Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Classification of formalin-fixed bladder cancer cells with laser tweezer Raman spectroscopy†
Nga Tsing
Tang‡
 ab,
Richard
Robinson
cd,
Richard D.
Snook
ab,
Mick
Brown
ab,
Richard
Robinson
cd,
Richard D.
Snook
ab,
Mick
Brown
 c,
Noel
Clarke
c,
Noel
Clarke
 cde and
Peter
Gardner
cde and
Peter
Gardner
 *ab
*ab
aDepartment of Chemical Engineering and Analytical Science, School of Engineering, University of Manchester, Manchester, M13 9PL, UK. E-mail: peter.gardner@manchester.ac.uk
bManchester Institute of Biotechnology, University of Manchester, 131 Princess Street, Manchester, M1 7DN, UK
cDivision of Cancer Sciences, University of Manchester, Manchester, M20 4GJ, UK
dDepartment of Urology, Salford Royal NHS Foundation Trust, Salford, M6 8HD, UK
eDepartment of Surgery, The Christie NHS Foundation Trust, Manchester, M20 4BX, UK
First published on 24th July 2023
Abstract
Bladder cancer is a common cancer that is relatively hard to detect at an early stage because of its non-obvious symptoms. It is known that bladder cells can be found in urine samples which potentially could be used for early detection of bladder cancer. Raman spectroscopy is a powerful non-invasive tool for accessing biochemical information of cells. Combined with laser tweezers, to allow isolation of single cells, Raman spectroscopy has been used to characterise a number of bladder cells that might be found in a urine sample. Using principal component-canonical variates analysis (PC-CVA) and k-fold validation, the results shows that the invasive bladder cancer cells can be identified with accuracy greater than 87%. This demonstrates the potential of developing an early detection method that identifies the invasive bladder cancer cells in urine samples.
1. Introduction
Bladder cancer accounts for 3% of all cancer incidence and 3% mortality in the UK, with a higher incidence in men,1 similar to the global figures of 3% and 2.1% respectively.2Bladder cancer is usually hard to detect at early stage as symptoms are not usually obvious or specific. However, 80% of bladder cancer patients will present with Haematuria, blood in the urine,3 which should be followed up by renal ultrasound, CT Urogram and/or cystoscopy. Whilst imaging has good sensitivity and specificity for diagnosis (CT Urography sensitivity of 79–93% and specificity of 83–99%), confirmation of bladder cancer requires invasive cystoscopy visualisation of the bladder wall and pathological validation via a transurethral resection of bladder tumour (TURBT).4,5 Currently there are no non-invasive urinary bladder cancer diagnostic biomarkers with high and stable sensitivity available in clinical practice.6 The gold standard for diagnosis of bladder cancer is reported to be cystoscopy, which is an invasive method that often needs to take the risks of infection, pain, and haematuria into account. Although cytology provides a non-invasive alternative diagnostic method, it also has limitations such as low sensitivity, and has high reliance on the analysis by pathologists.7 It has also been reported that immunocytological and/or cytological approaches were not be able to be used for the analysis due to insufficient number of cells.8 Fluorescence-Activated Cell Sorting (FACS) is another alternative. However, previous knowledge is required for targeting specific biomarkers for different types of cells and there are only four types of cytokeratins tested as potential urinary biomarkers.9 Therefore, it would be very helpful if a non-invasive diagnostic method without the need for previous knowledge could be developed with high sensitivity and specificity for bladder cancer.
Urine is one of the most non-invasive samples taken for diagnostic reasons and it has been shown that it contains mostly urethral cells, transitional epithelial cells and occasionally, in men, prostate cells. By isolating these cells of interest, measurement is amenable to spectroscopic investigation. Indeed, Raman spectroscopy is a powerful analysis tool for biological materials which can be used for accessing the biochemistry of biological samples in a label-free manner. With the ability to work with aqueous samples, it can be coupled with other techniques to perform a wider range of biochemical analysis and in the work presented here Raman spectroscopy is coupled with laser tweezers, to form laser tweezers Raman spectroscopy (LTRS).10
There have been a few preliminary studies using Raman spectroscopy to classify or identify bladder cancer cells in mixed-cell population. Canetta et al. used modulated Raman spectroscopy (MRS) and atomic force microscopy (AFM) to classify the human urothelial cells (SV-HUC-1) and the bladder cancer cell (MGH-U1) in urine sample and achieved high sensitivity and specificity at 83% and above.11,12 Kerr et al. used Raman microscopy to classify two bladder cancer cell lines, which are defined as low grade (RT-112) and high grade (T24) cell lines. The classification results show high sensitivity and specificity at 90% and above for these two cell lines.13
Apart from standard Raman spectroscopy and microscopy, studies on the classification of urological cells had also been done with using LTRS. Harvey et al. on a mixed population of prostate and bladder cells in either water or an artificial urine environment14 achieved sensitivity and specificity of 75% or above for the classifications of urological cells with different lengths of time of urine exposure up to 12 hours. Although there are fluctuations of these values with different lengths of urine exposure time, it still shows possibility of utilising the LTRS method for urological cancer cells detection from urine samples. Other studies by Casabella et al. demonstrated the use of a LTRS system with an automatic microfluidic device for single cell analysis in urological cell samples.15 Similar techniques were also reported by Dochow et al. where LTRS was used to analyse erythrocytes, leukocytes, acute myeloid leukaemia cells (OCI-AML3), and breast tumour cells BT-20 and MCF-7 in microfluidic glass channels.16 Two types of optical traps were used in the study: capillary based optical trap and microchip based optical trap, and k-fold cross validation of linear discriminant analysis was used for analysing the data. The results show that the accuracy on classifying these cells is improved in the microchip-based experiment when comparing to the capillary based experiment due to the choice of materials of the devices. The results of the microchip based optical trap method shows that the classification can achieve an accuracy of 86% or above. That study demonstrated the feasibility of Raman-activated cell sorting for classification of different types of single cells.16 Also, a study by Schie et al. shows that rapid acquisition of mean spectra of eukaryotic cells is one of the possible solutions to achieve high throughput by significantly reducing the acquisition time down to a few seconds,17 which can be combined with the Raman-activated cell sorting system.
In this work, the LTRS system was used for the classification of seven bladder cancer cell lines, including the two used by Kerr et al.,13 in which single cell isolation and spectrum acquisition can be done at the same time. The eventual aim of this study is to determine the feasibility of developing a urological cancer diagnosis method for bladder cancer that can separate the invasive feature of the cell lines.
2. Materials and methods
2.1 Purpose built laser tweezers Raman spectroscopy system
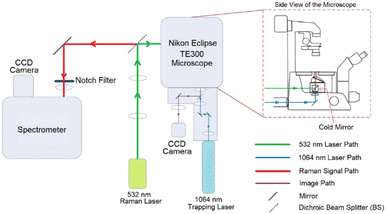
The newly configured purpose built LTRS system which was used for all the measurements presented in this work is shown in Fig. 1.The Raman system consists of a Horiba Scientific iHR-320 Imaging Spectrometer with focal length of 320 mm and f/4.1 aperture and a diffraction grating of 1200 lines per mm was coupled to a thermoelectrically cooled Horiba Syncerity charged coupled device (CCD) detector. The trapping part of the system consists of a Nikon Eclipse TE300 microscope equipped with a Plan Fluor 100× oil immersion objective which provides a high numerical aperture (NA) of 1.3 necessary for cell trapping.
A Laser Quantum's DPSS Ventus laser operating at 532 nm and capable of providing a maximum power of 110 mW at source was used for Raman excitation. A Laser Quantum's Diode Pumped Solid State (DPSS) Ventus laser operating at 1064 nm was used for cell trapping. The Raman laser power was set at 70 mW at source which was calculated to be reduced to around 41% at the sample by taking relative power measurements at source and objective. The reduction is caused by the overfilling of the objective and the complex optical structure of the system. The calculated approximate maximum Airy laser spot size for the Raman laser (532 nm) with the objective slightly overfilled is 0.50 μm which is significantly smaller than a cell. However, it had been proved that single point analyses in cells are representative for cell line classification with the fact that there is a certain degree of heterogeneity in a cell. Pavillon et al. suggested that a hybrid approach of rapid scanning across an area can provide a better picture of the overall information within a cell.18 However, a study conducted by Harvey demonstrated that cell size, and therefore laser spot size coverage, has only little correlation to the classification outcome.19 Also, similar study had also conducted by Kujdowicz et al. and Tang et al. demonstrated that despite variances across a single cell can be observed, these variances had insignificant effect on the overall outcome.20,21 An integration time of 30 s was chosen so that high signal-to-noise ratio (SNR) can be obtained when acquiring Raman spectra. For the trapping laser, trapping power was set to 760 mW at source, which was then attenuated to ∼127 mW at the sample.
2.2 Sample preparation
Table 1 states the origins of the cell lines, the culture media used and additional supplements present for specific cell lines. These cell lines are all transitional cell carcinoma (TCC) as listed in the table.
| Name | Origin of cell line | Culture media |
|---|---|---|
| T24 | ATCC collection – via the Translational Radiobiology Group, Paterson Institute for Cancer Research, University of Manchester | RPMI-1640 (Sigma-Aldrich, Poole, UK) |
| Grade 3, primary tumour untreated | ||
| TCC | ||
| Female | ||
| J82 | ATCC collection – via the Translational Radiobiology Group, Paterson Institute for Cancer Research, University of Manchester | EMEM (Sigma-Aldrich, Poole, UK) |
| Grade 3, stage T3 | ||
| Primary tumour treated | ||
| TCC | ||
| Male | ||
| 5637 | Obtained from Carcinogenesis group, Paterson Institute for Cancer Research, Cancer Research UK, Manchester | McCoy's 5A (Sigma-Aldrich, Poole, UK) |
| Grade 2, primary tumour | ||
| TCC | ||
| Male | ||
| RT-112 | ECACC collection | EMEM (Sigma-Aldrich, Poole, UK) 1% Non-essential amino acid (NEAA, Sigma-Aldrich, Poole, UK) |
| Grade 2 papillary, stage T2 | ||
| Primary tumour untreated | ||
| TCC | ||
| Female | ||
| UMUC-3 | ATCC collection – via the Translational Radiobiology Group, Paterson Institute for Cancer Research, University of Manchester | DMEM (Sigma-Aldrich, Poole, UK) |
| High grade cancer | ||
| TCC | ||
| Male | ||
| HT-1376 | ATCC collection – via the Translational Radiobiology Group, Paterson Institute for Cancer Research, University of Manchester | DMEM (Sigma-Aldrich, Poole, UK) |
| Grade 3 invasive, stage ≥ pT2 | ||
| Primary tumour untreated | ||
| TCC | ||
| Female | ||
| T24-CDDPR | Developed by the Iwamura Group from the School of Medicine, Kitasato University22 | RPMI-1640 (Sigma-Aldrich, Poole, UK) |
| Cisplatin resistant cell line derived from T24 cell line | ||
| Stepwise exposure of T24 cells to up to 40 μM of cisplatin |
2.2.5.1. Formalin fixation. When the cells reached 70–80% confluency, cells were trypsinised and washed with Dulbecco's phosphate-buffered saline (DPBS) twice before formalin fixation (10% formalin (neutral buffered, approximate 4% formaldehyde, Sigma-Aldrich)) and stored at 5 °C before Raman analysis.
2.2.5.2. Pre-measure preparation. Since cells measured in formalin will give a peak at ∼1040 cm−1 in Raman spectra, which is a direct spectral contaminant from formalin,24,25 formalin fixed samples have to be washed with DPBS before taking any Raman spectrum. On the day of Raman analysis, cells in formalin solution were washed with DPBS twice, resuspended in DPBS and measured within 24 hours. The reason for doing that is to get rid of the artefact that is brought by the formalin,26 and also minimise the fluorescence background during the Raman spectra acquisition. Four biological replicates were measured for each cell line.
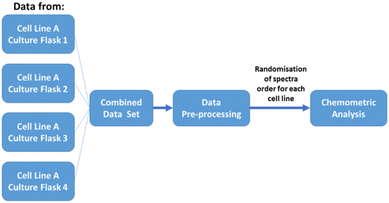
2.2.5.3. Biological replicates. Four biological replicates of each cell line were used, with each replicate derived from a different cell culture flask cultured under the same condition. Replicates (each contains 55 single-cell measurements) were measured on different days and times using the same instrument. During data analysis all the measured data were combined based on the type of cell line. Randomisation of the single cell spectra were also applied before any chemometric analysis. A schematic flow chart defining ‘biological replicates’ used in this work is shown in Fig. 2.
2.3 Raman cell spectra acquisition
During the Raman spectra acquisition, cells suspended in DPBS were placed into an open-top glass bottom dish. In order to trap a cell, agitation was applied until a cell falls into the optical trap. As mentioned in section 2.2.5 (iii), 55 cell spectra were measured from each set of replicates where the total measurement time is around 30 minutes per replicate. A new glass dish was used for each set of measurements to avoid any possible contamination. All replicates for each cell line were then combined into a bigger data set for further data processing and random sampling in later stages.2.4 Data pre-processing for the Raman results
The acquired spectral data needs to be pre-processed before carrying out any chemometric analysis. Data pre-processing can be simply divided into five parts explained in detail in the following sections.2.5 Chemometrics analysis of the Raman results
Principal component-canonical variates analysis (PC-CVA) was used as the major chemometric analysis method as in the clirspec-summer-school: CLIRSPEC-Summer-School-2015 (v1.0) package (https://doi.org/10.5281/zenodo.57398).28 It is a quadratic discriminant analysis method and is suitable for multi-class problems.29–31 Also, k-fold cross validation of PC-CVA was performed on the combined data set and results were presented as confusion matrices. As mentioned, 55 measurements were taken for each replicate per cell line, then all four replicates were combined to become a bigger data set (220 cells) for later data processing and analysis. After pre-processing the number of spectra per cell line was reduced. Considering the number of identities in the smallest data group (cell line), 70% of the spectra (120 spectra) were therefore taken from each cell line at each time to build 5 independent classifiers. This was done to avoid any bias brought by data size of each group.Random Forest was also used as a complementary classification method in which 80% of the spectra from the full data set was used as the training set i.e. 20% of the spectra from the full data set was used as the test set. Random Forest was picked as the supervised machine learning method in this work, as a complementary analysis with the PC-CVA method, which is treated as a quick exploratory analysis on new studies. Although PC-CVA is a good first approach as an extension of PCA, machine learning algorithm will be required when translating the work to solve real clinical problem. Random Forest will allow this to be done. Random Forest is a robust technique that does not involve any transformation of the data into other forms of presentation such as scores plots, while PCA and CVA do some form of eigenvalue eigenvector decomposition. This may lead to extensive change in the loadings and hence the output if extra data is added into the classification. Another major consideration of using Random Forest is its ability to do both regression and classification tasks on large data sets with high accuracy in predicting outcome.
Independent tests were also performed where using data from one of the replicates as the test set and using the remaining data as the training test. 500 trees (the number determined by the out of bag error rate curves) were used for building the classifier in all cases described in this work.
Candidates used for building these classifiers were selected by undersampling method which will retain all features and reduce the chance of overfitting of some classes.32,33
3. Results
3.1 Characterisation of the bladder cell lines
Characterisation of the cell lines involved in this work was performed, which is used for the investigation of the separation pattern obtained from the Raman results.3.2 Results of the classification of the six bladder cancer cell lines
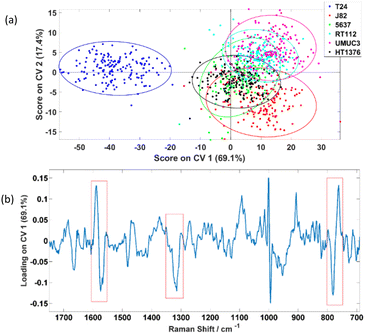
The PC-CVA scores plot for the first two CVs is shown in Fig. 5(a) which shows a distinctive separation between the T24 and the other five cell lines. Loadings on the corresponding CV 1 is shown Fig. 5(b), the amide II/lipid band at 1555–1600 cm−1, amide III peak at 1315 cm−1 and the tryptophan ring breathing at 764 cm−1 (indicated in red boxes) are weighting dominantly for separating the invasive T24 and the other non-invasive cell lines. However, the phenylalanine peak at ∼1000 cm−1 also shows up in the loading plot as significant features. Since the phenylalanine peak is very sharp, tiny differences in peak shape or peak shift will show up in loading plots. This inconsistency in phenylalanine peaks is constantly observed and had been reported by various studies. Casabella et al. concluded that one of the reasons of this observation is because of the photon flux fluctuation between two adjacent pixels on a detector, which is more apparent in sharp peak then board peaks.27 Instead of unavoidable instrumentation reasons, Li-Chan et al. reported that this phenomenon is caused by the conformation and macro-environment within a cell.34 Loadings plots on other CVs can be found in the ESI section 2.†
To further investigate the results obtained, 5-fold CVA cross-validation was applied to the data. Instead of just focusing on the first two CVs, this cross-validation method will consider all five CVs available for a 6-class problem i.e. classifying the six bladder cell lines in a 5-dimension manner during the analysis. Table 2 shows the classification results and it suggests that the six cell lines can be classified with average rates of correct classification range from 54.4% to 100.0%. Especially for the T24, the average correctly classified rate can be reached up to 100.0%. The resultant confusion matrices of the five folds are independently displayed in the ESI section 3.†
| True condition | |||||||
|---|---|---|---|---|---|---|---|
| T24 | J82 | 5637 | RT-112 | UMUC-3 | HT-1376 | ||
| Prediction | T24 | 100.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| J82 | 0.0% | 92.5% | 8.1% | 1.3% | 0.6% | 14.4% | |
| 5637 | 0.0% | 2.5% | 79.4% | 8.8% | 3.8% | 28.8% | |
| RT-112 | 0.0% | 1.3% | 6.3% | 81.3% | 3.1% | 0.6% | |
| UMUC-3 | 0.0% | 0.6% | 0.6% | 7.5% | 91.9% | 1.9% | |
| HT-1376 | 0.0% | 3.1% | 5.6% | 1.3% | 0.6% | 54.4% | |
One of the observations is that the classifier was also able to classify J82 with 92.5% accuracy which suggests there are significant differences between this cell line and the other cell lines, yet this is not showing up in the CV 1 and CV 2 dimensions. This raises a question on what feature(s) of cells is causing the separation observed in the PC-CVA scores plots. Therefore, further investigation was performed, and the results can be found in section 3.3.
| True condition | |||||||
|---|---|---|---|---|---|---|---|
| T24 | J82 | 5637 | RT-112 | UMUC-3 | HT-1376 | ||
| Prediction | T24 | 100.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| J82 | 0.0% | 87.1% | 0.0% | 2.9% | 0.0% | 11.8% | |
| 5637 | 0.0% | 3.2% | 96.4% | 5.7% | 6.3% | 0.0% | |
| RT-112 | 0.0% | 3.2% | 0.0% | 85.7% | 0.0% | 2.9% | |
| UMUC-3 | 0.0% | 0.0% | 0.0% | 5.7% | 93.8% | 0.0% | |
| HT-1376 | 0.0% | 6.5% | 3.6% | 0.0% | 0.0% | 85.3% | |
As shown in Table 3, the six bladder cell lines can be classified with high rates of correct classification ranging from 85.3% to 100.0%. Importantly the invasive T24 cell line was able to be classified with 100.0% accuracy. This is reasonably consistent with the classification results generated by k-fold PC-CVA, apart from the sensitivity of the HT-1376 cell line. The sensitivity of the HT-1376 cell line increase from 54.4% to 85.3% when classified by Random Forest.
The average classification results for the 6-class problem are shown in Table 4 where the confusion matrix for each independent test can be found in the ESI section 3.† The average results show that the sensitivities for the cell lines are not always high. Especially for 5637 and RT-112, they can only be classified with accuracies just above 30%. In a 6-class problem, decisions can be made if the trees’ voting percentage greater than 17%, but this is not convincing enough if aiming to bring this into clinical translation. The aim of this project is to build a model that is able to detect invasive cancers as early as possible, therefore achievement of an average sensitivity of 88% for the invasive T24 cell lines are more important.
| True condition | |||||||
|---|---|---|---|---|---|---|---|
| T24 | J82 | 5637 | RT-112 | UMUC-3 | HT-1376 | ||
| Prediction | T24 | 87.5% | 1.0% | 1.4% | 3.1% | 2.5% | 7.7% |
| J82 | 0.0% | 59.1% | 9.7% | 7.3% | 2.2% | 24.3% | |
| 5637 | 0.8% | 10.7% | 33.7% | 31.3% | 4.0% | 38.3% | |
| RT-112 | 8.3% | 5.0% | 14.4% | 37.4% | 14.2% | 7.8% | |
| UMUC-3 | 1.1% | 0.4% | 4.1% | 18.2% | 75.3% | 5.3% | |
| HT1376 | 2.3% | 23.8% | 36.8% | 2.8% | 1.9% | 16.6% | |
3.3 Results of the classification of the seven bladder cancer cell lines
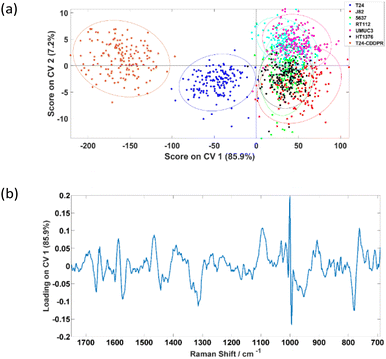
The PC-CVA scores plot for the classification of the seven cell lines is shown in Fig. 6(a), where it shows that the separation between the T24, T24-CDDPR and the other five cell lines is mainly on the CV 1 axis. Since the cisplatin sensitivity of the T24-CDDPR cell line should be a lot lower than the T24 cell line, separation on CV 1 is less likely to be due to the cisplatin resistance of the cell lines. It is more probable that there are other features that are dominant in both T24 and T24-CDDPR than the other cell lines.
5-Fold CVA was carried out using this combined set of data and the results are presented as average values taken form the five folds in Table 5, where the individual resultants are presented in the ESI section 4.†
| True condition | ||||||||
|---|---|---|---|---|---|---|---|---|
| T24 | J82 | 5637 | RT112 | UMUC3 | HT1376 | T24-CDDPR | ||
| Prediction | T24 | 98.1% | 0.6% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| J82 | 1.3% | 87.5% | 10.0% | 7.5% | 1.3% | 19.4% | 0.0% | |
| 5637 | 0.0% | 5.0% | 76.3% | 6.9% | 4.4% | 27.5% | 0.0% | |
| RT112 | 0.0% | 1.3% | 6.9% | 76.9% | 5.6% | 0.6% | 0.0% | |
| UMUC3 | 0.0% | 1.3% | 3.8% | 8.8% | 86.3% | 2.5% | 0.0% | |
| HT1376 | 0.0% | 4.4% | 3.1% | 0.0% | 2.5% | 50.0% | 0.0% | |
| T24-CDDPR | 0.6% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 100.0% | |
Table 5 shows that the invasive T24 and invasive resistant T24-CDDPR cell lines can be classified with very high accuracy of 98.1% and 100.0% respectively, hence LTRS can potentially be distinguishing the invasiveness of cell lines, and also able to identify the cisplatin sensitivities of the cell lines.
| True condition | ||||||||
|---|---|---|---|---|---|---|---|---|
| T24 | J82 | 5637 | RT-112 | UMUC-3 | HT-1376 | T24-CDDPR | ||
| Prediction | T24 | 96.9% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| J82 | 0.0% | 83.3% | 0.0% | 0.0% | 0.0% | 6.3% | 0.0% | |
| 5637 | 0.0% | 5.6% | 93.3% | 3.1% | 0.0% | 3.1% | 0.0% | |
| RT-112 | 0.0% | 5.6% | 0.0% | 93.6% | 0.0% | 0.0% | 0.0% | |
| UMUC-3 | 0.0% | 0.0% | 3.3% | 3.1% | 100.0% | 3.1% | 0.0% | |
| HT-1376 | 3.0% | 5.6% | 3.3% | 0.0% | 0.0% | 87.5% | 0.0% | |
| T24-CDDPR | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 100.0% | |
| True condition | ||||||||
|---|---|---|---|---|---|---|---|---|
| T24 | J82 | 5637 | RT-112 | UMUC-3 | HT-1376 | T24-CDDPR | ||
| Prediction | T24 | 88.4% | 1.6% | 0.0% | 5.0% | 1.6% | 5.1% | 3.4% |
| J82 | 0.7% | 64.7% | 7.8% | 8.3% | 2.0% | 25.5% | 0.0% | |
| 5637 | 0.8% | 8.9% | 40.7% | 28.9% | 4.5% | 38.3% | 0.0% | |
| RT-112 | 7.7% | 3.6% | 14.2% | 38.0% | 13.1% | 8.5% | 1.2% | |
| UMUC-3 | 0.8% | 0.6% | 2.7% | 17.0% | 75.9% | 6.4% | 0.6% | |
| HT-1376 | 1.6% | 20.7% | 34.6% | 2.8% | 2.9% | 16.2% | 0.0% | |
| T24-CDDPR | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 94.8% | |
The cell lines of interest, T24 and T24-CDDPR, were classified with accuracy of 88.4% and 94.8% respectively. However, the correct classification rates of the remaining cell lines vary from 16.2–75.9% which is relatively low. In some cases, the accuracies go below 10%. This is believed to be caused by the complexity of the classifiers, in the fact that it is a seven-class problem.
4. Discussion
The aim of this work was to assess the ability of LTRS to distinguish different types of formalin-fixed bladder cell line with varied phenotypes. Although there are a range of publications reporting the use of Raman spectroscopy on the classifications between cancerous and non-cancerous samples, with some including classification on grades and stages of tumours, it had also been reported that the percentage agreement on grading and staging of a range of cancers between pathologists vary in different type of cancer, including bladder cancer.35 From Fig. 5(a), it can be seen that the T24 cell line was significantly different to the other five cell lines. It was first suspected this difference was related to the cells’ invasiveness and/or cisplatin sensitivity. However, according to the characterisation results in Fig. 5(a), if separation was caused by the invasiveness of cells, J82 which is the second most invasive cell line following T24, should separate more towards the T24. Similar observations were acquired by Kujdowicz et al. that the two subtypes of T24 are highly distinguishable when analysing by Raman and FTIR spectroscopy.20 One other possible reason of causing this separation pattern is that the classifier is picking up changes in the Raman signal related to cells’ cisplatin sensitivity in which T24 is significantly more sensitive than other cell lines analysed. To elucidate this possibility further a cisplatin resistant T24 cell line, T24-CDDPR, was added into the analysis. There are significant changes in protein between the T24 and resistant T24-CDDPR cell lines according to the Agarose 2-DE and MS/MS analysis results by Taoka et al.,22 and according to the study by Sun et al.,36 the selection of cisplatin resistance in the T24 cell line enhanced proliferation, invasion and malignant behaviour. The introduction of cisplatin resistance in T24 and J82 leads to massive increase in MDR1 and HIF-1α and increase in aggressive behaviour. This is an indicative of these cells facing a more stem-like phenotype. Therefore it is reasonable to expect similar increase in aggressive invasion behaviour in the T24-CDDPR, even though it is a different cisplatin resistant T24 cell line. This concurs with our results show in Fig. 6(a) that T24-CDDPR is separated from both T24 and other cell lines. Although similar behaviours also observed in J82 and cisplatin resistant J82, T24 and cisplatin resistant T24 are still significantly more invasive then J82 cell lines according to the same study by Sun et al.36However, results in Fig. 6(a) shows that the separation along CV 1 is unlikely to be caused by the cisplatin sensitivity of the cell lines as the T24-CDDPR does not cluster with the J82, 5637, RT-112, UMUC-3, and HT-1376. It separates in the same direction with the T24 as a different data cluster on the negative CV 1. This indicates that the T24-CDDPR is very likely to have the invasive property as the T24 as they both separate towards the same direction on the CV 1. The distinction observed between T24-CDDPR and T24 is suspected to be caused by an enriched characteristic in the T24-CDDPR caused during the cisplatin resistant development. Yang et al. showed that the T24 cell line, overexpressed the long noncoding RNA (lncRNA) ASAP1-IT1, similar to bladder cancer tissues, which plays a role in maintaining cell stemness. Although J82, 5643 and UMUC3 cell lines do express ASAP1-IT1 it is at a significantly lower level than T24, and overexpression of ASAP1-IT1 in T24 induces a stem like phenotype.37 It is therefore possible that the separation of the T24 and the cisplatin resistant derivative T24-CDDPR is based upon their more stem like phenotype. Although the results acquired show that the invasiveness and drug resistance of these cells are extremely complex, in which a single component from chemometric analysis is not enough to explain the observations, clear separations between clusters of different type of cell lines were achieved. This is partly caused by the fact that there is lack of information on standardising the development of the drug-resistant T24 cell lines and corresponding study on their metabolisms, further analysis on these cell lines has to be done to confirm to test the hypothesis.
The ultimate aim of this study is to demonstrate the feasibility of using LTRS to identify different types of bladder cancer cells from urine samples. The samples used in this work presented were the formalin fixed cells, which is different from the cells that can be found in urine sample or in the in vivo condition. Despite this formalin fixation can preserve the biochemical information of cells and can significantly reduce the cells’ stress response to photochemical oxidative damage.38 This is very important when this study is aimed at performing preliminary test on whether the LTRS can be used for identifying different types of bladder cancer. Non-fixed cells and cells exposed in urine should then be used to mimic the real cells situation that can be found in urine samples in future research on this topic. However, this work with using formalin fixed cells will allow a starting point for demonstrating that LTRS can be used in real time capture and analysis of different phenotypic features of bladder cancer cells in a urine streamline.
5. Conclusions
This work presented has successfully demonstrated the use of LTRS system to classify different types of formalin fixed bladder cancer cells with high sensitivity when classifying with PC-CVA. Complementary classifications with using Random Forest were also performed and consistent results were obtained. However multiple replicates in Random Forest analysis will be needed to build a better model. As in an early stage of a pilot study of the development of using LTRS to detect bladder cancer cells in urine streamline, this work shows the positive outcomes on separating the cell line with invasive phenotype. It shows that phenotypic feature is more dominant than the internal variability within cell lines. Further work using a wider range of non-fixed urological cells and detailed study on the separation components is necessary if the technique is to be translated into clinical applications.Author contributions
N. T. T. performed the bulk of the experimental work. R. R. performed the experimental work on the invasion assay and cytotoxicity assay, with experimental design and project supervision by N. W. C. and M. D. B., N. T. T. wrote the main body of the manuscript. P. G. supervised the project and handled logistics. N. T. T., R. D. S., M. D. B. and P. G. were equally involved in experimental design. M. D. B. was involved in the communication and logistic of obtaining the drug resistant T24-CDDPR cell line and contributed to the writing of the manuscript. R. D. S. was involved in experimental design and input into Raman instrument configuration. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank the Genito Urinary Cancer Research Group for the donation of the cell lines for this study. Moreover, we would like to acknowledge Dr Kazumasa Matsumoto from the Kitasato University for the donation of the cisplatin resistant cell line T24-CDDPR.References
- Cancer Research UK, Prostate cancer incidence statistics, https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/prostate-cancer/incidence#heading-Two, (accessed 3 April 2023).
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- Cancer Research UK, Bladder cancer - Screening - Cancer Research UK, https://www.cancerresearchuk.org/about-cancer/bladder-cancer/getting-diagnosed/screening, (accessed 9 May 2017).
- C. Z. Zhu, H. N. Ting, K. H. Ng and T. A. Ong, J. Cancer, 2019, 10, 4038–4044 CrossRef CAS PubMed.
- S. J. Galgano, S. Rais-Bahrami, K. K. Porter and C. Burgan, Diagnostics, 2020, 10, 1–16 CrossRef PubMed.
- H. H. Lee and S. H. Kim, Transl. Cancer Res., 2020, 9, 6554–6564 CrossRef CAS PubMed.
- M. Matuszczak, A. Kiljańczyk and M. Salagierski, Int. J. Mol. Sci., 2022, 23, 8597 CrossRef CAS PubMed.
- B. J. Schmitz-Dräger, L. A. Tirsar, C. Schmitz-Dräger, J. Dörsam, Z. Mellan, E. Bismarck and T. Ebert, World J. Urol., 2008, 26, 31–37 CrossRef PubMed.
- R. Malinaric, G. Mantica, L. Lo Monaco, F. Mariano, R. Leonardi, A. Simonato, A. Van der Merwe and C. Terrone, Int. J. Environ. Res. Public Health, 2022, 19, 9648 CrossRef CAS PubMed.
- R. D. Snook, T. J. Harvey, E. Correia Faria and P. Gardner, Integr. Biol., 2009, 1, 43–52 CrossRef CAS PubMed.
- E. Canetta, M. Mazilu, A. C. De Luca, A. E. Carruthers, K. Dholakia, S. Neilson, H. Sargeant, T. Briscoe, C. S. Herrington and A. C. Riches, J. Biomed. Opt., 2011, 16, 037002 CrossRef PubMed.
- E. Canetta, A. Riches, E. Borger, S. Herrington, K. Dholakia and A. K. Adya, Acta Biomater., 2014, 10, 2043–2055 CrossRef CAS PubMed.
- L. T. Kerr, A. Adams, S. O. Dea, K. Domijan, I. Cullen and B. M. Hennelly, Photon. Solut. Better Health Care, 2014, 9129, 1–8 Search PubMed.
- T. J. Harvey, E. Correia Faria, A. Henderson, E. Gazi, A. D. Ward, N. W. Clarke, M. D. Brown, R. D. Snook and P. Gardner, J. Biomed. Opt., 2008, 13, 064004-1–064004-12 CrossRef PubMed.
- S. Casabella, P. Scully, N. Goddard and P. Gardner, Analyst, 2016, 141, 689–696 RSC.
- S. Dochow, C. Krafft, U. Neugebauer, T. Bocklitz, T. Henkel, G. Mayer, J. Albert and J. Popp, Lab Chip, 2011, 11, 1484–1490 RSC.
- I. W. Schie, R. Kiselev, C. Krafft and J. Popp, Analyst, 2016, 141, 6387–6395 RSC.
- N. Pavillon and N. I. Smith, J. Biomed. Opt., 2015, 20, 016007 CrossRef PubMed.
- T. J. Harvey, The Development of Vibrational Spectroscopic Cytology for Prostate Cancer Diagnosis, University of Manchester, 2008 Search PubMed.
- M. Kujdowicz, W. Placha, B. Mech, K. Chrabaszcz, K. Okoń and K. Malek, Cancers, 2021, 13, 1–20 Search PubMed.
- N. T. Tang, R. D. Snook, M. D. Brown, B. A. Haines, A. Ridley, P. Gardner and J. L. Denbigh, Molecules, 2020 DOI:10.3390/molecules25071652.
- Y. Taoka, K. Matsumoto, K. Ohashi, S. Minamida, M. Hagiwara, S. Nagi, T. Saito, Y. Kodera and M. Iwamura, Biomed. Res., 2015, 36, 253–261 CrossRef CAS PubMed.
- C. A. Hart, M. Brown, S. Bagley, H. Sharrard and N. W. Clarke, Br. J. Cancer, 2005, 92, 503–512 CrossRef CAS PubMed.
- Z. Huang, A. McWilliams, S. Lam, J. English, D. I. McLean, H. Lui and H. Zeng, Int. J. Oncol., 2003, 23, 649–655 Search PubMed.
- M. G. Shim and B. C. Wilson, Photochem. Photobiol., 1996, 63, 662–671 CrossRef CAS PubMed.
- F. Lyng, E. Gazi and P. Gardner, in Biomedical Applications of Synchrotron Infrared Microspectroscopy: A Practical Approach, The Royal Society of Chemistry, 2011, pp. 145–191 Search PubMed.
- S. Casabella, Development of automated analysis and sorting of single cells using Laser Tweezers Raman Spectroscopy, University of Manchester, 2015 Search PubMed.
- A. Henderson, CHI Toolbox, 2016.
- A. M. C. Daviesa and T. Fearnb, Spectrosc. Eur., 2008, 20, 18–20 Search PubMed.
- L. Nørgaard, R. Bro, F. Westad and S. B. Engelsen, J. Chemom., 2006, 20, 425–435 CrossRef.
- C. Peltier, M. Visalli and P. Schlich, Food Qual. Prefer., 2015, 40, 326–333 CrossRef.
- G. E. A. P. A. Batista, R. C. Prati and M. C. Monard, A Study of the Behavior of Several Methods for Balancing Machine Learning, Training Data, 2004 Search PubMed.
- B. W. Yap, K. A. Rani, H. A. A. Rahman, S. Fong, Z. Khairudin and N. N. Abdullah, An Application of Oversampling, Undersampling, Bagging and Boosting in Handling Imbalanced Datasets, 2014, pp. 13–22 Search PubMed.
- E. Li-Chan, J. M. Chalmers and P. R. Griffiths, Applications of Vibrational Spectroscopy in Food Science, John Wiley & Sons, 2010 Search PubMed.
- E. Compérat, A. Oszwald, G. Wasinger, D. E. Hansel, R. Montironi, T. van der Kwast, J. A. Witjes and M. B. Amin, World J. Urol., 2022, 40, 915–927 CrossRef PubMed.
- Y. Sun, Z. Guan, L. Liang, Y. Cheng, J. Zhou, J. Li and Y. Xu, Oncol. Rep., 2016, 35, 1549–1556 CrossRef CAS PubMed.
- L. Yang, Y. Xue, J. Liu, J. Zhuang, L. Shen, B. Shen, J. Yan and H. Guo, Neoplasma, 2017, 64, 847–855 CrossRef CAS PubMed.
- T. J. Harvey, C. Hughes, A. D. Ward, E. Correia Faria, A. Henderson, N. W. Clarke, M. D. Brown, R. D. Snook and P. Gardner, J. Biophotonics, 2009, 2, 47–69 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00119a |
| ‡ Current affiliation: The Open Innovation Hub for Antimicrobial Surfaces, Surface Science Research Centre, Department of Chemistry, University of Liverpool, Liverpool, L69 3BX, UK. |
| This journal is © The Royal Society of Chemistry 2023 |