Open Access Article
Open Access ArticleGlucose sandwich assay based on surface-enhanced Raman spectroscopy†
Tingting
Zhang
a,
Rui
Lu
 *a,
Gongying
Wang
a,
Xiuyun
Sun
a,
Jiansheng
Li
*a,
Gongying
Wang
a,
Xiuyun
Sun
a,
Jiansheng
Li
 a and
Boris
Mizaikoff
a and
Boris
Mizaikoff
 *bc
*bc
aJiangsu Key Laboratory of Chemical Pollution Control and Resources Reuse, School of Environmental and Biological Engineering, Nanjing University of Science and Technology, Nanjing 210094, People's Republic of China. E-mail: rlu@njust.edu.cn
bInstitute of Analytical and Bioanalytical Chemistry, Ulm University, Albert Einstein Allee 11, 89081 Ulm, Germany
cHahn-Schickard, Ulm Sedanstrasse 14, 89077 Ulm, Germany. E-mail: boris.mizaikoff@uni-ulm.de
First published on 17th July 2023
Abstract
A facile and sensitive glucose sandwich assay using surface-enhanced Raman scattering (SERS) has been developed. Glucose was captured by 3-aminopheyonyl boronic acid (APBA) modified Ag nanoparticles decorated onto a polyamide surface. Then, Ag nanoparticles modified with 3-amino-6-ethynylpicolinonitrile (AEPO) and APBA were used as SERS tags. APBA forms specific cis-diol compounds with glucose molecules avoiding interference by other saccharides and biomolecules in urine enabling its selective detection. As the actual Raman reporter, AEPO exhibited a distinctive SERS peak in the Raman silent region, thus increasing the sensitivity of the glucose detection to 10−11 M. Additionally, the developed SERS assay was reusable, and its applicability in artificial urine samples demonstrated future clinical utility confirming the potential of this innovative technology as a diagnostic tool for glucose sensing.
1. Introduction
D-Glucose is both a necessity and a energy source for bodily fluids (such as brain fluid or human blood). Changes in glucose concentrations in physiological or pathological events indicate a biological dysfunction.1–3 Diabetes is a common metabolic disorder that is defined by elevated blood sugar levels.4 It can result in a variety of complications, including perivascular diseases, nephropathy, retinopathy and atherosclerosis.5 Diabetes mellitus has evolved into one of the most serious diseases, posing a global threat to human health. According to the International Diabetes Federation data, 642 million people worldwide will have diabetes by 2040 as a result of an increase in energy-dense diets and an aging population.6 As a consequence, accurate monitoring of blood glucose levels is essential to prevent diabetes-related complications and to aid diabetes treatment. The determination of glucose concentrations in bodily fluids is critical for diabetes mellitus monitoring and make an early clinical diagnosis.7,8 As a result, many sensing methods have been developed to detect glucose with high sensitivity and selectivity, including electrochemical techniques,9–12 colorimetric methods,13 fluorescence sensing,14 surface-enhanced Raman spectroscopy (SERS).15,16 However, most electrochemical techniques rely on glucose enzyme-based sensors, which need sophisticated enzyme fixation and are easily susceptible to environmental influences.17 Additionally, due to the strong interaction between boric acid and diol to form borate esters, fluorescence-based sensors are predominantly composed of boronic acid modified with chromophores. While these sensors are highly sensitive for detecting glucose in bodily fluid samples, they lack a characteristic analyte response.18 Furthermore, directly sensing glucose at the physiological level without interference remains a challenge. A straightforward, enzyme-free assay is urgently required.19 Therefore, it is critical to develop a novel method for glucose detection.SERS has been widely applied in the detection of a broad variety of chemical compounds and biomolecules due to its superior Raman signal enhancement capacity, resulting in the development of extremely sensitive molecular sensors at the micro level.20 Due to the strong surface plasmon resonance of nanostructured surfaces, the Raman signals of molecules on them can be amplified by several orders of magnitude (usually 106–108),5,21–23 therefore improving the inherent low sensitivity of Raman spectroscopy. Many studies have been conducted on the detection of glucose using SERS.24–26 Zhang et al.27 proposed an innovative Ag nanorods@Al2O3 structure SERS substrates capable of detecting glucose at concentrations as low as 0.1 mM using SERS by increasing hot spots and tuning resonance between Ag nanopillars and Ag nanorods. However, glucose naturally emits weak Raman signals due to its small Raman scattering cross-section, measuring only 5.6 × 10−30 cm2 per molecule per steradian, which is approximately 14 orders of magnitude smaller than its fluorescence, and glucose's Raman emission is easily obscured by strong background noises from the surrounding environment, limiting glucose detection sensitivity and selectivity. At present, Raman-label detection is the most frequently employed approach for identifying biological molecular targets. In comparison to direct detection (label-free), Raman reporters with larger Raman scattering cross sections can generate strong distinctive SERS signals, improving glucose selectivity and enabling the measurement of receptor/glucose response signal variations.28
To overcome this limitation, one method is to utilize carbohydrate recognition molecules, such as boronic acid,26 to capture glucose molecules onto the nanostructured surface. This increases the concentration of captured glucose, hence improving the detection sensitivity. Boronic acid and its derivatives have been demonstrated to selectively form trans–cis-diol complexes with glucose molecules in a selective manner. The quantitative analysis of glucose was performed by SERS functionalized with a variety of boronic acid derivatives.4,29–31 For example, to monitor glucose with high specificity in complicated aqueous media, Li's group22 investigated a conceptually novel “selective capture and controllable detection” nanoreactor. To form stable boronic acid-diol complexes, syn-periplanar hydroxyl groups should be present to allow saccharides in the α-furanose form to attach preferentially. There are two separate sets of syn-periplanar diol groups present in α-D-glucofuranose. Thus, glucose can form covalent bands in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio with two boronic acids. There is substantial evidence that cyclic borates formed between glucose and boronic acid enable us to perceive glucose more selectively than other saccharides. Malini Olivo et al.29 quantify the concentration of glucose by utilizing its capacity to form a bidentate glucose-boronic complex. This technology was further improved by substituting an alkyne-functionalized boronic acid for the boronic acid, which offers a novel glucose binding mechanism on a highly sensitive SERS substrate, overcoming obstacles associated with detecting specific glucose in bio-fluids.32–34 However, the SERS spectra of the glucose molecule and functional groups of the carbohydrate recognition molecules, such as the boronate (1370 cm−1) and aromatic (1580 cm−1) group in phenylboronic acid, lie in the 400–1800 cm−1 region, which is susceptible to interference from inherent biomolecules’ Raman signal.23,32,35 As a result, constructing a simple, reliable, and highly sensitive SERS-based glucose sensor remains a challenge.
2 ratio with two boronic acids. There is substantial evidence that cyclic borates formed between glucose and boronic acid enable us to perceive glucose more selectively than other saccharides. Malini Olivo et al.29 quantify the concentration of glucose by utilizing its capacity to form a bidentate glucose-boronic complex. This technology was further improved by substituting an alkyne-functionalized boronic acid for the boronic acid, which offers a novel glucose binding mechanism on a highly sensitive SERS substrate, overcoming obstacles associated with detecting specific glucose in bio-fluids.32–34 However, the SERS spectra of the glucose molecule and functional groups of the carbohydrate recognition molecules, such as the boronate (1370 cm−1) and aromatic (1580 cm−1) group in phenylboronic acid, lie in the 400–1800 cm−1 region, which is susceptible to interference from inherent biomolecules’ Raman signal.23,32,35 As a result, constructing a simple, reliable, and highly sensitive SERS-based glucose sensor remains a challenge.
In order to further improve the selectivity, we presented a simple and sensitive method for glucose sandwich detection (Scheme 1) that captured glucose via 3-aminopheyonyl boronic acid (APBA) modified on polyamide-Ag. SERS tags were added to independent Ag nanoparticles utilizing 3-amino-6-ethynylpicolinonitrile (AEPO) and APBA. Ag on polyamide film is modified with boronic acid in order to selectively capture compounds with a cis-diol structure. On the one hand, as a Raman reporter, AEPO exhibits a unique Raman peak at 1977 cm−1 in a biologically silent region (1800–2800 cm−1) where the majority of endogenous molecules, including glucose, do not exhibit Raman scattering, resulting in a higher sensitivity than other SERS glucose sensing methods.36,37 On the other hand, the Raman reporter exhibits exceptional Raman scattering, which significantly improves the sensitivity of glucose determination. Meanwhile, simple C≡C compounds were used instead of complex compounds, simplifying the detection conditions. Additionally, Ag loaded on polyamide has a high degree of uniformity, which eliminates the issues associated with Ag nanoparticles aggregating in solution and the hot spot being non-uniform. This straightforward, sensitive and selective glucose sandwich assay based on SERS has the potential to be evolved into a glucose diagnostic tool. More importantly, the Raman peak location can be adjusted by changing the marker molecule, hence facilitating glucose detection.
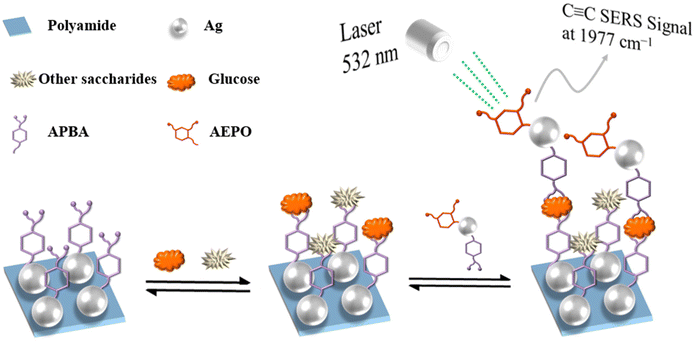 | ||
| Scheme 1 Schematic illustration of selective glucose detection mechanism via the developed sandwich SERS assay. | ||
2. Experimental section
2.1. Materials
Hydrochloric acid (HCl), sodium hydroxide (NaOH), polyamide 6 powder, ammonium hydroxide, ethanol, urea, potassium chloride (KCl), sodium chloride (NaCl), sodium sulfate (Na2SO4), ammonium chloride (NH4Cl), potassium dihydrogen phosphate (KH2PO4), calcium chloride dihydrate (CaCl2·2H2O), creatinine, ovalbumin, sodium borohydride (NaBH4), silver nitrate (AgNO3), formic acid, albumin from human serum, hemoglobin from bovine and trisodium citrate dihydrate were obtained from Sinopharm Chemical Reagent Co., Ltd. (3-Aminopheyonyl) boronic acid (APBA) and 3-Amino-6-ethynylpicolinonitrile (AEPO) were acquired from Bide Pharmatech Ltd. Galactose, ribose, arabinose, rhodamine 6G (R6G) and polyvinyl pyrrolidone K30 (PVP) were purchased from Sigma-Aldrich. Maltose, fructose, rhamnose and D-glucose were bought from Shanghai Macklin Biochemical Co., Ltd. All solutions were prepared in Milli-Q ultrapure water and all chemicals were used without further purification.2.2. Instrumentation
A Quanta 250 FEG microscope (Thermo Fisher Scientific, USA) was used to perform field emission scanning electron microscopy (FE-SEM) and energy dispersive spectrometry (EDS) measurements on the samples. The contact angles were measured by DSA30 (Krüss, Hamburg, Germany). X-ray photoelectron spectroscopy (XPS) measurements were conducted using a Thermo ESCALAB 250.2.3. Synthesis of 30 nm Ag NPs
First, 4 nm Ag seed solution was synthesized according to previous reference. Malini Olivo et al.29 quantify the concentration of glucose by utilizing its capacity to form a bidentate glucose-boronic complex. This technology was further improved by substituting an alkyne-functionalized boronic acid for the boronic acid, which offers a novel glucose binding mechanism on a highly sensitive SERS substrate, overcoming obstacles associated with detecting specific glucose in bio-fluids.38 A citrate solution of 0.2 g was dissolved in 95 mL of deionized water and incubated at 70 °C for 15 minutes. After that,1.7 mL of AgNO3 (1 wt%) was added, followed by 2 mL of freshly prepared NaBH4 (1 wt%) aqueous solution, which was quickly added and constantly stirred at 70 °C for 1 hour. After cooled to room temperature, deionized water was added to bring the dispersion volume to 100 mL. For the preparation of 30 nm Ag NPs, 2 mL of citrate solution was added to 75 mL of deionized water and heated for 15 minutes at 130 °C. The solution was then vigorously stirred for 1 hour at 125 °C with 10 mL of the prepared 4 nm seed solution and 1.7 mL of fresh aqueous AgNO3. The solution was then added 2 mL of citrate solution and 1.7 mL of AgNO3, stirred for 1 hour, and then repeated. Finally, it was cooled to ambient temperature before adding deionized water to make the dispersion volume 100 mL. The concentration of synthesized nanoparticles was around 4.75 × 10−9 M.392.4. Fabrication of an ordered polyamide-Ag flexible thin film
The pH of Ag nanoparticles solutions was adjusted to pH 6.0 using 0.1 M HCl. The polyamide flexible thin films were immersed in a prepared Ag nanoparticles solution for 18 hours to complete the self-assembly process. Following that, the polyamide-Ag flexible thin film was immersed in 0.l M PVP for 6 hours to ensure that Ag remained stable on the film substrate and to avoid non-specific adsorption. They were cleaned and cut into a specific size for the next experiments.2.5. Modified Ag nanoparticles with APBA/AEPO
To fabricate a modified probe, 100 μL of 0.1 mM APBA and 100 μL of 0.1 mM AEPO were added to 9.8 mL of 30 nm-Ag nanoparticles solution, and the mixed solutions were allowed to stir for 6 hours at room temperature before aging for 12 hours without stirring in dark. Finally, the unbound APBA and AEPO molecules were removed from the APBA/AEPO-modified Ag nanoparticles by centrifugation at 6000 rpm for 3 minutes and rinsed with deionized water twice.2.6. SERS measurements
Rhodamine 6G (R6G) was chosen as the probe molecule. To evaluate the SERS performance of manufactured polyamide-Ag flexible thin film, a series of R6G solutions (10−9 M) have been used. The polyamide-Ag flexible thin films were immersed in 3 mL of aqueous R6G solutions at various concentrations for R6G detection. The SERS substrates were removed six hours later and dried with high purity nitrogen.The glucose concentration was determined using the same protocol. To complete the adsorption process, the flexible thin film was immersed in glucose solution for six hours and then rinsed with ultrapure water to remove unbound glucose molecules. Subsequently, the flexible thin film was then immersed in a solution of APBA/AEPO-modified Ag nanoparticles that had been pre-adjusted to a pH of 8.50 by 0.1 M HCl or NaOH. Six hours later, the substrate was rinsed with water to remove any residual Ag nanoparticles, and they were detected using a Raman spectrometer. For comparison, the same procedures were used for other saccharides.
Raman spectra were acquired using a confocal Raman spectrometer (Aramis, Horiba, Japan) excited with a 532 nm Ar-ion laser excitation at a power level of 0.54 mW at the excitation port. The laser beam was precisely focused using a 50× long-focus objective, and the laser spot was 1 μm, the data acquisition time for glucose and R6G was 10 seconds. The error bar is calculated from the measurement data collected from six different randomly chosen points on the target substrates.
2.7. Reusability
Put the thin film of the synthesized sandwich structure into an aqueous solution with a weak acidic pH (pH = 5) and incubated at room temperature for 6 h to relieve the binding of glucose and APBA on the film. The remaining polyamide-Ag-APBA would be washed and reused.3. Results and discussion
3.1. Characterization of polyamide-Ag flexible thin film
Polyamide-Ag and polyamide-Ag-APBA film were synthesized and characterized by SEM, as shown in Fig. S1.† A uniform dispersion of nanoparticles with a diameter of around 30 nm was observed, and the Ag morphology remained unchanged after APBA functionalization. Then, X-ray energy dispersive spectroscopy (EDS) was used to analyze the components of polyamide-Ag and polyamide-Ag-APBA film, as illustrated in Fig. S2.† A uniform distribution of boron elements was observed in polyamide-Ag-APBA, indicating that the APBA was successful in bonding with Ag nanoparticles. By comparison, it was determined that the number of Ag nanoparticles increases significantly after the sandwich structure is formed, which is due to the interaction between Ag-APBA/AEPO and glucose. The polyamide-Ag-APBA was further analyzed by XPS (Fig. S3†). According to the survey scan spectrum of polyamide-Ag-APBA, there are distinct peaks of C 1s, O 1s, B 1s, and Ag 3d evident further demonstrating that APBA is successfully bonded to the Ag nanoparticles surface. The contact angles reveal that polyamide-Ag, polyamide-Ag-APBA, and polyamide-Ag-APBA-PVP have a high degree of hydrophilicity (Fig. S4†). To summarize, the polyamide-Ag-APBA preparation was indeed successful.3.2. Self-assembled polyamide-Ag-APBA film and Ag NP-based SERS tags
APBA was self-assembled on a polyamide-Ag thin film. Strong SERS signals at 698, 787, 996, 1019, 1070, and 1552 cm−1 were observed, which were attributed to the characteristic vibrational modes of APBA (Fig. S5†). The bands at 698, 787, 996, 1019, 1070, 1552 cm−1 are attributed to C–C ring bending, C–H out-of-plane bending, C–C in-plane bending, C–H in-plane bending, C–C in-plane bending coupled with non-totally symmetric ring stretching, and totally symmetric ring stretching, respectively.Fig. S6† shows the SERS spectra of Ag nanoparticles modified with APBA, AEPO, and their combinations. The SERS spectral characteristics of the APBA-modified Ag nanoparticles were virtually identical to those of the polyamide-Ag-APBA film. The SERS peaks for AEPO-modified Ag nanoparticles were rather strong at 1468, 1384, 1156, 1226, 1576 and 1977 cm−1. The Raman shifts at 1468, 1384 and 1156, 1226 cm−1 are corresponded to the N=C and N–C stretching vibrations of AEPO.30 The Raman shifts at 1070 and 1977 cm−1 correspond to the vibrations of AEPO's totally symmetric ring stretching and alkynyl stretching, respectively. To prepare the SERS tags, which produce remarkably intense Raman signals, as well as the second carbohydrate reporters, Ag nanoparticles were modified with AEPO and APBA mixture at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Ag-AEPO/APBA) then were subsequently applied to the sample.
1 (Ag-AEPO/APBA) then were subsequently applied to the sample.
3.3. SERS detection of glucose sandwich assay
After incubation with various concentrations of aqueous glucose solutions, the polyamide-Ag-APBA film was washed to remove unbound glucose. Through the formation of a cyclic borate between the boronic acid of APBA and one set of syn-periplanar diol groups of glucose, the boronic acid of APBA captured glucose in the polyamide-Ag-APBA film. After additionally incubation with the colloidal solution of Ag/AEPO-APBA, the glucose-captured polyamide-Ag-APBA film was washed to eliminate unbound SERS tags. Ultimately, the polyamide-Ag-APBA film captured the SERS tags via the glucose sandwich structure between them, resulting in the formation of dual cyclic borates. Raman signals obtained with SERS tags were predominantly from AEPO, and the intensity of peak at 1977 cm−1 rose with glucose concentration increased.According to the results shown in Fig. S7,† the limit of detection (LOD) for histamine was estimated to be 8.34 × 10−12 M (S/N = 3),40,41 with each characteristic peak of the probe molecule being identified unambiguously. Simultaneously, 10−6–10−10 M glucose was prepared to establish a linear relationship, and the correlation coefficient was 0.9886, demonstrating that glucose concentration can be detected accurately by our substrate.
Glucose monitoring for diabetes is generally performed using urine or blood samples; the former is not a substitute for blood glucose monitoring but rather a supplement that can provide extremely valuable information in situations where blood glucose monitoring is not accessible, affordable, or desirable. Due to the complexity of real urine, we formulated artificial urine spiked with glucose, based on previous literature with slight modifications, to evaluate the excellent SERS performance of the polyamide-Ag hybrid substrate. The composition of artificial urine is shown in Table S1.†
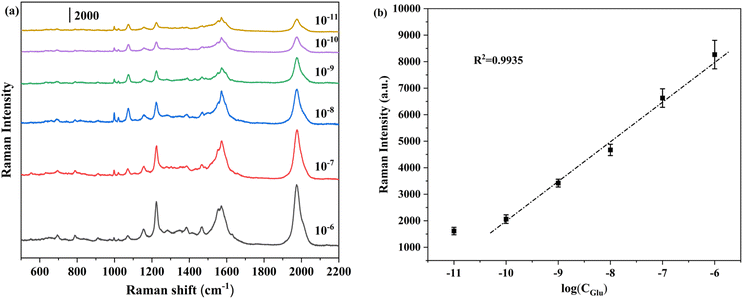
Fig. 1 illustrate the relationship between various glucose concentrations and peak intensity of probe molecule in artificial urine. The LOD was estimated to be 7.62 × 10−12 M (S/N = 3), and each characteristic peak of probe molecule can be easily identified. Simultaneously, glucose of 10−6–10−10 M was used to establish a linear relationship, and the correlation coefficient value was 0.9935, confirming that our substrate could still capable of accurately determining glucose in actual samples. When glucose was added at a concentration of 10−6–10−10 M, the peak intensity of probe molecules was not different from that of in aqueous solution. This means that the real sample has no effect on the Raman intensity of the probe within a certain concentration range. The comparison of specific data was shown in Table S2.†
3.4. Selectivity of glucose assay
For confirming the specific glucose detection properties of this strategy, a mixture of nine different biomolecules including monosaccharides (glucose, fructose, galactose, ribose, rhamnose, arabinose, and maltose) and proteins (albumin, and hemoglobin) has been added simultaneously to the solution. In the following, glucose was analysed via the sandwich detection strategy based on SERS in the presence of these additional biomolecules (10−6 M of each analyte). For comparison, the SERS peak intensity at 1977 cm−1 (in the presence of other biomolecules) is shown in Fig. S8.† The results indicate that the SERS signal at 1977 cm−1 was significantly enhanced in the presence of glucose, whereas the signatures for other biomolecules remained at the noise level. Other monosaccharides and proteins exhibited only modest SERS signals, which is attributed to their reduced tendency to form dual cyclic borates and an associated sandwich structure. Although certain saccharides have a higher affinity for monoboronic acids and can form 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes with them, glucose can form 1
1 complexes with them, glucose can form 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complexes with diboronic acids. That is, whereas all of those saccharides interacted with the self-assembled APBA monolayers, Ag/AEPO-APBA bonded preferentially to the surface-captured glucose through another set of syn-periplanar diol groups. These results indicate that the probe performs well as a specific material for glucose detection and has a good selectivity for glucose detection.
2 complexes with diboronic acids. That is, whereas all of those saccharides interacted with the self-assembled APBA monolayers, Ag/AEPO-APBA bonded preferentially to the surface-captured glucose through another set of syn-periplanar diol groups. These results indicate that the probe performs well as a specific material for glucose detection and has a good selectivity for glucose detection.
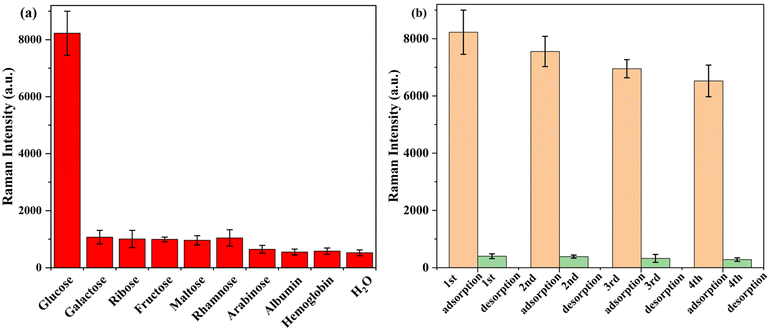
The SERS intensity of glucose and other biomolecules (10−6 M) in artificial urine at 1977 cm−1 is depicted in Fig. 2(a). The results indicate that when glucose is present, the SERS signal at 1977 cm−1 is significantly enhanced, while other biomolecules are present, the peak intensity is at the noise level. These results demonstrate that the probe remains extremely selective for the detection of glucose in real samples.
This agreement of all data reveals that the devised method was effective at avoiding interference from other biomolecules. Therefore, this facile and sensitive glucose sandwich assay based on SERS that does not require the use of enzymes has the potential to be developed into a diagnostic tool for glucose level assessment.
3.5. Stability and SERS performance of polyamide-Ag hybrid substrate
To evaluate the stability of the polyamide-Ag, the same batch of SERS substrate was manufactured for storage and subsequent detection, and the SERS performance was evaluated after three weeks of room temperature storage. As illustrated in Fig. S9,† Raman spectra of 10−9 M R6G were collected, and an average of at least six spots from each base was randomly selected. After several weeks of storage, each characteristic band of 10−9 M R6G was clearly visible and the Raman signal was relatively consistent. The stability of SERS substrate was assessed according to the intensity of characteristic peak at 609 cm−1, as shown in Fig. S9(b).† After three weeks of long-term storage, the substrate's SERS performance remained rather stable, and Raman signal intensity did not exhibit a noticeable declining trend.For evaluating the reproducibility of the substrate, Raman spectra were randomly selected for 10−9 M R6G from 30 different locations at the polyamide-Ag substrate. As illustrated in Fig. 3, the intensities of the characteristic R6G Raman peaks were relatively consistent at different locations. An evaluation of the characteristic 609 cm−1 spectral band was performed, and the relative standard deviation (RSD) of the Raman intensity at 30 randomly selected sites at the same substrate was 8.73%, which indicates that the fabricated SERS substrate was sufficiently reproducible. According to equation (S1†), the enhancement factor (EF) was 3.95 × 105 (details of the calculation are provided in Table S3†). As shown in Fig. S10,† the intensity of AEPO Raman characteristic peaks measured at various locations were relatively consistent. The characteristic spectral bands of 1977 cm−1 were evaluated, and the RSD of Raman intensity of 20 randomly selected sites on the same substrate was 5.32%. These advantages are a result of the preparation strategy we designed. Firstly, a polyamide polymer film was employed to transport Ag nanoparticles in order to avoid stacking or aggregation caused by the disordered self-assembly of Ag nanoparticles during the reaction process. Secondly, the long-chain molecular structure of polyamide also contributes to the monolayer distribution of seeds on the surface of nanorods, hence limiting variation in the distribution hot spots over the substrate.
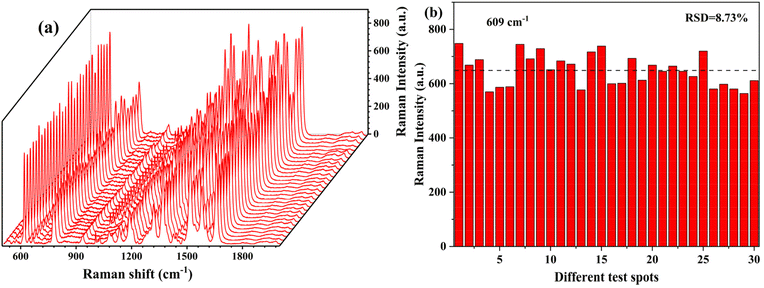 | ||
| Fig. 3 (a) SERS spectra of and (b) peak intensities at 609 cm−1 of 10−9 M R6G obtained from 30 random points on the same substrate. | ||
3.6. Reusability
Further investigation was conducted on the reusability of manufactured polyamide-Ag, and the results are illustrated in Fig. 2(b). The peak intensity decreased slightly (20%) after four cycles. These results proved the stability and reusability of the synthesized polyamide-Ag during the detection process.3.7. Comparison with other adsorbents
In addition, Table S4† compares the developed method to previously reported SERS methods for glucose detection (in ESI†). The data indicated that the LOD of this study was superior to that of other reported Raman methods. Most importantly, the method is unique in its identification of the cis-diol component due to the introduction of the boronic acid compound. Furthermore, glucose can be recovered by adjusting the pH without altering other variables or adding additional substances. On the other hand, simple C≡C compounds were used instead of complex compounds, which simplifying the detection conditions.4. Conclusions
We demonstrated the utility, specificity and sensitivity of SERS-based glucose sandwich assays by uniquely utilizing polyamide-Ag-APBA films and SERS tags at Ag nanoparticles modified with AEPO and APBA. This straightforward sandwich assay demonstrates superior selectivity for glucose vs. other saccharides. AEPO at the Ag nanoparticles surface during the SERS measurements exhibited significantly stronger characteristic SERS peaks that were distinct from those of APBA, and therefore served as the actual Raman reporter. The Raman reporter significantly increased the sensitivity of the glucose detection while eliminating interferences from other similar biomolecules. Simultaneously, the polyamide-Ag-APBA film exhibits excellent stability, reproducibility, and reusability. After three weeks of long-term storage, the SERS performance of the substrate remained constant, and the Raman signal intensity did not exhibit a discernible decline. The relative standard deviation of the Raman intensity at 30 random loci using the same base was 8.73%. After four cycles, the peak intensity of the Raman reporter decreased somewhat at approx. 20%. In summary, this simple, selective, and sensitive non-enzymatic SERS-based glucose sandwich assay has the potential to be evolved into a diagnostic tool for detecting relevant glucose concentrations in biofluids.Author contributions
Tingting Zhang: writing – original draft, experiment, formal analysis. Rui Lu: conceptualization, funding acquisition, writing – review & editing, supervision. Gongying Wang: experiment, data curation. Xiuyun Sun: investigation. Jiansheng Li: conceptualization, project administration. Boris Mizaikoff: writing – review & editing, supervision.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
The authors wish to thank the Natural Science Foundation of Jiangsu Province (BK20191294), the Fundamental Research Funds for the Central Universities (No. 30922010603), the open fund of Information Materials and Intelligent Sensing Laboratory of Anhui Province (IMIS202109), State Key Laboratory of Applied Microbiology Southern China (Grant No. SKLAM012-2021) and Anhui Provincial Key Laboratory of Environmental Pollution Control and Resource Reuse (2022EPC04) for the partial support of this work.References
- X. Sun and T. D. James, Chem. Rev., 2015, 115, 8001–8037 CrossRef CAS PubMed.
- M. Wei, Y. Qiao, H. Zhao, J. Liang, T. Li, Y. Luo, S. Lu, X. Shi, W. Lu and X. Sun, Chem. Commun., 2020, 56, 14553–14569 RSC.
- J. C. Pickup, F. Hussain, N. D. Evans, O. J. Rolinski and D. J. Birch, Biosens. Bioelectron., 2005, 20, 2555–2565 CrossRef CAS PubMed.
- D. Yang, S. Afroosheh, J. O. Lee, H. Cho, S. Kumar, R. H. Siddique, V. Narasimhan, Y. Z. Yoon, A. T. Zayak and H. Choo, Anal. Chem., 2018, 90, 14269–14278 CrossRef CAS PubMed.
- A. Ahmed and R. Gordon, Nano Lett., 2012, 12, 2625–2630 CrossRef CAS PubMed.
- K. Ogurtsova, J. D. da Rocha Fernandes, Y. Huang, U. Linnenkamp, L. Guariguata, N. H. Cho, D. Cavan, J. E. Shaw and L. E. Makaroff, Diabetes Res. Clin. Pract., 2017, 128, 40–50 CrossRef CAS PubMed.
- J. Qiao, H. Wu, H. Wei, L. Mao, T. Wang and L. Qi, Anal. Chem., 2020, 92, 4445–4450 CrossRef CAS PubMed.
- T. Maric, G. Mikhaylov, P. Khodakivskyi, A. Bazhin, R. Sinisi, N. Bonhoure, A. Yevtodiyenko, A. Jones, V. Muhunthan, G. Abdelhady, D. Shackelford and E. Goun, Nat. Methods, 2019, 16, 526–532 CrossRef CAS PubMed.
- C. Deng, J. Chen, X. Chen, C. Xiao, L. Nie and S. Yao, Biosens. Bioelectron., 2008, 23, 1272–1277 CrossRef CAS PubMed.
- R. Ahmad, M. Khan, N. Tripathy, M. I. R. Khan and A. Khosla, J. Electrochem. Soc., 2020, 167, 107504 CrossRef CAS.
- H. Lee, Y. J. Hong, S. Baik, T. Hyeon and D. H. Kim, Adv. Healthcare Mater., 2018, 7, 1701150 CrossRef PubMed.
- D. W. Hwang, S. Lee, M. Seo and T. D. Chung, Anal. Chim. Acta, 2018, 1033, 1–34 CrossRef CAS PubMed.
- N. Lu, M. Zhang, L. Ding, J. Zheng, C. Zeng, Y. Wen, G. Liu, A. Aldalbahi, J. Shi, S. Song, X. Zuo and L. Wang, Nanoscale, 2017, 9, 4508–4515 RSC.
- A. L. Hu, Y. H. Liu, H. H. Deng, G. L. Hong, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Biosens. Bioelectron., 2014, 61, 374–378 CrossRef CAS PubMed.
- X. Gu, H. Wang, Z. D. Schultz and J. P. Camden, Anal. Chem., 2016, 88, 7191–7197 CrossRef CAS PubMed.
- W.-C. Lee, E. H. Koh, D.-H. Kim, S.-G. Park and H. S. Jung, Sens. Actuators, B, 2021, 344, 130297 CrossRef CAS.
- J. Axthelm, S. H. C. Askes, M. Elstner, G. Upendar Reddy, H. Gorls, P. Bellstedt and A. Schiller, J. Am. Chem. Soc., 2017, 139, 11413–11420 CrossRef CAS PubMed.
- J. Axthelm, H. Gorls, U. S. Schubert and A. Schiller, J. Am. Chem. Soc., 2015, 137, 15402–15405 CrossRef CAS PubMed.
- W. Zhai, L. Male and J. S. Fossey, Chem. Commun., 2017, 53, 2218–2221 RSC.
- A. Campion and P. Kambhampati, Chem. Soc. Rev., 1998, 27, 241–250 RSC.
- D. Wang, W. Zhu, M. D. Best, J. P. Camden and K. B. Crozier, Nano Lett., 2013, 13, 2194–2198 CrossRef CAS PubMed.
- Z. Y. Li and Y. Xia, Nano Lett., 2010, 10, 243–249 CrossRef CAS PubMed.
- K. A. Willets and R. P. Van Duyne, Annu. Rev. Phys. Chem., 2007, 58, 267–297 CrossRef CAS PubMed.
- K. E. Shafer-Peltier, C. L. Haynes, M. R. Glucksberg and R. P. Van Duyne, J. Am. Chem. Soc., 2003, 125, 588–593 CrossRef CAS PubMed.
- X. Sun, Anal. Chim. Acta, 2022, 1206, 339226 CrossRef CAS PubMed.
- K. V. Kong, Z. Lam, W. K. Lau, W. K. Leong and M. Olivo, J. Am. Chem. Soc., 2013, 135, 18028–18031 CrossRef CAS PubMed.
- Y. Kang, X. Xue, W. Wang, Y. Fan, W. Li, T. Ma, F. Zhao and Z. Zhang, J. Phys. Chem. C, 2020, 124, 21054–21062 CrossRef CAS.
- X. Teng, F. Chen, Y. Gao, R. Meng, Y. Wu, F. Wang, Y. Ying, X. Liu, X. Guo, Y. Sun, P. Lin, Y. Wen and H. Yang, Anal. Chem., 2020, 92, 3332–3339 CrossRef CAS PubMed.
- K. V. Kong, C. J. Ho, T. Gong, W. K. Lau and M. Olivo, Biosens. Bioelectron., 2014, 56, 186–191 CrossRef CAS PubMed.
- D. Sun, X. Liu, S. Xu, Y. Tian, W. Xu and Y. Tao, Chem. Res. Chin. Univ., 2018, 34, 899–904 CrossRef CAS.
- B. Sharma, P. Bugga, L. R. Madison, A. I. Henry, M. G. Blaber, N. G. Greeneltch, N. Chiang, M. Mrksich, G. C. Schatz and R. P. Van Duyne, J. Am. Chem. Soc., 2016, 138, 13952–13959 CrossRef CAS PubMed.
- N. Piergies, E. Proniewicz, Y. Ozaki, Y. Kim and L. M. Proniewicz, J. Phys. Chem. A, 2013, 117, 5693–5705 CrossRef CAS PubMed.
- J. Ye, Y. Chen and Z. Liu, Angew. Chem., Int. Ed., 2014, 53, 10386–10389 CrossRef CAS PubMed.
- X. Bi, X. Du, J. Jiang and X. Huang, Anal. Chem., 2015, 87, 2016–2021 CrossRef CAS PubMed.
- D. N. Reinemann, A. M. Wright, J. D. Wolfe, G. S. Tschumper and N. I. Hammer, J. Phys. Chem. A, 2011, 115, 6426–6431 CrossRef CAS PubMed.
- D. Liang, Q. Jin, N. Yan, J. Feng, J. Wang and X. Tang, Adv. Biosyst., 2018, 2, 1800100 CrossRef.
- J. Wu, D. Liang, Q. Jin, J. Liu, M. Zheng, X. Duan and X. Tang, Chemistry, 2015, 21, 12914–12918 CrossRef CAS PubMed.
- Y. Li, R. Lu, J. Shen, W. Han, X. Sun, J. Li and L. Wang, Analyst, 2017, 142, 4756–4764 RSC.
- L. Mandrile, I. Cagnasso, L. Berta, A. M. Giovannozzi, M. Petrozziello, F. Pellegrino, A. Asproudi, F. Durbiano and A. M. Rossi, Food Chem., 2020, 326, 127009 CrossRef CAS PubMed.
- W. Parys and A. Pyka-Pająk, Processes, 2022, 10, 971 CrossRef CAS.
- T. Zhang, X. Xin, A. Li, T. Xu, L. Li, C. Liu, W. Li, J. Li, Y. Li and R. Lu, Analyst, 2022, 147, 4026–4039 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00481c |
| This journal is © The Royal Society of Chemistry 2023 |