Open Access Article
Open Access ArticleIn vivo Raman spectroscopy in the diagnostics of colon cancer
Markéta
Fousková
 *a,
Jan
Vališ
*a,
Jan
Vališ
 a,
Alla
Synytsya
a,
Alla
Synytsya
 a,
Lucie
Habartová
a,
Lucie
Habartová
 a,
Jaromír
Petrtýl
a,
Jaromír
Petrtýl
 b,
Luboš
Petruželka
b,
Luboš
Petruželka
 c and
Vladimír
Setnička
c and
Vladimír
Setnička
 a
a
aDepartment of Analytical Chemistry, University of Chemistry and Technology, Prague, Technická 5, 166 28, Prague 6, Czech Republic. E-mail: marketa.fouskova@vscht.cz
b4th Department of Internal Medicine, General University Hospital in Prague and 1st Faculty of Medicine, Charles University in Prague, U Nemocnice 2, 128 08, Prague 2, Czech Republic
cDepartment of Oncology, General University Hospital in Prague and 1st Faculty of Medicine, Charles University in Prague, U Nemocnice 2, 128 08, Prague 2, Czech Republic
First published on 2nd May 2023
Abstract
Early detection and accurate diagnosis of colorectal carcinoma are crucial for successful treatment, yet current methods can be invasive and even inaccurate in some cases. In this work, we present a novel approach for in vivo tissue diagnostics of colorectal carcinoma using Raman spectroscopy. This almost non-invasive technique allows for fast and accurate detection of colorectal carcinoma and its precursors, adenomatous polyps, enabling timely intervention and improved patient outcomes. Using several methods of supervised machine learning, we were able to achieve over 91% accuracy in distinguishing colorectal lesions from healthy epithelial tissue and more than 90% classification accuracy for premalignant adenomatous polyps. Moreover, our models enabled the discrimination of cancerous and precancerous lesions with a mean accuracy of almost 92%. Such results demonstrate the potential of in vivo Raman spectroscopy to become a valuable tool in the fight against colon cancer.
Introduction
Raman spectroscopy has been in the spotlight of the new methods of non-invasive cancer diagnostics for several decades. In addition to potential non-invasiveness, the advantages of the technique include high sensitivity to the biochemical composition of the analyzed tissue and achievable automation.1 Furthermore, compared to the complementary vibrational technique, infrared (IR) spectroscopy, Raman spectroscopy offers lower sensitivity to the presence of water in the tissue sample, which otherwise strongly absorbs mid-IR radiation and thus overlaps the more valuable signals of other biochemical components contained in the tissue.2 These benefits have resulted in the spread of high throughput studies of histological3 or cytological4,5 slides that can be effortlessly diagnosed employing methods of machine or deep learning, while reaching high levels of diagnostic accuracy, some of which are even on the verge of translation into the clinical environment.6The ultimate aim in this field, however, is to recognize tissue pathologies in real time, and moreover, without the need for invasive sampling. This goal can be achieved with the implementation of fiber-optic devices – such an approach might accelerate the diagnostic process for many epithelial cancers and their precancerous lesions, such as colorectal cancer. The incidence and mortality of this disease are expected to increase not only as an indirect consequence of the COVID-19 pandemic7,8 and related preventive protective measures which in many cases resulted in inferior access to elective and preventive care. The most recent number of newly diagnosed colon cancer cases per year globally was estimated at more than 1.9 million, with more than 900 thousand expected deaths9 despite the existence of preventive testing. Patients can, however, avoid the established screening techniques, such as fecal occult blood testing, or a preventive colonoscopy out of anticipated discomfort. On the other hand, most biomarkers of early disease that are currently employed lack sensitivity.10 The infallible golden standard of tissue diagnostics, hematoxylin and eosin staining by histopathologists poses the problems of invasive collection of samples, a prolonged period between the biopsy and obtaining the results, and the restricted information about the tissue components. What is more, the results’ objectivity may be affected by the human factor.
The situation might be improved by the use of in vivo Raman spectroscopy. Compared to conventional diagnostic techniques, this type of vibrational spectroscopy can deliver results within the scope of colonoscopy without prolonging the patient distress, while achieving similar levels of sensitivity and specificity. Moreover, it provides more detailed information about the biochemical composition of the tissue.
Raman spectroscopy was found to be effective in tissue diagnostics of colorectal cancer in studies focusing on ex vivo samples,11–16 while others aimed to turn to in vivo analyses and succeeded in testing specialized optical fiber systems.17–19
The first use of an in vivo Raman probe to diagnose precancerous colonic polyps showed 95% accuracy on a set of 19 patients.20 The in vivo study by Ding et al.21 focused on the Raman spectroscopic properties of colon tissue of various anatomical sites and the effect of age, BMI, and other physiological factors on the resulting spectra. A study by Bergholt et al.22 combined the information from fingerprint and high wavenumber regions of the in vivo Raman spectra to differentiate between hyperplastic polyps and precancerous adenomatous polyps of the colon and rectum. They achieved diagnostic sensitivity of 91%, demonstrating the significant potential of Raman spectroscopy in the diagnosis of colorectal pathologies.
Therefore, the purpose of our work was to evaluate the use of near-IR in vivo Raman spectroscopy in the diagnostics of colorectal carcinoma and its precancerous lesions – benign adenomatous polyps – with a set of various supervised machine learning classification methods and their respective ensembles.
Experimental
Participants
The subjects were recruited between July 2020 and June 2022 at the 4th Internal Clinic – Gastroenterology and Hepatology of General University Hospital in Prague from patients requiring preventive or curative colonoscopies. A signed informed consent form was obtained from all subjects who participated in the study. All experiments were conducted in accordance with the guidelines of the Declaration of Helsinki and approved by Ethics Committee of the 1st Faculty of Medicine and the General University Hospital in Prague (IRB00002705, project NU20-09-00229, 18 June 2020).Altogether, 317 participants were recruited (median age 64 years, interquartile range 51–71 years; 124 (39.1%) female, 193 (60.9%) male), out of which 243 were healthy subjects (median age 64 years (IQR 52–71), mean BMI 26.5), 64 patients were diagnosed with benign epithelial tumors (median age 65 years (IQR 47–72), mean BMI 27.8) and 10 patients suffered from colorectal adenocarcinoma (median age 72 years (IQR 61–76), mean BMI 27.7). The diagnoses of the patients were confirmed by histological examination of biopsied samples of the tissue analyzed in vivo.
Methods
All in vivo spectroscopic analyses were conducted via a custom-built combination of a fiber-optic microprobe (EmVision, US) with a portable HT Raman spectrometer (EmVision, US) equipped with the thermoelectrically cooled iVac 316 CCD camera (Oxford Instruments, UK). The excitation source of radiation was presented by a narrow-linewidth laser (Beijing RealLight Technology, CN) with a wavelength of 785 nm. All in vivo Raman spectra were collected with an integration time of 2·3 s and a power of 20–25 mW at the tip of the probe. The custom-made fiber-optic microprobe consisted of 11 low-hydroxyl silica collection fibers (d = 200 μm each) surrounding one excitation fiber (d = 300 μm), all enclosed in a nylon casing. The distal part of the microprobe (l = 2.0 m) was long enough to reach even the caecum through the working channel of the endoscope. The construction of the probe tip enabled contact analyses, which prevented shifts in the position of the microprobe during the collection of Raman spectra. To minimize the loss of laser power in the microprobe, a fiber collimator F220SMA/FC-780 (Thorlabs, US) was introduced between the light source and the excitation fiber of the probe.Preprocessing and statistical methods
The collected in vivo Raman spectra of colorectal tissue were algorithmically preprocessed using a custom Python script consisting of the following steps. First, the spectra were truncated to the region of 500–2000 cm−1, subsequently, the spectral baseline elevated by tissue autofluorescence was corrected using the novel morphological baseline removal technique, the BubbleFill algorithm.23 The minimum bubble width was set to 150 cm−1. The spectra were then truncated again, so that only the region of 900–1800 cm−1 remained. The data were smoothed by fast Fourier transformation filtering (cut-off frequency of 1/(25 cm−1)) and normalized to the sum of a unit vector to maximize the relevant spectral features.The spectral data preprocessed as described above were used to compute the mean and difference spectra in order to reveal the differences indicating potential spectral biomarkers of carcinogenesis or other abnormal conditions in the analyzed colorectal tissue. Spectral band maxima were identified and assigned to their vibrational modes according to the literature.
The diagnostic groups of the preprocessed spectra were paired to enable binary classification using several machine learning methods. Python24 (version 3.9.5) was used for the preprocessing and analysis of the spectral dataset with the following libraries: pandas25,26 (version 1.2.5) and numpy27 (version 1.22.4) for data manipulation, scipy28 (version 1.9.0) and orplib23 (version 0.1.1) for spectral preprocessing, and scikit-learn29 (version 1.1.3) and imbalanced-learn30 (version 0.9.1) for final data analyses. The figures were created with OriginPro graphing software (version 2019b, OriginLab, US).
Principal component analysis – linear discriminant analysis
The first machine learning method to be employed was LDA combined with PCA for dimensionality reduction. The first step of the pipeline consisted of scaling – removing the mean and scaling to unit variance. The number of components used in the final PCA–LDA model was optimized employing a grid search cross-validation (CV) function for each model, beginning with coarse steps up to 70 principal components, followed by a search using a finer data step around the discovered score local maxima. The model with the best F1 score of 10-fold CV was chosen for classification.Support vector machine classifier
Before employing ensemble machine learning strategies, the effectivity of separate models was assessed. The first method of choice was the support vector machine classifier (SVM) applied to the whole spectral region (900–1800 cm−1). The optimized parameters of the model included the type of kernel and the value of C. The model with the best mean F1 score of a 10-fold CV was selected for classification.Decision tree classifier
The second algorithm to be used on its own was the decision tree classifier (DT). Only the maximum depth of the tree and the minimum number of samples per leaf were optimized using the grid search function. The optimal parameters were also selected based on the resulting F1 score of the 10-fold CV.Adaptive boosting of decision trees
The standalone classification algorithms were followed by boosted ensemble models, the first being adaptive boosting of decision trees classifiers (DT AdaBoost) from the scikit-learn package with DTs as its base estimating classifier. The optimal combination of hyperparameter values (maximum depth, minimum samples per leaf, and number of DTs) was found by a grid search based on a 10-fold CV F1 score.Imbalanced adaptive boosting of decision tree and support vector machine classifiers
The same procedure was performed with EasyEnsemble – bagged AdaBoost from the imbalanced-learn package, tackling imbalance in datasets by under- or oversampling of data in classes with an uneven number of samples. An F1 score-based hyperparameter optimization was carried out for both ensemble classifiers with chosen base estimators, SVM and DT (SVM AdaBoost IB and DT AdaBoost IB, respectively).The classification performance characteristics (sensitivity, specificity, overall accuracy, AUROC, precision, Cohen's kappa, and F1 score) were calculated from the 10-fold CV for all the resulting classification models to enable effective comparison of the feasibility of the methods for our spectral diagnostic dataset.
Results and discussion
In vivo Raman spectra of colorectal tissue
Altogether, 330 in vivo Raman spectra of colorectal tissue from a population of 317 patients were collected. The spectra of each of the three diagnostic types of tissue were averaged and the results are shown as a mean ± one standard deviation in Fig. 1.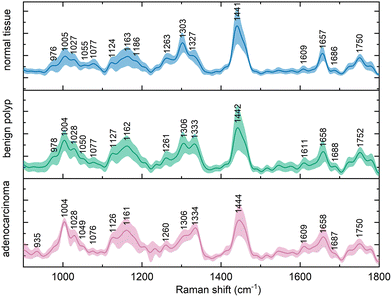 | ||
| Fig. 1 Average (mean ± standard deviation) in vivo Raman spectra (λex = 785 nm, normalized) of normal colorectal mucosa, benign epithelial polyps, and colorectal adenocarcinoma. | ||
The observed variance within these spectral representations of the tissue diagnostic groups might be increased as a result of functional difference of anatomical parts of the large intestine, such as between any part of colon and rectum. These variations in the spectra were, however, not as substantial as the differences between the diagnostic groups, which was in agreement with previous findings in the literature.21,31
The Raman bands in the spectra were identified and their origin was assigned according to the literature, the overview is presented in Table 1. The paired group means were subsequently subtracted to disclose the differences in the spectra (Fig. 2). This procedure revealed typical features in the spectra of each diagnostic group and unveiled spectral regions of lower intragroup and higher intergroup difference, potential spectral biomarkers. The trends in the obtained difference spectra showed similarities for both spectra resulting from the comparison of normal and diseased tissue. In vivo spectra of diseased tissue exhibited an increase in the spectral regions around 1005, 1028, 1126, 1162, and 1333 cm−1.
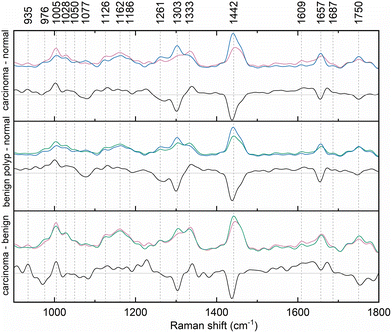 | ||
| Fig. 2 Difference of paired mean Raman spectra of normal colorectal mucosa (blue), benign epithelial polyp (green), and adenocarcinoma (pink) collected in vivo. | ||
| Band position (cm−1) | Intensity trend in carcinoma | Intensity trend in benign lesion | Vibrational mode | Assignment15,33,35,37–40 |
|---|---|---|---|---|
| 935 | + | ν (CC) | Protein, α-helix, Pro, Val | |
| 976 | − | ν (CC) | Collagen backbone | |
| 1005 | + | + | δ (CC) breathing | Phe |
| 1028 | + | + | δ (CH) in plane | Phe |
| 1050 | − | − | ν (CN), ν (CO) | Pro (collagen) |
| 1077 | − | − | ν (CC), ν (CO) | Glucose, triglycerides, lipids |
| 1126 | + | + | ν (CN), ν (CC) skeletal of acyl backbone | Proteins, phospholipids, lipids |
| 1162 | + | + | ν (CN), ν (CC), | Proteins (Tyr – collagen) |
| 1178 | + | + | δ (C–H), ν (CO–O–C) | Lipids, Phe, Tyr – collagen, nucleic bases C and G |
| 1261 | − | − | Amide III, δ (CH2) in plane | Proteins, lipids, phospholipids |
| 1306 | − | − | δ (CH2) twist, amide III (NH) | Lipids, Phe, Trp, α-helix |
| 1333 | + | + | δ (CH), δ (CH3CH2) wagging | A, G of DNA/RNA and proteins (Trp, Pro) |
| 1442 | − | − | δ (CH2) scissoring | Lipids, triglycerides and proteins |
| 1609 | + | + |
δ (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) C) |
Phe, Tyr, Trp, nucleic bases C, T |
| 1657 | − | − | Amide I (ν (CO), ν (CN), δ (NH)), ν (CO) | Proteins, α-helix/random chain, lipids |
| 1687 | + | + | Amide I | β-Sheet structure of proteins |
| 1750 | − | − |
ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
Lipids, phospholipids |
Conversely, the relative intensity of regions around 1261, 1306, 1442, 1657, and 1750 cm−1 was decreased. Fig. 2 documents slight wavenumber shifts in the local intensity extremes of the subtraction spectra in comparison to the original mean class spectra. However, by calculating the second derivate of the difference spectra, according to literature, it was verified that main distinctions between the diagnostic groups of the spectra laid predominantly in the relative band intensities of the mean spectra.32
The key differences between the spectra of normal tissue and those of diseased tissue specifically resulted from the changing ratio of lipid and protein components in the tissue mainly occurring as a result of cell proliferation or carcinogenesis.33,34 The majority of the bands with decreased intensity in the spectra of diseased tissue can be assigned to vibrational modes of functional groups and residues found in lipids and phospholipids,35 which might suggest a raised energy demand of the proliferating cells in the affected tissue, as the patient groups were BMI-matched. Other explanations may be alterations in lipid metabolism and abnormal composition of cell membranes in the abnormal tissue.36
The difference spectra of the two types of diseased tissue, benign polyps and cancerous lesions, enabled us to reveal the subtle distinctions between the two, which are key for predicting the malignant turn in the polypous lesions which might have been considered benign for a long time. The relative intensity of the bands with maxima around 935, 1005, and 1333 cm−1 showed an increase for cancerous lesions, whereas that of bands at 976, 1303, 1442, and 1750 cm−1 was lower.
Such distinctions encourage the hypothesis that in vivo Raman spectra of colorectal carcinoma can be discriminated even from the corresponding spectra of benign adenomatous polyps.
Classification of normal tissue and cancerous lesions
A subset of preprocessed spectra consisting of samples of normal tissue (n = 249, 95.8% of the subset) and adenocarcinoma (n = 11, 4.2%) was analyzed with PCA–LDA. The number of components was optimized based on the F1 score and amounted to 35 components describing 98.6% of the total variability. The PCA–LDA model showed high levels of accuracy (99.2%) and specificity (100.0%), whereas in contrast, its sensitivity was lower (81.8%, Table 2).| PCA–LDA | SVM | DT | DT AdaBoost | DT AdaBoost IB | SVM AdaBoost IB | |
|---|---|---|---|---|---|---|
| Accuracy | 0.992 | 0.981 | 0.988 | 0.996 | 0.769 | 0.758 |
| Sensitivity | 0.818 | 0.545 | 1.000 | 0.909 | 1.000 | 1.000 |
| Specificity | 1.000 | 1.000 | 0.988 | 1.000 | 0.759 | 0.747 |
| AUROC | 0.998 | 0.990 | 0.996 | 0.990 | 0.991 | 0.954 |
| Precision | 1.000 | 1.000 | 0.786 | 1.000 | 0.155 | 0.149 |
| Cohen's kappa | 0.896 | 0.697 | 0.785 | 0.950 | 0.137 | 0.130 |
| F1 score | 0.900 | 0.706 | 0.880 | 0.952 | 0.268 | 0.259 |
The SVM model (polynomial kernel, C = 1) of the identical dataset exhibited a lower classification accuracy (98.1%) due to a reduced sensitivity of 54.5% (Table 2).
The DT classifier (maximum depth = 5, minimum of samples per leaf = 3) on the other hand, showed a sensitivity of 100%; therefore, its accuracy increased to 98.8% (Table 2).
The first boosted classifier used was an adaptive boosting of decision trees, the hyperparameters of which were optimized based on the F1 score to 100 base estimators with a minimum of samples per leaf and maximum depth values of 1. This model showed an improvement in accuracy, specificity, precision, kappa value, and F1 score (Table 2).
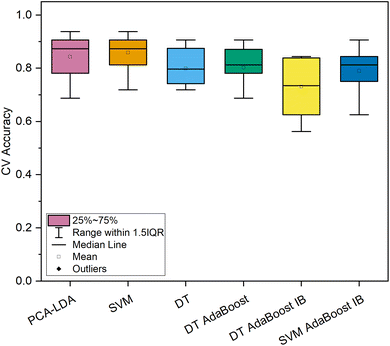
In contrast, both machine learning strategies specializing in imbalanced datasets reached lower levels of most qualitative metrics. For the DT-based ensemble model, hyperparameters were optimized to 10 base estimators with minimally 3 samples per leaf and a maximum depth of 3. This model reached a high level of sensitivity (100.0%) for the spectra of cancerous tissue; however, the model's precision was exceptionally low (15.5%). Similar results were obtained by the SVM-based imbalanced adaptively boosted model. With optimized hyperparameters (n = 5, C = 10, polynomial kernel), this model reached 100.0% sensitivity for the carcinoma spectra, whereas the precision was decreased even in comparison with the model based on the DT classifier (14.9%, Table 2). Comparing the repeated 10-fold CV accuracies of the spectral dataset of normal and cancerous colorectal tissue (Fig. 3), the PCA–LDA model, the SVM model, and the DT-based AdaBoost model reached similarly high values (>95% on average). Moreover, the ranges of classification accuracies for the ten subsets within 1.5 IQR were rather narrow compared to the rest of the classification models (Fig. 3), therefore, these three machine learning methods seem appropriate for the most imbalanced of the data subsets (249 vs. 11 Raman spectra). This is either evidence of the potential stability of the obtained results of a further developing dataset or possibly a sign of slight overfitting of the model's most numerous category. Altogether, most of the highest classification performance characteristics for the discrimination of the subset of the spectra of normal and cancerous tissue were reached for the DT-based AdaBoost model (Table 2).
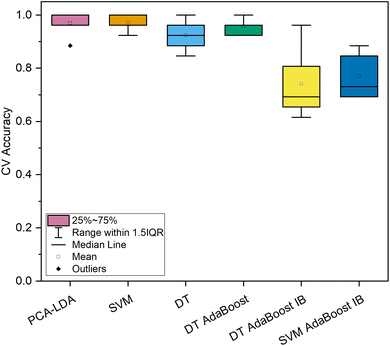 | ||
| Fig. 3 Classification accuracy of 10-fold cross-validation of machine learning models for the dataset of in vivo Raman spectra of normal (n = 249) and cancerous (n = 11) colorectal epithelial tissue. | ||
Classification of normal tissue and benign epithelial tumors
The most abundant and balanced subset of in vivo Raman spectra, that of normal tissues (n = 249, 78.1%) and benign adenomatous polyps (n = 70, 21.9%) was analyzed using the same set of machine learning methods as the subset of spectra of normal and cancerous tissues; PCA–LDA, SVM, DT, and three boosted ensemble models. The summary of their performance characteristics is provided in Table 3.| PCA–LDA | SVM | DT | DT AdaBoost | DT AdaBoost IB | SVM AdaBoost IB | |
|---|---|---|---|---|---|---|
| Accuracy | 0.897 | 0.959 | 0.906 | 0.962 | 0.824 | 0.881 |
| Sensitivity | 0.657 | 0.829 | 0.671 | 0.843 | 0.943 | 0.886 |
| Specificity | 0.964 | 0.996 | 0.972 | 0.996 | 0.791 | 0.880 |
| AUROC | 0.911 | 0.981 | 0.847 | 0.966 | 0.971 | 0.929 |
| Precision | 0.836 | 0.983 | 0.870 | 0.983 | 0.559 | 0.674 |
| Cohen's kappa | 0.638 | 0.869 | 0.672 | 0.879 | 0.509 | 0.613 |
| F1 score | 0.736 | 0.899 | 0.758 | 0.908 | 0.702 | 0.765 |
The PCA–LDA was conducted utilizing 40 principal components (98.7% variability). The model was able to accurately classify 96.4% of the control samples; however, it was able to detect only 65.7% of the polyps correctly (Table 3).
The SVM model (C = 5, polynomial kernel) presented an improvement in both sensitivity (82.9%) and specificity (99.6%) in comparison with the PCA–LDA model. Furthermore, the model precision was as high as 98.3% (Table 3).
The simple DT classifier did not prove very beneficial in the analysis of this data subset. With the optimized hyperparameters (maximum depth = 5, minimum samples per leaf = 1), the model reached a classification accuracy of 90.6% even though the sensitivity level was 67.1% (Table 3). The hyperparameters of the DT-based AdaBoost model were optimized at the F1 score resulting in values of a DT maximum depth of two, a minimum of samples per DT leaf of three, and the number of DTs selected as 100. The model was able to discriminate between the spectra of normal tissue and those of adenomatous polyps with high levels of most classification evaluation metrics. The overall accuracy of the model was as high as 96.2%, mostly due to its specificity level of 99.6% (Table 3).
The last models for the discrimination of normal tissue and adenomatous polyps of the colon and rectum were DT- and SVM-based imbalanced boosted ensemble models, both of which demonstrated a worse performance than their base classification algorithms in most metrics. The DT parameters were selected as a maximum depth of three and a minimum samples per leaf of one. The base SVM models were fitted with a C value of 1.0 and a polynomial kernel. Both final imbalanced models were constructed from ten base classifiers. The DT-based model performed better in classifying spectra of polyps (sensitivity of 94.3%), in contrast, its specificity was reduced to 79.1%. The performance of the SVM-based imbalanced model was similar for both diagnostic groups, its sensitivity and specificity reached 88.6% and 88.0%, respectively (Table 3).
The evaluation of the resulting repeated CV classification accuracies for the spectral subset of normal tissue and adenomatous polyps displayed a higher variance for all algorithms in comparison to the models discriminating the Raman spectra of normal and cancerous colorectal tissue (Fig. 3 and 4), possibly due to the higher number of samples in the diseased tissue category, and moreover more subtle differences between two types of non-cancerous tissues. In connection with these facts, the repeated average classification accuracies were reduced (73–86%, Fig. 3 and 4).
Overall, for the most numerous subset of spectra, the most potent classification strategy seemed to be SVM if the main objective is diagnosing the smallest amount of false positives or the imbalanced AdaBoost based on DTs, which enabled the capture of the polyps with a minimal amount of false negatives.
Classification of benign epithelial tumors and cancerous lesions
The last subset of data consisted of the in vivo spectra of two types of diseased colorectal tissue, benign adenomatous polyps (n = 70, 86.4%) and colorectal adenocarcinoma (n = 11, 13.6%). The evaluation metrics of the models’ performance are summarized in Table 4.| PCA–LDA | SVM | DT | DT AdaBoost | DT AdaBoost IB | SVM AdaBoost IB | |
|---|---|---|---|---|---|---|
| Accuracy | 1.000 | 0.975 | 0.963 | 0.951 | 0.802 | 0.802 |
| Sensitivity | 1.000 | 0.818 | 0.818 | 0.909 | 1.000 | 1.000 |
| Specificity | 1.000 | 1.000 | 0.986 | 0.957 | 0.771 | 0.771 |
| AUROC | 1.000 | 1.000 | 0.987 | 0.950 | 0.993 | 0.971 |
| Precision | 1.000 | 1.000 | 0.900 | 0.769 | 0.407 | 0.407 |
| Cohen's kappa | 1.000 | 0.886 | 0.803 | 0.729 | 0.375 | 0.375 |
| F1 score | 1.000 | 0.900 | 0.857 | 0.833 | 0.579 | 0.579 |
The number of principal components of the PCA–LDA model was chosen as 60 (99.9% variability). The resulting model was able to classify all the spectra in their proper diagnostic groups, thus achieving 100% sensitivity, specificity, and accuracy (Table 4).
The SVM model (C = 1, polynomial kernel) was fitted to the same dataset, but as not all of its performance characteristics reached the same levels as in the case of the PCA–LDA model, they should not be considered low. The SVM sensitivity of 81.8% and specificity of 100.0% resulted in an overall classification accuracy of 97.5% (Table 4).
The simple DT tree classifier (maximum depth = 3, minimum of samples per leaf = 3) also performed adequately for this dataset. The model classification accuracy amounted to 96.3% thanks to a higher specificity of 98.6% (Table 4).
The optimal combination of hyperparameters of a DT-based AdaBoost classifier was found by a grid search to be a maximum depth of DT of 3 and only 1 sample per leaf at the least. One hundred classifiers turned out to provide the best F1 score of the CV classification and were therefore selected for the final ensemble model. The resulting model enabled the discrimination of spectra of cancerous and non-cancerous lesions with a sensitivity, specificity, and overall accuracy of 90.9%, 95.7%, and 95.1% respectively (Table 4). The hyperparameters of the imbalanced version of the AdaBoost classifiers were found using a similar strategy. For the DT-based model, the combination consisted of the maximum depth of three and the minimum samples per leaf of one. The number of DTs was selected as 10. The SVM-based imbalanced model was calculated with five base classifiers (C = 1, polynomial kernel). The classification evaluation metrics for both models excelled in their sensitivity to cancerous lesions, both reached a level of 100.0% (Table 4).
In contrast, their specificity was only 77.1%, and accordingly, their accuracy was only 80.2% (Table 4).
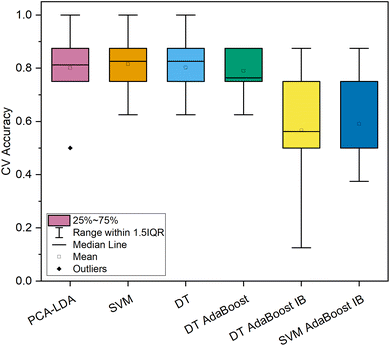
Assessing the repeated 10-fold CV classification accuracy of the spectral subset of cancerous and benign lesions of the colon and rectum (Fig. 5) it was discovered that the PCA–LDA, SVM, DT, and standard DT-based AdaBoost models showed comparable resulting accuracy scores (79.9–81.5% on average) even with a similar level of variance (Fig. 5), as opposed to the imbalanced machine learning models, the accuracy of which varied significantly among the 10 folds of CV and their mean accuracy did not exceed 60%.
Though the classification efficiency metrics are encouraging for all analyzed subsets of the data, it must be admitted, that the sample set of cancerous lesions is rather limited, therefore the dataset needs to be expanded in this direction. Otherwise, the significance of the classification might be considered low, as the obtained models can be prone to overfitting, usually to correctly classify the samples of the more numerous classes.41 Such a phenomenon was observed in the classification models of both data subsets containing the spectra of cancerous lesions. A difference was recognized in repeated CV accuracy and, moreover, in sensitivity. In contrast, the value of sensitivity was considerably higher in the models tackling the class imbalance – DT- and SVM-based AdaBoost for both datasets. The specificity and consequently also the classification accuracy and other performance characteristics of these complex models were lowered in comparison to traditional ones, such as PCA–LDA and SVM. We hypothesize that with an augmented dataset the difference in balanced and imbalanced model performance will be addressed. A sign confirming this assumption was observed for the dataset of in vivo Raman spectra of normal tissue and adenomatous polyps, the difference between the models’ sensitivities and specificities was not as variable.
Another limitation of our proposed diagnostic method presents the difficulty of collecting the spectra of less accessible lesions, for example behind a tissue fold in the direction of the proceeding endoscope during colonoscopy. Such lesions require highly skilled and experienced endoscopists for successful spectra collection. This practical problem, however, may be overcome with training motivated by the possibility of obtaining real-time diagnostic results.
Conclusions
We have demonstrated the efficiency of in vivo Raman spectroscopy combined with both traditional and novel ensemble machine learning methods for the real-time diagnostics of colorectal carcinoma and its precursor lesions – benign adenomatous polyps, which can be performed during a diagnostic colonoscopy without any significant extension of its duration.We have described the main features of the in vivo Raman spectra that contribute to the discrimination of the three diagnostic groups and assigned their originating biochemical changes in the tissue.
Using PCA–LDA, SVM, DT, DT-based AdaBoost, and DT- and SVM-based AdaBoost with adapted sampling, we have reached high levels of diagnostic accuracy (91.2% on average, 75.8–100%) for all analyzed subsets of data. What is more, the methods provided stable results for all the subsets; the average accuracy was 91.4% for the set of spectra of normal and cancerous tissue, 90.5% for normal and benign lesions, and 91.6% for cancerous and benign lesions. Such equally leveled results illustrate the potential of our approach in the field of instant endoscopic diagnostics, where it should be applied after an expansion of the dataset and a thorough multicenter validation.
Author contributions
Markéta Fousková: writing – original draft, conceptualization, software, validation, formal analysis, investigation, data curation, visualization, Jan Vališ: investigation, writing – review and editing Alla Synytsya: conceptualization, investigation, funding acquisition, writing – review and editing, Lucie Habartová: investigation, writing – review and editing, Jaromír Petrtýl: investigation, resources, Luboš Petruželka: conceptualization, supervision, funding acquisition, resources, Vladimír Setnička: supervision, resources, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grant no. NU20-09-00229 provided by the Ministry of Health of the Czech Republic.References
- H. J. Butler, L. Ashton, B. Bird, G. Cinque, K. Curtis, J. Dorney, K. Esmonde-White, N. J. Fullwood, B. Gardner, P. L. Martin-Hirsch, M. J. Walsh, M. R. McAinsh, N. Stone and F. L. Martin, Nat. Protoc., 2016, 11, 664–687 CrossRef CAS PubMed.
- C. Krafft and V. Sergo, Spectroscopy, 2006, 20, 195–218 CrossRef CAS.
- T. C. Hollon, B. Pandian, A. R. Adapa, E. Urias, A. V. Save, S. S. S. Khalsa, D. G. Eichberg, R. S. D'Amico, Z. U. Farooq, S. Lewis, P. D. Petridis, T. Marie, A. H. Shah, H. J. L. Garton, C. O. Maher, J. A. Heth, E. L. McKean, S. E. Sullivan, S. L. Hervey-Jumper, P. G. Patil, B. G. Thompson, O. Sagher, G. M. McKhann, R. J. Komotar, M. E. Ivan, M. Snuderl, M. L. Otten, T. D. Johnson, M. B. Sisti, J. N. Bruce, K. M. Muraszko, J. Trautman, C. W. Freudiger, P. Canoll, H. Lee, S. Camelo-Piragua and D. A. Orringer, Nat. Med., 2020, 26, 52 CrossRef CAS PubMed.
- D. Traynor, S. Duraipandian, R. Bhatia, K. Cuschieri, C. M. Martin, J. J. O'Leary and F. M. Lyng, J. Biophotonics, 2019, 12, e201800377 CrossRef PubMed.
- K. O'Dwyer, K. Domijan, A. Dignam, M. Butler and B. M. Hennelly, Cancers, 2021, 13, 4767 CrossRef PubMed.
- M. Paraskevaidi, J. M. Baker, J. H. Butler, J. H. Byrne, P. V. T. Chakkumpulakkal, L. Christie, S. Crean, P. Gardner, C. Gassner, G. S. Kazarian, K. Kochan, M. Kyrgiou, M. G. K. Lima, L. P. Martin-Hirsch, E. Paraskevaidis, S. Pebotuwa, A. J. Adegoke, A. Sala, M. Santos, J. Sulé-Suso, G. Tyagi, M. Walsh and B. Wood, Appl. Spectrosc. Rev., 2021, 56, 804–868 CrossRef CAS.
- E. Williams, J. C. Kong, P. Singh, S. Prabhakaran, S. K. Warrier and S. Bell, Aust. N. Z. J. Surg., 2021, 91, 2091–2096 CrossRef PubMed.
- A. Sud, M. E. Jones, J. Broggio, C. Loveday, B. Torr, A. Garrett, D. L. Nicol, S. Jhanji, S. A. Boyce, F. Gronthoud, P. Ward, J. M. Handy, N. Yousaf, J. Larkin, Y. E. Suh, S. Scott, P. D. P. Pharoah, C. Swanton, C. Abbosh, M. Williams, G. Lyratzopoulos, R. Houlston and C. Turnbull, Ann. Oncol., 2020, 31, 1065–1074 CrossRef CAS PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- C. Coghlin and G. I. Murray, Proteomics: Clin. Appl., 2015, 9, 64–71 CAS.
- X. Wu, S. Li, Q. Xu, X. Yan, Q. Fu, X. Fu, X. Fang and Y. Zhang, Jpn. J. Appl. Phys., 2021, 60, 067001 CrossRef CAS.
- B. Brozek-Pluska, J. Musial, R. Kordek and H. Abramczyk, Int. J. Mol. Sci., 2019, 20, 3398 CrossRef CAS PubMed.
- A. Synytsya, A. Vaňková, M. Miškovičová, J. Petrtýl and L. Petruželka, Diagnostics, 2021, 11, 2048 CrossRef CAS PubMed.
- E. Widjaja, W. Zheng and Z. Huang, Int. J. Oncol., 2008, 32, 653–662 CAS.
- A. Synytsya, M. Judexová, D. Hoskovec, M. Miškovičová and L. Petruželka, J. Raman Spectrosc., 2014, 45, 903–911 CrossRef CAS.
- S. Sato, R. Sekine, H. Kagoshima, K. Kazama, A. Kato, M. Shiozawa and J.-I. Tanaka, J. Anus Rectum Colon, 2019, 3, 84–90 CrossRef PubMed.
- R. Sekine, S. Sato, J.-I. Tanaka, H. Kagoshima, T. Aoki and M. Murakami, Showa Univ. J. Med. Sci., 2018, 30, 381–389 CrossRef.
- M. A. Short, W. B. Wang, I. T. Tai and H. S. Zeng, J. Biophotonics, 2016, 9, 44–48 CrossRef CAS PubMed.
- J. J. Wood, C. Kendall, J. Hutchings, G. R. Lloyd, N. Stone, N. Shepherd, J. Day and T. A. Cook, Colorectal Dis., 2014, 16, 732–738 CrossRef CAS PubMed.
- A. Molckovsky, L. Song, M. G. Shim, N. E. Marcon and B. C. Wilson, Gastrointest. Endosc., 2003, 57, 396–402 CrossRef PubMed.
- H. Ding, A. W. Dupont, S. Singhal, L. D. Scott, S. Guha, M. Younes, Y. Q. Ye and X. H. Bi, J. Raman Spectrosc., 2017, 48, 902–909 CrossRef CAS.
- M. S. Bergholt, K. Lin, J. F. Wang, W. Zheng, H. Z. Xu, Q. W. Huang, J. L. Ren, K. Y. Ho, M. Teh, S. Srivastava, B. Wong, K. G. Yeoh and Z. W. Huang, J. Biophotonics, 2016, 9, 333–342 CrossRef CAS PubMed.
- G. Sheehy, F. Picot, F. Dallaire, K. Ember, T. Nguyen, K. Petrecca, D. Trudel and F. Leblond, J. Biomed. Opt., 2023, 28, 025002 Search PubMed.
- G. Van Rossum and F. Drake Jr., Python reference manual, 1995 Search PubMed.
- W. McKinney , Proceedings of the 9th Python in Science Conference, 2010, 445, 56–61.
- The pandas development team, pandas-dev/pandas: Pandas, 2021, DOI:10.5281/zenodo.3509134.
- C. R. Harris, K. J. Millman, S. J. van der Walt, R. Gommers, P. Virtanen, D. Cournapeau, E. Wieser, J. Taylor, S. Berg, N. J. Smith, R. Kern, M. Picus, S. Hoyer, M. H. van Kerkwijk, M. Brett, A. Haldane, J. F. del Río, M. Wiebe, P. Peterson, P. Gérard-Marchant, K. Sheppard, T. Reddy, W. Weckesser, H. Abbasi, C. Gohlke and T. E. Oliphant, Nature, 2020, 585, 357–362 CrossRef CAS PubMed.
- P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser, J. Bright, S. J. van der Walt, M. Brett, J. Wilson, K. J. Millman, N. Mayorov, A. R. J. Nelson, E. Jones, R. Kern, E. Larson, C. J. Carey, İ. Polat, Y. Feng, E. W. Moore, J. VanderPlas, D. Laxalde, J. Perktold, R. Cimrman, I. Henriksen, E. A. Quintero, C. R. Harris, A. M. Archibald, A. H. Ribeiro, F. Pedregosa, P. van Mulbregt, A. Vijaykumar, A. P. Bardelli, A. Rothberg, A. Hilboll, A. Kloeckner, A. Scopatz, A. Lee, A. Rokem, C. N. Woods, C. Fulton, C. Masson, C. Häggström, C. Fitzgerald, D. A. Nicholson, D. R. Hagen, D. V. Pasechnik, E. Olivetti, E. Martin, E. Wieser, F. Silva, F. Lenders, F. Wilhelm, G. Young, G. A. Price, G.-L. Ingold, G. E. Allen, G. R. Lee, H. Audren, I. Probst, J. P. Dietrich, J. Silterra, J. T. Webber, J. Slavič, J. Nothman, J. Buchner, J. Kulick, J. L. Schönberger, J. V. de Miranda Cardoso, J. Reimer, J. Harrington, J. L. C. Rodríguez, J. Nunez-Iglesias, J. Kuczynski, K. Tritz, M. Thoma, M. Newville, M. Kümmerer, M. Bolingbroke, M. Tartre, M. Pak, N. J. Smith, N. Nowaczyk, N. Shebanov, O. Pavlyk, P. A. Brodtkorb, P. Lee, R. T. McGibbon, R. Feldbauer, S. Lewis, S. Tygier, S. Sievert, S. Vigna, S. Peterson, S. More, T. Pudlik, T. Oshima, T. J. Pingel, T. P. Robitaille, T. Spura, T. R. Jones, T. Cera, T. Leslie, T. Zito, T. Krauss, U. Upadhyay, Y. O. Halchenko and Y. Vázquez-Baeza, Nat. Methods, 2020, 17, 261–272 CrossRef CAS PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss and V. Dubourg, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- G. Lemaître, F. Nogueira and C. K. Aridas, J. Mach. Learn. Res., 2017, 18, 559–563 Search PubMed.
- M. S. Bergholt, W. Zheng, K. Lin, J. F. Wang, H. Z. Xu, J. L. Ren, K. Y. Ho, M. Teh, K. G. Yeoh and Z. W. Huang, Anal. Chem., 2015, 87, 960–966 CrossRef CAS PubMed.
- J. Grdadolnik, Vib. Spectrosc., 2003, 31, 279–288 CrossRef CAS.
- K. W. Short, S. Carpenter, J. P. Freyer and J. R. Mourant, Biophys. J., 2005, 88, 4274–4288 CrossRef CAS PubMed.
- H. J. Byrne, K. M. Ostrowska, H. Nawaz, J. Dorney, A. D. Meade, F. Bonnier and F. M. Lyng, in Optical Spectroscopy and Computational Methods in Biology and Medicine, ed. M. Baranska, Springer Netherlands, Dordrecht, 2014, 355–399 Search PubMed.
- G. Socrates, Infrared and Raman Characteristic Group Frequencies: Tables and Charts, Wiley, 2004 Search PubMed.
- W. Szlasa, I. Zendran, A. Zalesińska, M. Tarek and J. Kulbacka, J. Bioenerg. Biomembr., 2020, 52, 321–342 CrossRef CAS PubMed.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2007, 42, 493–541 CrossRef CAS.
- I. U. Rehman, Z. Movasaghi and S. Rehman, Vibrational Spectroscopy for Tissue Analysis, Taylor & Francis Group, Baton Rouge, USA, 2012 Search PubMed.
- A. Rygula, K. Majzner, K. M. Marzec, A. Kaczor, M. Pilarczyk and M. Baranska, J. Raman Spectrosc., 2013, 44, 1061–1076 CrossRef CAS.
- J. Depciuch, B. Klębowski, M. Stec, R. Szatanek, K. Węglarczyk, M. Baj-Krzyworzeka, M. Parlińska-Wojtan and J. Baran, Int. J. Mol. Sci., 2020, 21, 1826 CrossRef CAS PubMed.
- Q. Kang, L. Shi, M. Zhou, X. Wang, Q. Wu and Z. Wei, IEEE Trans. Neural Networks Learn. Syst., 2018, 29, 4152–4165 Search PubMed.
| This journal is © The Royal Society of Chemistry 2023 |