanska, J. Morawiec, M. Blasinska-Morawiec, Z. Morawiec, J. Piekarski, D. Nejc, R. Brodowski, A. Zaryczanska, M. Sobjanek, R. J. Nowicki, W. Owczarek, M. Slowinska, K. Wrobel, A. Bieniek, A. Wozniacka, M. Skibinska, J. Narbutt, W. Niemczyk, K. Ciazynski and A. Lesiak, Sci. Rep., 2021, 11, 4337 CrossRef CAS PubMed.
- K. Seretis, V. Thomaidis, A. Karpouzis, D. Tamiolakis and I. Tsamis, Dermatol. Surg., 2010, 36, 15–22 CrossRef PubMed.
- C. Global Burden of Disease Study, Lancet, 2015, 386, 743–800 CrossRef PubMed.
- V. Samarasinghe, V. Madan and J. T. Lear, Expert Rev. Anticancer Ther., 2011, 11, 763–769 CrossRef PubMed.
- J. Chen, I. Ruczinski, T. J. Jorgensen, G. Yenokyan, Y. Yao, R. Alani, N. J. Liegeois, S. C. Hoffman, J. Hoffman-Bolton, P. T. Strickland, K. J. Helzlsouer and A. J. Alberg, J. Natl. Cancer Inst., 2008, 100, 1215–1222 CrossRef PubMed.
- K. Eberhardt, C. Stiebing, C. Matthaus, M. Schmitt and J. Popp, Expert Rev. Mol. Diagn., 2015, 15, 773–787 CrossRef CAS PubMed.
- V. Ahlgrimm-Siess, T. Cao, M. Oliviero, R. Hofmann-Wellenhof, H. S. Rabinovitz and A. Scope, Arch. Dermatol., 2010, 146, 694–695 Search PubMed.
- M. Varaka, M. Z. Vardaki, G. Gaitanis, I. D. Bassukas and N. Kourkoumelis, Appl. Sci., 2022, 12, 8277 CrossRef CAS.
- W. B. Wang, J. H. Zhao, M. Short and H. S. Zeng, J. Biophotonics, 2015, 8, 527–545 CrossRef PubMed.
- M. Gniadecka, H. C. Wulf, N. N. Mortensen, O. F. Nielsen and D. H. Christensen, J. Raman Spectrosc., 1997, 28, 125–129 CrossRef CAS.
- N. Kourkoumelis, I. Balatsoukas, V. Moulia, A. Elka, G. Gaitanis and I. D. Bassukas, Int. J. Mol. Sci., 2015, 16, 14554–14570 CrossRef CAS PubMed.
- S. Devpura, K. N. Barton, S. L. Brown, O. Palyvoda, S. Kalkanis, V. M. Naik, F. Siddiqui, R. Naik and I. J. Chetty, Med. Phys., 2014, 41, 050901 CrossRef PubMed.
- I. P. Santos, R. van Doorn, P. J. Caspers, T. C. B. Schut, E. M. Barroso, T. E. C. Nijsten, V. N. Hegt, S. Koljenovic and G. J. Puppels, Br. J. Cancer, 2018, 119, 1339–1346 CrossRef PubMed.
- M. Z. Vardaki, B. Gardner, N. Stone and P. Matousek, Analyst, 2015, 140, 5112–5119 RSC.
- S. Mosca, P. Dey, M. Salimi, B. Gardner, F. Palombo, N. Stone and P. Matousek, Anal. Chem., 2021, 93, 6755–6762 CrossRef CAS PubMed.
- M. Z. Vardaki, H. G. Schulze, K. Serrano, M. W. Blades, D. V. Devine and R. F. B. Turner, Transfusion, 2021, 61, 2159–2168 CrossRef CAS PubMed.
- M. Z. Vardaki, H. G. Schulze, K. Serrano, M. W. Blades, D. V. Devine and R. F. B. Turner, Spectrochim. Acta, Part A, 2022, 276, 121220 CrossRef CAS PubMed.
- M. Z. Vardaki, D. V. Devine, K. Serrano, N. Simantiris, M. W. Blades, J. M. Piret and R. F. B. Turner, Appl. Spectrosc., 2020, 74, 223–232 CrossRef CAS PubMed.
- M. D. Keller, E. Vargis, N. D. Granja, R. H. Wilson, M. A. Mycek, M. C. Kelley and A. Mahadevan-Jansen, J. Biomed. Opt., 2011, 16, 077006 CrossRef PubMed.
- N. Stone, R. Baker, K. Rogers, A. W. Parker and P. Matousek, Analyst, 2007, 132, 899–905 RSC.
- J. W. Su, Q. Wang, Y. Tian, L. Madden, E. M. L. Teo, D. L. Becker and Q. Liu, Biomed. Opt. Express, 2019, 10, 6114–6128 CrossRef CAS PubMed.
- N. K. Afseth, M. Bloomfield, J. P. Wold and P. Matousek, Appl. Spectrosc., 2014, 68, 255–262 CrossRef CAS PubMed.
- M. Z. Vardaki, K. Seretis, G. Gaitanis, I. D. Bassukas and N. Kourkoumelis, Appl. Sci., 2021, 11, 9498 CrossRef.
- C. A. Lieber, S. K. Majumder, D. Billheimer, D. L. Ellis and A. Mahadevan-Jansen, J. Biomed. Opt., 2008, 13, 024013 CrossRef PubMed.
- C. A. Lieber, S. K. Majumder, D. L. Ellis, D. D. Billheimer and A. Mahadevan-Jansen, Lasers Surg. Med., 2008, 40, 461–467 CrossRef PubMed.
- C. A. Patil, H. Kirshnamoorthi, D. L. Ellis, T. G. van Leeuwen and A. Mahadevan-Jansen, Lasers Surg. Med., 2011, 43, 143–151 CrossRef PubMed.
- J. Schleusener, P. Gluszczynska, C. Reble, I. Gersonde, J. Helfmann, J. W. Fluhr, J. Lademann, J. Rowert-Huber, A. Patzelt and M. C. Meinke, Exp. Dermatol., 2015, 24, 767–772 CrossRef PubMed.
- M. Morhac, J. Kliman, V. Matousek, M. Veselsky and I. Turzo, Nucl. Instrum. Methods Phys. Res., Sect. A, 1997, 401, 113–132 CrossRef CAS.
- B. Escofier and J. Pages, Comput. Stat. Data Anal., 1994, 18, 121–140 CrossRef.
- J. Pages, Rev. Colomb. Estad., 2004, 27, 1–26 Search PubMed.
- R. Jain, D. Calderon, P. R. Kierski, M. J. Schurr, C. J. Czuprynski, C. J. Murphy, J. F. McAnulty and N. L. Abbott, Anal. Chem., 2014, 86, 3764–3772 CrossRef CAS PubMed.
- A. E. Hendrickson and P. O. White, Br. J. Statist. Psychol., 1964, 17, 65–70 CrossRef.
- F. L. Silveira, M. T. T. Pacheco, B. Bodanese, R. A. Zangaro and L. Silveira, Photonic Therapeutics and Diagnostics Ix, 2013, vol. 8565 Search PubMed.
- L. Lim, B. Nichols, M. R. Migden, N. Rajaram, J. S. Reichenberg, M. K. Markey, M. I. Ross and J. W. Tunnell, J. Biomed. Opt., 2014, 19, 117003 CrossRef PubMed.
- J. H. Zhao, H. S. Zeng, S. Kalia and H. Lui, Dermatol. Clin., 2017, 35, 495–504 CrossRef CAS PubMed.
- M. Gniadecka, H. C. Wulf, O. F. Nielsen, D. H. Christensen and J. Hercogova, Photochem. Photobiol., 1997, 66, 418–423 CrossRef CAS PubMed.
- H. T. M. Nguyen, Y. Zhang, A. J. Moy, X. Feng, K. R. Sebastian, J. S. Reichenberg, M. C. Fox, M. K. Markey and J. W. Tunnell, Photonics, 2021, 8, 282 CrossRef CAS.
- P. Meksiarun, B. B. Andriana, H. Matsuyoshi and H. Sato, Sci. Rep., 2016, 6, 37068 CrossRef CAS PubMed.
- A. Nijssen, T. C. B. Schut, F. Heule, P. J. Caspers, D. P. Hayes, M. H. A. Neumann and G. J. Puppels, J. Invest. Dermatol., 2002, 119, 64–69 CrossRef CAS PubMed.
- F. Silver, R. Shah, M. Richard and D. Benedetto, J. Tissue Eng. Regen. Med., 2019, 5, 61–66 Search PubMed.
- X. Feng, M. C. Fox, J. S. Reichenberg, F. C. P. S. Lopes, K. R. Sebastian, M. K. Markey and J. W. Tunnell, Biomed. Opt. Express, 2019, 10, 104–118 CrossRef CAS PubMed.
- M. J. Choi and H. I. Maibach, Am. J. Clin. Dermatol., 2005, 6, 215–223 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00684k |
| This journal is © The Royal Society of Chemistry 2023 |