Open Access Article
Open Access ArticleDNA compaction enhances the sensitivity of fluorescence-based nucleic acid assays: a game changer in point of care sensors?†
Sujesh
Sudarsan‡
a,
Anusha
Prabhu‡
a,
Dinesh
Prasad
 b and
Naresh Kumar
Mani
b and
Naresh Kumar
Mani
 *ac
*ac
aMicrofluidics, Sensors and Diagnostics (μSenD) Laboratory, Department of Biotechnology, Manipal Institute of Technology, Manipal Academy of Higher Education, Manipal, Karnataka 576104, India. E-mail: naresh.mani@manipal.edu; maninaresh@gmail.com
bDepartment of Bioengineering and Biotechnology, Birla Institute of Technology, Mesra, Ranchi 835215, India
cManipal Centre for Infectious Diseases, Prasanna School of Public Health, Manipal Academy of Higher Education, Manipal, Karnataka 576104, India
First published on 1st April 2023
Abstract
Fluorescence-based nucleic acid assays frequently exhibit a feeble signal at low analyte concentrations, necessitating complex, expensive methods such as the development of sequence-specific oligo tags, molecular beacons, and chemical modifications to maintain high detection sensitivity. Hence, there is growing interest in accomplishing fluorescence enhancement in nucleic acid assays using robust and cost-effective strategies. The study exploits the use of two compaction agents, PEG 8000 and CTAB, to compact the ITS-2 amplicon of the fungus Candida albicans and evaluates the effect of both of these agents on the fluorescence intensity of SYTO-9 labelled nucleic acids. Conventional fluorometric measurements showed that both CTAB and PEG 8000 enhanced the emission intensity by ∼1.2- and 2-fold, respectively. Furthermore, we leveraged paper-based spot tests and distance-based assays to validate the effect of DNA compaction for enhancing sensitivity in the point-of-care context. The spot assay performed on paper with compacted samples showed an increase in the emission intensity of SYTO-9 and this was manifested by an elevated G channel intensity in the order of PEG 8000 compacted > CTAB compacted > amplified. Moreover, in the distance-based assay, the PEG 8000 compacted sample was found to migrate farther compared to CTAB compacted and amplified DNA samples at amplicon concentrations, 15 μg ml−1 and 39.65 μg ml−1. The limit of detection (LOD) for PEG 8000 and CTAB compacted samples on both paper-spot and distance-based assays were found to be 0.4 μg ml−1 and 0.5 μg ml−1, respectively. Hence our work provides an overview of employing DNA compaction as an approach for enhancing the sensitivity of fluorescence-based point-of-care nucleic acid assays without the need for cumbersome sensitivity enhancement methods.
Introduction
Several investigations undertaken over the past 16–25 years have revealed an increase in the prevalence of infectious diseases worldwide.1 Although they significantly affect millions of lives every year, the impact of these infections on human well-being is often overlooked. Over 150 million severe instances of infections occur each year worldwide, resulting in nearly 1.7 million fatalities.2 The mortality rate associated with these infections beats 50%, despite the availability of multifarious medicines, which is of special concern. Aside from the emergence of such infectious diseases, it is anticipated that over 3 million individuals globally are exposed to fungal infections caused by multiple strains of Candida.3 The opportunistic fungal infection, candidiasis, is prevalent in women, preterm neonates, and immunocompromised individuals. Candidiasis may exist either as superficial infections such as vaginal or oral candidiasis,4 or invasive candidiasis such as candidemia5 or disseminated candidiasis.2 Though the discovery of anti-microbial agents was one of the major advances in medicine in the 20th century, delayed disease detection and the growth of drug-resistant pathogen populations have created a significant global health issue. Pathogen detection is, therefore, more important than ever for proper disease management and related treatments.6The conventional methods for detecting microorganisms primarily depend on culture-based techniques, where the samples are isolated and cultured in selective media.7 The cultured samples are then examined under a microscope or tested biochemically. Although the aforementioned techniques are inexpensive, reliable, and effective, they are less sensitive and demand expert interventions and long assay stints.7 However, many of such drawbacks of conventional methods can be resolved by molecular-based methods, particularly if they depend on polymerase chain reaction (PCR).8 PCR-based assays are significantly faster and more precise than conventional methods. But the inherent drawbacks of using PCR include the requirement of an expensive and sophisticated thermal cycler, as it requires a precise control of temperature over multiple cycles of denaturation, annealing, and extension, inhibition from various substances in the reaction mixture, high chances of getting false positives due to non-specific binding of primers, prolonged turnaround times and requirement of trained professionals.9
Recently, the benefits of Loop-Mediated Isothermal Amplification (LAMP) on PADs are opening new avenues for point-of-care nucleic acid testing in regions where resources are scarce.10 LAMP has emerged as a low-cost alternative to PCR simplifying the hardware requirements while enabling visual detection of amplicons. It is highly specific as it involves five distinct primers and extends the target sequence by a specific strand displacing enzyme, “Bst DNA polymerase”, at a steady temperature of 60–65 °C for 1–2 h.6 Furthermore, the method is easily deployable, because it eliminates the need for a thermal cycler and enables the amplification to be conducted in a dry block heater or incubator. To date, several in vitro studies have demonstrated that LAMP amplification could detect low copies of the target sequence (∼1–10 copies of DNA).11 Furthermore, it has also been reported that LAMP is more tolerant than PCR to the inhibitors present in the sample, indicating that LAMP may be a more suitable amplification technique for samples with potential inhibitor contamination.12,13
Despite the advantages of LAMP, its heavy dependency on indirect detection techniques, such as turbidity and non-specific dyes, often leads to false positive results.14 To overcome these limitations, researchers have investigated the sequence-specific detection of LAMP amplicons by adding DNA intercalating fluorescent or colorimetric dyes straight into the tubes.15 Different pathogenic microbes including bacteria, fungi, viruses, and parasites have been detected using real-time LAMP integrated with fluorescent and colorimetric dyes. In a study conducted by Cao et al., the authors described the development of a rapid, sensitive, and quantitative RealAmp method for U. maydis detection using the dye, SYTO-9.16 The detection sensitivity of the RealAmp assay was 200 times higher than that of detection through conventional PCR.16 Yang et al. have reported a LAMP method developed for the detection of astrovirus using an in vitro RNA transcript with the colorimetric dye, hydroxynaphthol blue (HNB).17 The detection limit of LAMP using the in vitro RNA transcript was 3.6 × 10 copies per μL which is as sensitive as PCR. The bioluminescent real-time reporter (BART) of LAMP and a smartphone were recently integrated to quantify the Zika virus and spatiotemporally map the illness.18
Although there have been various reports suggesting that fluorescence-based LAMP amplification assays are robust and reproducible in detecting various pathogens, these methods encounter many challenges. The major drawbacks of fluorescence-based LAMP assays include fluorescence quenching by the components in solution,19 photon-induced chemical damage to the fluorophores (photobleaching)20 and scattering of excitation light leading to minimal signal readouts.21 However, researchers have tried different alternatives to enhance the fluorescence readout emerging as a signal from LAMP amplification. By integrating LAMP, Ding et al. presented a cleavable, energy-transfer-tagged probe for rapid, sensitive, and accurate nucleic acid detection.18 The developed Cleavable Hairpin Beacon (CHB) probe is a single fluorophore-tagged oligo comprising 5 consecutive ribonucleotides that are cleaved by ribonuclease to begin DNA amplification and generate enhanced fluorescence signals.18 A ribonuclease-dependent cleavable beacon primer (CBP) enabling the fluorescence LAMP recognition of single nucleotide mutation was earlier developed by Ding et al.18 The aforementioned approaches are expensive, tedious, labour intensive and demand long assay time, and they are also prone to failures due to non-specific oligo preparations, primer dimer formation, stability issues, etc.
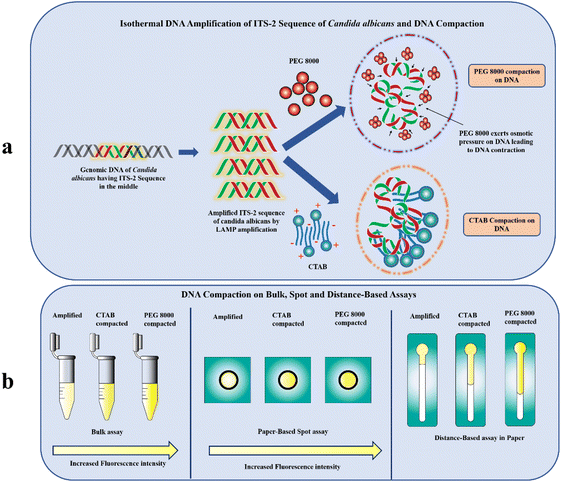
In this paper, we have explored the process of “compaction”, a transition between an elongated form and a concealed arrangement of nucleic acid strands (Fig. 1a). Though compaction of nucleic acids has been performed primarily on the single molecule level22–25 to demonstrate different transcriptional processes such as gene therapy, in vitro gene regulation and DNA manipulation (as a gene protection strategy to generate nanomaterials with well-defined size),26 its significance in improving the sensitivity of fluorescently-labelled nucleic acid-based assays on Microfluidic Paper-based Analytical Devices (μPADs) has not been explored by the researchers. Our study exploits the use of two compaction agents ‘PEG 8000’ and ‘CTAB’ to compact the ITS-2 LAMP amplicon of the fungus Candida albicans and evaluates the effect of the compaction on the fluorescence intensity of SYTO-9 labelled amplicons using simple paper-based spot assay (Fig. 1b). The rationalization behind the enhanced fluorescence by CTAB is attributed to the cooperative binding interaction of the cationic surfactant with DNA.27 A lower surfactant concentration is preferred for DNA compaction as it accounts for a change in the physico-chemical factor that eventually promotes surfactant aggregation around the SYTO-9 labelled DNA.27 When the CTAB concentration is close to the critical micelle concentration (CMC), the aggregated/micellized CTAB combines with the DNA to produce DNA/CTAB compact aggregates.28 These aggregates alter the local environment of SYTO-9 labelled DNA restricting its free movement inside the condensed complex. The fluorescence decay from the excited vibrational levels of SYTO-9 is reduced leading to enhanced fluorescence.
Another possible method for compacting DNA is triggering unfavourable interactions between DNA monomers and the solvent by lowering the solvent's dielectric constant.29 The compaction of this kind proceeds through limiting the volume accessible to DNA (crowding effect) by neutral polymers such as PEG and there is no intermediate formed between the coil–globule transition otherwise known as the all-or none mode of compaction.29 The ensemble clustering of PEG in the vicinity of DNA, would result in their abrupt contraction29 and eventually stable state of the fluorescent probe. This when excited with light having specific excitation maxima would give rise to an enhanced emission intensity with much lesser decay (Fig. 1b). Further, we have developed distance-based assays on paper to evaluate the effect of compaction on the migration distance of the SYTO-9 labelled DNA amplicons (Fig. 1b). This variation in the distance is due to the interfacial interaction of SYTO-9 with the cellulose surface.30 Strikingly, this approach paves the way for developing a simple and easy-to-use fluorescence intensification strategy utilizing DNA compaction. To the best of our knowledge, this is the first study employing DNA compaction for the sensitivity enhancement of fluorescently labelled nucleic acid assays on paper-based diagnostics.
Results and discussion
DNA extraction from Candida albicans and LAMP amplification of the ITS-2 region
In the present study, the genomic DNA from Candida albicans was extracted using the Trizol reagent for the LAMP amplification of the ITS-2 sequence. The method was found to be reliable and robust compared to the HipurA kit-based nucleic acid extraction method (Table 1). The mean concentration of the extracted DNA from Candida albicans was 35.56 μg ml−1 with the mean absorbance at 260 nm being 0.472. Similarly, the purity of the DNA extracted from the fungus was determined to be 1.81. The quality of amplified products was further assessed using gel electrophoresis (ESI Fig. 3†) and a UV-visible spectrophotometer (ESI Fig. 4†) Fig. 2 depicts the DNA Ladder (lane 1) and two amplified ITS-2 sequence of C. albicans (lane 2 and lane 3). Besides, the mean concentration measured for the amplicon using a UV-visible spectrophotometer revealed that there was a 55-fold increase in the concentration of ITS-2 (ESI Fig. 4†). Both the results corroborate an effective amplification of the target sequence of C. albicans.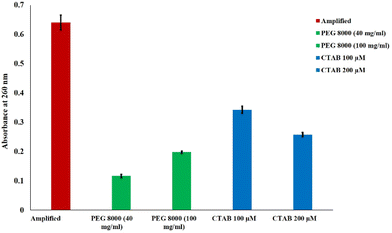 | ||
| Fig. 2 Absorbance measured for amplified, PEG 8000 (40 mg ml−1) compacted, PEG 8000 (100 mg ml−1) compacted, CTAB (100 μM) compacted and CTAB (200 μM) compacted ITS-2 sequence of Candida albicans. | ||
| Method of DNA extraction | Concentration (μg ml−1) | Purity |
|---|---|---|
| HipurA yeast genomic DNA extraction | 19.6 | 1.64 |
| Trizol method | 35.56 | 1.81 |
Estimating the interaction between DNA and condensation agents using absorbance measurements
Due to the existence of phosphate groups, DNA is regarded as a polyelectrolyte with a strong negative charge.31 Its structure is the result of hydrophobic and electrostatic interactions between various residues within them.32 Upon mixing with neutral polymers such as PEG and the cationic surfactant such as CTAB, DNA changes from a semiflexible coil to a more compacted state through a process known as DNA compaction.25,26,33–35 The double helix is folded during this process with a loss of conformational entropy, electrostatic repulsions between negatively charged phosphates in the DNA and strong compaction.36,37 The rationalization behind the DNA compaction by the neutral polymer PEG is that, highly charged DNA in aqueous solutions is thought to make thermodynamically unfavourable interactions with the polymer.38 As a result, when PEG is added, the solvent character for DNA alters, initiating the attraction between different segments of DNA.39 As a consequence, the abrupt contraction of the DNA coil, known as, “coil–globule transition” takes place at a critical PEG concentration.40Similarly, compaction induced by CTAB has been demonstrated by Dias et al. According to Dias et al., the polarity of the hydrophilic headgroup of a majority of the cationic complexes is monovalent, and it is not possible to induce DNA compaction by a single surfactant molecule.41 However, due to their amphiphilic nature, the self-assembly of surfactants occurs in the proximity of DNA and causes DNA to be compacted41 The surfactants generate micellar complexes due to hydrophobic interactions on the surface of nucleic acids at a particular surfactant concentration known as the critical association concentration (CAC).27 It has been known that the interaction of the cationic surfactant with DNA is highly cooperative in nature. Therefore, when CTAB is added, the surfactant molecules self-assemble on the DNA leading to the formation of a nucleation core due to attractive interactions between various DNA components that expands along the molecular chain resulting in the polymer being compacted by ion correlation effects (ESI Fig. 1†).42
In this work, UV absorbance studies were performed to confirm the DNA compaction by the neutral polymer, PEG 8000 and the cationic surfactant, CTAB. Two concentrations of CTAB (100 μM and 200 μM) and PEG 8000 (40 mg ml−1 and 100 mg ml−1) were considered for the compaction studies. Fig. 2 presents the UV-visible spectrophotometric results of 39.65 μg ml−1 amplicon, CTAB compacted amplicon, and PEG 8000 compacted amplicon. The absorbance measured for the amplicon was 0.624, whereas the samples compacted by PEG 8000 at concentrations of 40 mg ml−1 and 100 mg ml−1 were found to be 0.117 and 0.198 respectively. Furthermore, samples treated with CTAB at concentrations of 100 μM and 200 μM showed an absorbance of 0.343 and 0.258 respectively. The observation from these experiments is the dramatic reduction in the recorded absorbance for the compacted samples compared to the control having no compaction agents.43 This observation is in agreement with the previous study conducted by Carlstedt et al.44 The observed decrease in the absorbance measured at 260 nm in the case of CTAB compacted samples is attributed to the self-assembly of the CTAB micelles in the proximity of the DNA, which makes the UV light inaccessible for the conjugated double bonds in the core of the compacted complex.45 Similarly, in the presence of the neutral polymer PEG 8000, the double-stranded DNA is compacted into tight condensates due to poor solvent quality, which hinders the absorption of UV light by conjugated double bonds in the purine and pyrimidine rings of the DNA situated at the core of the condensate.45 We also noticed that the absorbance measured for the amplicon treated with 100 mg ml−1 of PEG 8000 was slightly high (0.198) compared to the amplicon treated with 40 mg ml−1 of PEG 8000 (0.117). This increase in the absorbance represents the decondensation behaviour of amplicons from the developed condensates at a higher polymer concentration.46 Based on the absorbance results, the selected concentrations of PEG 8000, and CTAB for fluorescence sensitivity enhancement utilizing compaction were 40 mg ml−1 and 200 μM, respectively.
Fluorescence-based measurement of DNA compaction by PEG 8000 and CTAB
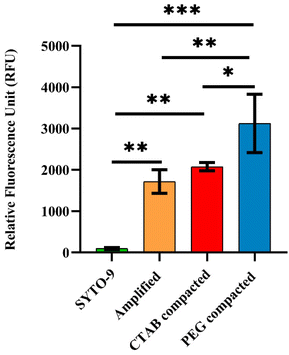
Conventional fluorometric measurement was carried out in a 96-well plate using a microplate reader to evaluate the effect of PEG 8000 and CTAB on the fluorescence intensity of the SYTO-9 labelled amplicon. The relative fluorescence intensities of compacted samples were compared to the control having 39.65 μg ml−1 of the amplicon (Fig. 3). It is evident from Fig. 3 that the relative fluorescence intensity (RFU) of SYTO-9 is 100 RFU and that for the amplicon without any compaction agents is 1715.76 RFU. Whereas, the relative fluorescence intensities of CTAB and PEG 8000 compacted amplicon were 2077.29 RFU and 3124 RFU respectively.We speculate that the reason for the enhancement in the fluorescence intensity (∼1.2-fold) upon compaction with CTAB is attributed to its interaction with SYTO-9 labelled DNA. It is well-known that the cationic surfactant CTAB self assembles into complexes of nanoscopic dimensions in the vicinity of DNA.42 When the CTAB concentration is close to the critical micelle concentration (CMC), the aggregated/micellized CTAB reacts with DNA to form DNA–CTAB compact aggregates47 altering the local environment of SYTO-9 labelled DNA. The fluorescence decay from the excited vibrational levels of SYTO-9 is therefore reduced because the free movement of SYTO-9 labelled DNA is constrained inside the condensed complex.47 The improvement in the emission intensity is the result of a more stabilized excited state of SYTO-9 in the proximity of CTAB.47
According to Grueso et al., when a surfactant such as CTAB is added to ethidium bromide labelled DNA, displacement of the probe occurs from its position between the bases of the polynucleotide to the aqueous solvent.48 This movement is detected by a decrease in the emission intensity of the dye. However, contrary to the widely accepted dye displacement theory, since SYTO-9 possesses high affinity to DNA through multiple binding modes including charge interactions with the phosphate backbone, intercalation and binding to the groove of the double helix, is not displaced upon addition of cationic surfactants.49 Moreover, the rigid nature of SYTO-9, when bound to double-stranded DNA contributes to an enhanced fluorescence.50
Upon treating SYTO-9 labelled DNA amplicon with PEG 8000, we have noticed a 2-fold increase in the fluorescence intensity. The fluorescence intensity attained through compaction by PEG 8000 (p < 0.01) was found to be statistically significant compared to the control. Generally, nucleic acids maintain their coil shape when PEG levels are low, and they compact into a small globular conformation when PEG levels are elevated.51 The reason for the compaction of DNA macromolecules in the PEG solution is attributed to the effect of the immiscibility of stiff chains (DNA) and flexible chains (PEG), known as “stiff-flexible chain incompatibility” which was first proposed in the paper of Flory and Abe.52 For stiff polyelectrolytes such as DNA, the addition of flexible polymers such as PEG induces perfect segregation between the polymer and the DNA.53 The ensemble clustering of PEG molecules around the DNA exerts an osmotic pressure on the DNA making it to be contracted53 which in turn, stabilizes the labelled SYTO-9 on DNA. The resulting stable state of the probe, when excited with light having specific excitation maxima, emits fluorescence with enhanced intensity and lesser fluorescence decay.
To confirm that fluorescence enhancement is due to DNA compaction and not due to any artifacts, we have also measured the fluorescence of PEG 8000 and CTAB in the presence of SYTO-9 (without DNA amplicons) and the results revealed a less fluorescence intensity of 201.23 RFU and 84.06 RFU respectively (ESI Fig. 2†). These findings are in line with the fact that the fluorescent probe SYTO-9 does not produce fluorescence with the surfactants in the absence of amplicons.49 However, when analysing the fluorescence of control samples, PEG 8000 containing the dye, SYTO-9 showed a slightly elevated peak. The result indicated the emissive behaviour of PEG 8000 owing to the presence of lone pairs of electrons in the heteroatoms (Oxygen) of PEG 8000. The delocalization of electrons organizes as clusters or vesicles resulting in the overlapping of electron clouds, lowering the energy gaps and forming more rigidified conformations leading to fluorescence emission as per the Clusterization Triggered Emission (CTE) principle.54
Spot assay
To validate the utility of DNA compaction mediated fluorescence enhancement for point-of-care applications, we leveraged paper-based spot assays. In the last several years, different kinds of μPADs have been exploited for the colorimetric detection of species ranging from small molecules to biological macromolecules.55 The method is widely accepted due to its ease of operation and clear signal output, but the practical use of this method is limited by the high chance of contamination leading to false positives,56 users viewpoint and colour perception to decide the diagnosis,56 interference of comorbidities in the colour formation, low sensitivity57 and strong reliance on temperature and pH.56 Nevertheless, fluorescence-based nucleic acid assays are yet another optical method, which exhibits inherently much higher sensitivity than colorimetric methods. In addition, these methods possess high specificity due to the unique optical properties of the probe, better signal to noise ratio (SNR) and ability to measure the analyte concentration by means of fluorescence intensity and decay times.58 However, it is very difficult to perform fluorometric measurements for low concentrations of nucleic acids directly on μPADs for two reasons. Firstly, aggregates formed during paper processing cause an uneven distribution of scattering centres, which results in a minimal excitation of fluorescent probes and eventually minimal signal readout.57 Secondly, LED-based transilluminators, which researchers use for the excitation of nucleic acid probes on μPADs are not as efficient as monochromators. Consequently, the aforementioned limitations of the fluorescence assay on μPADs extremely demand the requirement of cost-effective sensitivity enhancement approaches.Our study incorporates “DNA compaction” to enhance the fluorescence intensity of nucleic acid assays on μPADs. From the spot assay given in Fig. 4, it is evident that compacted amplicons show increased fluorescence compared to the control sample. The increasing order of fluorescence intensity observed for different samples in the spot assay is PEG 8000 compacted amplicon > CTAB compacted amplicon > amplified > amplified control. These findings would further open the door to the development of low-cost, highly sensitive, and specific devices with a rapid fluorescence readout. Additionally, this strategy could be used to overcome the challenges associated with performing fluorometric experiments on μPADs for low nucleic acid concentrations, due to the scattering of the excitation light emerging from the light source owing to the opaque nature of paper and the inadequate excitation capacity of blue-light transilluminators. On that account, it allows the researchers to perform these assays even with least possible fluorescence excitation settings.
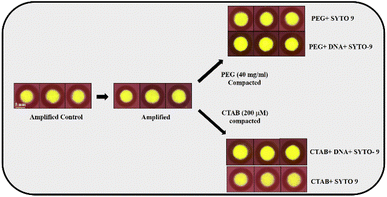 | ||
| Fig. 4 Spot assay performed on amplified, PEG 8000 (40 mg ml−1) compacted, and CTAB (200μM) compacted samples containing 39.65 μg ml−1 of amplicons. | ||
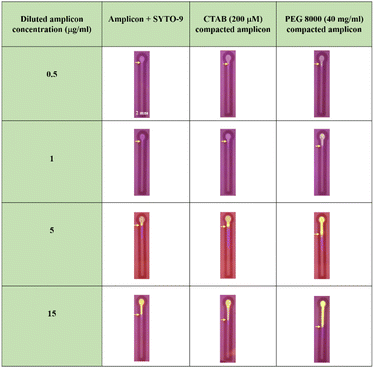
To assess the key impact of compaction on different concentrations of amplicons, PEG 8000 (40 mg ml−1) and CTAB (200 μM) were added in the presence of the fluorescent probe SYTO-9. Spot assay images of the amplified, CTAB compacted and PEG 8000 compacted samples containing different concentrations of amplicons are shown in Fig. 5. A colour change, from blue to green by the fluorescent probe SYTO-9 upon exposure to the blue-light transilluminator was observed for three different samples. Interestingly, the compacted samples by CTAB and PEG 8000 showed increased fluorescence in all the chosen concentrations of amplicons compared to the control.
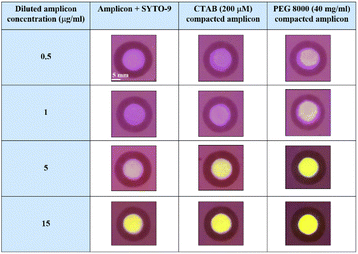 | ||
| Fig. 5 Spot assay performed on amplified, PEG 8000 (40 mg ml−1) compacted, and CTAB (200μM) compacted samples containing varying concentrations of amplicons ranging from 0.5 μg ml−1–15 μg ml−1. | ||
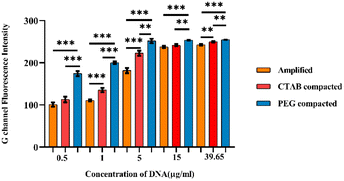
Further, G-channel intensities of all the samples were plotted as a graph shown in Fig. 6. In all the amplicon concentrations considered for the study, the PEG 8000 compacted amplicon showed better fluorescence followed by the CTAB compacted and amplified sample. The fluorescence intensity exhibited by the CTAB compacted amplicon was found to be statistically significant at amplicon concentrations 1 μg ml−1 (p < 0.001), 5 μg ml−1 (p < 0.001) and 39.65 μg ml−1 (p < 0.01), whereas the PEG 8000 compacted amplicon showed statistical significance at all the selected concentrations of amplicons. Besides, in lower concentrations of amplicons (i.e., at 0.5 μg ml−1, 1 μg ml−1, and 5 μg ml−1) the difference in fluorescence intensities between these three samples was considerably higher compared to higher concentrations (15 μg ml−1 and 39.65 μg ml−1). The reason for this minute difference in the intensity at higher amplicon concentrations is attributed to the extent of scattering of the light source during its passage through cellulose paper and inefficiency of the transilluminator to excite the labelled probe. This was manifested by a constant peak observed at amplicon concentrations, 15 μg ml−1 and 39.65 μg ml−1. Additionally, the limit of detection (LOD) of our developed paper-based spot assay was determined based on the visibility of the colour exhibited by lower concentrations of compacted amplicon samples in the presence of SYTO-9. The LODs were found to be 0.4 μg ml−1 and 0.5 μg ml−1 for PEG 8000 compacted and CTAB compacted amplicons respectively (ESI Fig. 5†). In a study conducted by Vesty et al., different DNA extraction techniques were performed for the saliva samples of patients with mild sub and supragingival dental plaque caused by pooled fungal communities.59 Based on the normalized qubit concentrations measured for pooled DNA samples extracted using different methods, the concentrations of Candida DNA in the saliva were found in the range of 0.05 μg ml−1–11 μg ml−1 (>50% of the total DNA concentration obtained for the pooled fungal samples contain DNA of C. albicans).59 As the LODs of our developed paper-based spot assay device fall within the mid-range of these clinical concentrations, these devices can be used to diagnose moderate to severe Candida infections.
Distance-based assay
In resource-constrained settings, distance-based nucleic acid assays using microfluidic paper-based analytical devices (μPAD) and other cellulose-based assays60–66 offer great potential for the rapid and early diagnosis of diseases and detecting analytes. These devices measure the distance as a digital readout to quantify the concentration of nucleic acids.67 In this work a microfluidic channel on paper was created using the inkjet printing of paraffin, a hydrophobic substance, that upon heating enters and blocks the perforations of the cellulose paper. We have created a hydrophilic channel (2 mm width) formed between two imprinted hydrophobic lines. Upon introduction, the sample wicks up into the hydrophilic channels due to the capillary wicking action. Fig. 7 depicts the distance-based assay performed for the control SYTO-9 (control), 39.65 μg ml−1 of amplicons, CTAB compacted amplicon, and PEG 8000 compacted amplicon. As evident from the figure, the migration distance of samples increases from left to right.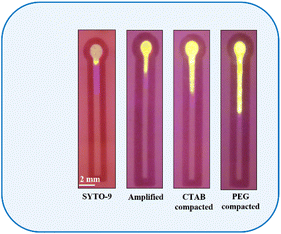 | ||
| Fig. 7 Distance travelled by SYTO-9, amplified, CTAB (200 μM) compacted and PEG 8000 (40 mg ml−1) compacted samples containing 39.65 μg ml−1 of the amplicon. | ||
The main reason behind the change in the migration distance is due to the interfacial interaction of fluorophores specifically electrostatic interaction, hydrogen bonding and non-specific adsorption with cellulose surface.30 SYTO-9 is a fluorescent dye which is categorized as “unsymmetrical cyanine dyes”. These compounds strongly bind to double-stranded DNA in multiple ways, including charge interactions with the phosphate backbone, intercalation, and adhering to the grooves of the DNA double helix.50 Moreover, the structures of these dyes are flexible in aqueous solutions but rigid when attached to nucleic acids.50 Hence, the binding affinity of SYTO-9 to double-stranded DNA is high compared to that of ethidium bromide and SYBR Green 1.
From Fig. 7, it is inferred that, in the absence of double-stranded amplicon, SYTO-9 exhibits a firm electrostatic attraction and hydrogen bonding on the cellulose surface, resulting in the strong retention of the dye at the sample loading zone leading to a reduced migration distance. However, when double-stranded amplicons are added, due to the strong intercalation of SYTO-9 to the amplicon, the retention ability of SYTO-9 on the cellulose is reduced leading to a longer migration distance. Alternatively, upon the introduction of CTAB to DNA, the CTAB forms rod-like micelles through self-assembly, resulting in the formation of a flexible CTAB–DNA complex. The high DNA stiffness as a result of this complex formation and the fact that CTAB forms rod-like micelles in the vicinity of DNA, the interaction between the dye, SYTO-9, and the cellulose is hindered41 Thus, the dye will be transported through the hydrophilic channel. Similarly, the rationale for the migration distance observed for PEG 8000 treated amplicon is described by the segregation of PEG 8000 molecules from DNA strands.53 The segregated PEG 8000 imposes an additional osmotic pressure leading to an abrupt contraction of the DNA to its globular state with an ensemble clustering of neutral polymer molecules around it.39 This hinders the attractive interaction between the dye, SYTO-9, and the cellulose surface leading to the longest migration distance. Additionally, to investigate the effect of compaction agents on the migration distance, we introduced CTAB and PEG 8000 into a distance-based channel in the presence of SYTO-9 as shown in Fig. 8. Our observations revealed that, both CTAB and PEG 8000 resulted in a reduced migration distance compared to the test samples, indicating that this effect was solely due to the association of agents with the fluorophore-labelled DNA.
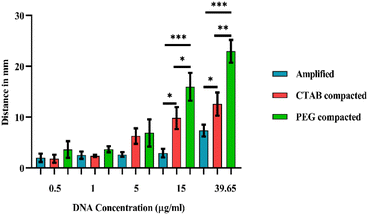
Further, PEG 8000 (40 mg ml−1) and CTAB (200 μM) were added to different concentrations of SYTO-9 labelled amplicons (in the distance-based channel) to evaluate the major effects of compaction on the sample migration distance. Strikingly, at amplicon concentrations, 15 μg ml−1 and 39.65 μg ml−1, PEG 8000 compacted amplicons were moved farther compared to CTAB compacted and amplified sample (Fig. 9). In contrast, the compacted samples did not exhibit a significant increase in the migration distance compared to the control at lower concentrations of amplicons (0.5 μg ml−1, 1 μg ml−1 and 15 μg ml−1). Based on the graph represented in Fig. 10, it is also evident that the migration distance of all three samples (amplified, CTAB compacted, and PEG 8000 compacted) from the sample loading zone increases with the increasing concentration of DNA.
This further indicates, when DNA concentration is high in the solution, the fluorescent probe, SYTO-9 adheres to a large number of double-stranded amplicons. This, in turn, alleviates the interaction of SYTO-9 with cellulose and moves through the hydrophilic channel. In addition, the effect of compaction on DNA further promotes this process by obstructing all the possibilities of confinement of the dye on the cellulose surface. The statistical analysis performed on the migration distance covered by amplified, CTAB compacted and PEG 8000 compacted samples has shown that, CTAB compacted samples (p < 0.05) and PEG 8000 compacted samples (p < 0.001) were significant compared to the amplified sample at amplicon concentrations of 15 μg ml−1 and 39.65 μg ml−1, whereas the results were not significant at lower amplicon concentrations (0.5 μg ml−1, 1 μg ml−1 and 5 μg ml−1). The usefulness of the device in assessing the extent of DNA compaction by migration distances covered by the PEG 8000 and CTAB compacted sample is demonstrated by measuring the limit-of-detection (LOD). As illustrated in the ESI Fig. 5,† the limit of detection (LOD) on the migration distance covered by compacted amplicon samples were analysed and these were found to be 0.4 μg ml−1 and 0.5 μg ml−1 for PEG 8000 compacted and CTAB compacted amplicon samples respectively. Similar to paper-based spot assays, these devices have great potential for diagnosing and monitoring moderate to severe Candida infections, since the limit of detection (LOD) of the distance-based assay falls within the mid-range of the clinical concentrations.
Combinatorial effect of CTAB and PEG 8000 on fluorescence sensitivity enhancement and DNA migration
To assess the synergistic effect of compaction agents, CTAB and PEG 8000 on the fluorescence sensitivity enhancement, varying concentrations of amplicons were prepared and treated with a mixture of 200 μM of CTAB and 40 mg ml−1 of PEG 8000 in presence of SYTO-9. The G channel intensities of all the samples were measured and recorded as depicted in the ESI Fig. 6.† Interestingly, the observed results revealed that the samples treated with a mixture of CTAB and PEG 8000 showed a higher fluorescence intensity compared to those treated only with CTAB or PEG 8000 at lower amplicon concentrations (0.5 μg ml−1 and 1 μg ml−1). Besides, at intermediate to higher concentrations of amplicons (5 μg ml−1, 15 μg ml−1 and 39.65 μg ml−1), G channel intensities were found to be constant at ∼250–254, which is attributed to the inefficiency of the blue light transilluminator to excite the labelled SYTO-9.Similarly, the combination of CTAB and PEG 8000 was assessed on the migration distances of varying concentrations of amplicons (0.5 μg ml−1, 1 μg ml−1, 5 μg ml−1, 15 μg ml−1, and 39.65 μg ml−1). The results revealed that the amplicon samples treated with a mixture of CTAB and PEG 8000 showed an increase in the migration distance compared to those treated only with PEG 8000 or CTAB (ESI Fig. 6†). The observation is attributed to the synergistic condensation effect on DNA in the vicinity of both CTAB and PEG 8000, which hinders the interfacial interaction of labelled SYTO-9 with cellulose in paper. Overall, the combination of CTAB and PEG 8000 could potentially provide a more effective way for compacting DNA and thereby enhance the sensitivity of SYTO-9 labelled DNA detection assays on both paper-based spot and distance-based assays.
Experimental
Grade 1 Whatman(R) filter paper (thickness 180 μm and pore size, 11 μm) was obtained from GE Life Sciences. Paraffin was procured from Merck, India. n-Heptane was obtained from SRL, India and colorants (red) were obtained from local candle manufacturers. The Trizol reagent was purchased from Invitrogen, Thermo Fisher Scientific. LAMP reagents were obtained from New England Biolabs. SYTO-9 was purchased from Thermo Fisher Scientific, and CTAB and PEG 8000 were procured from SRL, India. The Candida albicans (ATCC 24433) culture was collected from the Department of Microbiology, Kasturba Medical College, Manipal.Preparation of the in-house formulation using paraffin wax
The preparation of the in-house formulation using paraffin for the fabrication of paper devices was initially described by Prabhu et al.7 This was made by mixing paraffin, n-heptane, and the colorant. The paraffin was primarily made in n-heptane [6.5% (w/v)]. The resulting solution was then supplemented with 20 μL of the colorant and gently vortexed.Inkjet printing of paper-based devices
To print the hydrophobic formulation, an HP Deskjet 1112 printer was utilized by substituting the printer's original ink with the in-house formulation after filling the cartridge with a syringe. Before filling the ink formulation and loading it into the printer, the cartridge underwent thorough cleaning with n-heptane and isopropyl alcohol. The CorelDraw X6 software was used to design the distance-based pattern of the hydrophobic barrier for the paper devices,60,68,69 which were subsequently printed 20 times on a single side of Whatman(R) filter paper (grade 1). The cartridge was frequently replenished using the syringe when depleted. Following printing, the paper sheet was exposed to a hot air oven at 100 °C for 15 min to enable the paraffin penetration into the paper pores. The width of the minimum hydrophobic barrier attained was 2 mm, while the hydrophilic channel width was set at 5 cm.Growth of fungi
The fungus Candida albicans was collected from the Department of Microbiology, Kasturba Medical College, Manipal. Candida albicans was grown by inoculating the fungus in SDB broth overnight at 37 °C. The grown culture of Candida albicans (OD ∼ 0.8) was taken for DNA isolation.DNA extraction from Candida albicans by the Trizol reagent
The genomic DNA from the fungus, Candida albicans was extracted using the Trizol method by adhering to the protocol provided by Invitrogen (Thermo Fisher Scientific).70 1 ml of the fungal culture grown in SDB was pelleted by centrifugation and discarded the supernatant. 750 μL of the Trizol reagent was added to the pellet. The sample was homogenized by inversion and incubated for 5 min to permit the complete dissociation of the nucleoproteins complex. 150 μL of chloroform was added for the lysis of the cells. The resulting sample was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The DNA in the interphase was precipitated by adding 100% ethanol and washed with 0.1 M sodium citrate in 10% ethanol (pH 8.5). The DNA was further suspended in 8 mM NaOH and stored at −20 °C for amplification.
000 rpm for 15 min. The DNA in the interphase was precipitated by adding 100% ethanol and washed with 0.1 M sodium citrate in 10% ethanol (pH 8.5). The DNA was further suspended in 8 mM NaOH and stored at −20 °C for amplification.
Isothermal amplification of the ITS-2 region of Candida albicans
The conserved sequence of Candida albicans, the internal transcribed spacer-2 (ITS-2) sequence was amplified by loop-mediated isothermal amplification (LAMP) described by Nagamine et al.71 The primer sequence for Candida albicans ITS-2 was obtained from the previous literature (ESI Table 1†). All the samples were prepared in triplicates. The LAMP reaction was carried out with a final volume of 50 μL comprising five different primers (Proteogen Biosciences), dNTPs (New England Biolabs, USA), 1× isothermal amplification buffer (New England Biolabs, USA), MgSO4 (New England Biolabs, USA), 8 U of Bst 2.0 Warm Start DNA polymerase (New England Biolabs, USA) and 2 μL of the DNA template. The reaction was run at 65 °C for 60 minutes and was terminated at 80 °C for 5 min. For post-LAMP analysis, agarose gel electrophoresis and absorbance measurement were performed on the amplified LAMP product.The association between DNA and compaction agents
The compaction of the amplified IT2-2 sequence of Candida albicans was performed using compaction agents CTAB and PEG 8000. Separate stock solutions of PEG 8000 (500 mg ml−1) and CTAB (2 mM) were prepared by dissolving CTAB and PEG 8000 in water. Stock solutions of compaction agents were stored at room temperature for compaction studies. The minimum concentration of DNA required for the compaction was identified from the previous literature and diluted accordingly. Two concentrations of CTAB (100 μM and 200 μM) and PEG 8000 (40 mg ml−1 and 100 mg ml−1) have been considered for compaction studies. The compaction mixture was prepared by mixing appropriate concentrations of the agents to 39.65 μg ml−1 of amplicon. The samples were allowed to equilibrate at room temperature for 15 min.Absorbance measurements
Absorbance measurements were conducted to confirm the compaction of amplicon by CTAB PEG 8000 using an Eppendorf Bio spectrophotometer (Germany). The absorbance spectra of amplified, PEG 8000 compacted and CTAB compacted amplicons were recorded at a wavelength of 260 nm. Equal concentrations of PEG 8000 and CTAB were used as a blank for the compacted samples for nullifying the background noise. Nuclease-free water was taken as a blank for the amplified sample. A standard quartz cell with a 1 mm path length was used for the absorbance measurement.Fluorescence measurements
Steady-state fluorescence spectroscopy was explored to investigate the fluorescence enhancement of nucleic acid assays by PEG 8000 and CTAB. DNA condensation using CTAB and PEG 8000 was visualized using a fluorescent Dye SYTO-9 [(2.5 μM), Excmax = 485 nm and Emimax = 498 nm]. For fluorometric measurements, a Tecan fluorescence microplate reader (Infinite® M Nano) with a 96-well plate was used. The excitation and emission bandwidths were chosen to be 9 nm and 20 nm, respectively, to avoid spectral overlap. The gain value set was 100%. The amplified sample with SYTO-9 was taken as a control for the compacted samples. Besides, for eliminating the background noise generated by compaction agents, respective agents with SYTO-9 (without amplicon) were kept for the fluorometric analysis. The fluorescence intensity in terms of RFU for different samples was recorded and plotted as a graph.Spot assay
The fabrication of paper spot devices has been performed using the protocol given in our previous study.7 The fabricated spot device having a radius of 5 mm was used for spot assay analysis. In all the compaction mixtures, the concentration of amplicon and SYTO-9 were set at 39.65 μg ml−1 and 25 μM respectively. 20 μl of the sample containing PEG 8000 (40 mg ml−1), amplicon and SYTO-9 was added onto the spot device. Similarly, for compaction by CTAB, 200 μM of CTAB was added to the amplicon containing SYTO-9. Nuclease-free water was used to make up the required volume. A blue light transilluminator (BioBee, India) having a dye excitation between 420 nm to 500 nm and emission within 520 nm was used for exciting the fluorophore. All the samples were prepared in triplicates in nuclease-free water. Further, to analyse the effect of compaction on the fluorescence intensity of different concentrations of amplicons, specifically, 0.5 μg ml−1, 1 μg ml−1, 5 μg ml−1, and 15 μg ml−1, varying concentrations of amplicons were prepared by dilution with nuclease-free water in the presence of 25 μM of SYTO-9. The amplified sample without the condensing agents PEG 8000 and CTAB was used as the control. The samples were prepared in triplicates. The images of the spot devices were taken after one minute of sample addition to the spot device by keeping the camera (Canon EOS 80D DSLR) at a fixed distance and in a low-light setup. The images of the fluorometric test samples, including the controls, were split into red (R), green (G), and blue (B) channels in the Image J software.72 The mean intensity of the test zone was then determined by selecting a smaller circular region of the zone in the G-channel image.Distance-based assay
For distance-based analysis, a distance-based device with a channel length of 50 mm and width of 2 mm was fabricated. In all the compaction mixtures, the concentration of the amplicon and SYTO-9 were set at 39.65 μg ml−1 and 25 μM respectively. The device was added with a sample volume of 20 μl containing PEG 8000 (40 mg ml−1), amplicon and SYTO-9. Similarly, for the compaction by CTAB, the compaction mixture containing 200 μM of CTAB was added to the amplicon containing SYTO-9. Nuclease-free water was used to make up the volume. The fluorophore was excited using a blue light transilluminator (BioBee, India) with a dye excitation between 420 nm to 500 nm and emission within 520 nm. All the samples were prepared in triplicates.Similarly, for analysing the influence of compaction on the migration distance of different concentrations of amplicons, the amplified sample was diluted to 0.5 μg ml−1, 1 μg ml−1, 5 μg ml−1, and 15 μg ml−1 using nuclease-free water in the presence of 25 μM of SYTO-9. As a control, the amplicon without the compaction agents, CTAB and PEG 8000 was used. The sample was prepared in triplicates. The images of the migration distance of samples were taken after one minute of the sample addition to the devices by keeping the camera (Canon EOS 80D DSLR) at a fixed distance and in a low-light setup. These images were processed using ImageJ software for distance calculations. Scale measurements for the distance-based assay were set using the ImageJ software by capturing an image of a 30 cm scale at the same fixed distance. Later, a known distance was set as 5 mm in the scale measurement option in the Image J software.71 The same distance (in pixels) and known distance of 5 mm were kept for the subsequent distance analysis.71
Statistical analysis
Experimental data obtained from conventional fluorescence measurement, paper spot assay and distance-based assay were represented as the mean ± standard deviation (SD). In the conventional fluorescence measurement, the statistical significance between SYTO-9 and amplified, SYTO-9 and compacted (CTAB compacted and PEG compacted), amplified and compacted samples were computed using the one-way ANOVA test with Tukey's post-hoc analysis using the GraphPad Prism 8 software. Similarly, the statistical significance of the G channel intensity values and migration distance obtained for amplified and compacted samples (CTAB compacted and PEG compacted) at different amplicon concentrations was determined using the one-way ANOVA test with Tukey's post-hoc analysis. Statistical significance was considered at p < 0.05.Combinatorial effect of CTAB and PEG 8000 on fluorescence sensitivity enhancement and DNA migration
To investigate the combinatorial effect of CTAB and PEG 8000 on fluorescence sensitivity enhancement, the amplicons were diluted to different concentrations, specifically 0.5 μg ml−1, 1 μg ml−1, 5 μg ml−1, 15 μg ml−1, and 39.65 μg ml−1. Furthermore, the amplicons were treated with a mixture of 200 μM of CTAB and 40 mg mL−1 of PEG 8000 in the presence of 25 μM of the SYTO-9 dye. The samples were allowed to equilibrate at room temperature for 15 min. The prepared samples were added by spotting 20 μL onto the fabricated spot and distance-based devices. For the excitation of the fluorophore, a blue light transilluminator with an excitation range of 420 nm to 500 nm and an emission range of 520 nm was used. The images of spot and distance-based devices were captured and analysed following the method described previously.Conclusions
To summarize, our research demonstrates the utility of DNA condensation as a simple, reliable, and easy-to-use method for enhancing the fluorescence intensity of nucleic acid assays without necessitating cumbersome and expensive sensitivity enhancement techniques. The conventional fluorescence measurements carried out during the study with compaction agents CTAB and PEG 8000 have shown an increased fluorescence of the SYTO-9 labelled ITS-2 sequence of Candida albicans. The self-assembly of CTAB micelles at its critical micelle concentration (CMC) results in the development of CTAB–DNA compact aggregates, which in turn alters the local environment of SYTO-9 labelled DNA and reduces the fluorescence decay improving the emission intensity. Alternatively, the rationale for the enhancement in the fluorescence intensity upon compaction with PEG 8000 is attributed to the immiscibility of stiff chains (DNA) and flexible chains (PEG), known as “stiff-flexible chain incompatibility”. For stiff polyelectrolytes such as DNA, the addition of flexible polymers such as PEG induces perfect segregation between the polymer and the DNA. The ensemble clustering of PEG molecules around the DNA exerts an osmotic pressure making the DNA contracted, which in turn, stabilizes the bound SYTO-9 and contributes to an increase in the emission intensity.Furthermore, to demonstrate the efficacy of the observed results for point-of care applications, the compaction assays were performed on paper-based spot and distance-based devices. The results of the spot assays indicated that the compaction of nucleic acids delivered increased sensitivity. Besides, the difficulty in executing fluorometric assays for low nucleic acid concentrations on μPADs due to the opaque behaviour of paper and insufficient excitation capability of a blue-light transilluminator could be rectified using this approach. An increasing trend for the migration distance was observed in the distance-based channel in the order PEG 8000 compacted > CTAB compacted > amplified at amplicon concentrations, 15 μg ml−1 and 39.65 μg ml−1. The increasing distance corresponds to the compaction mediated-elution of the dye from the electrostatic and hydrogen bonding interactions with the cellulose surface. Our success in verifying the robustness of the distance-based assay as a point-of care approach to quantify nucleic acids would assist in screening infections allowing critical treatment to be devoted to those in need. Consequently, these strategies would enable researchers to perform fluorescence assays for intermediate to high analyte concentrations without the need for complex oligo preparation and other molecular and chemical-based methods. Additionally, the ease-of-operation makes it ideal for use in resource-limited settings with the advantage of having improved analytical sensitivity in clinical diagnostics. We envisage improvement and further exploration of better fluorescence intensification strategies based on nucleic acid adherence on the surface of μPADs aimed toward integrating image analysis methods through smartphones to reduce the assay time and ease of operation.
Author contributions
Conceptualization: NKM and AP. Data curation: NKM, AP, and SS. Formal analysis: NKM, AP, and SS. Funding acquisition: NKM and AP. Investigation: NKM, AP, and SS. Methodology: NKM, AP, SS, and DP. Project administration: NKM. Resources: NKM and DP. Software: NKM, AP, and SS. Supervision: NKM. Validation: SS, AP, and DP. Visualization: NKM. Writing – original draft: SS, AP, and NKM. Writing – review & editing: NKM and DP.Conflicts of interest
There are no conflicts to declare.Acknowledgements
NKM acknowledges the financial support received from the Science and Engineering Research Board (SERB), Department of Science and Technology, Govt of India, under the Core Research Grant (CRG) Scheme (file number CRG/2020/003060). NKM specially thanks Dr. (Cdr). Anil Rana, Director, MIT for his kind support to establish Microfluidics, Sensors and Diagnostics (μSenD) Laboratory. NKM and AP acknowledge the financial support from the Vision Group on Science and Technology, Government of Karnataka, under the SMYSR and RGS/F Scheme (sanction letter no: KSTePS/VGST/SMYSR-2016–17/GRD-595/2017–18, KSTePS/VGSTRGS/F/GRD No.711/2017–18). AP acknowledges the Indian Council of Medical Research (ICMR) for providing a Senior Research Fellowship (file no: 5/3/8/91/ITR-F/2020). SS acknowledges the Indian Council of Medical Research (ICMR) for providing a Senior Research Fellowship (file no: 5/3/8/67/ITR-F/2022-ITR). All authors thank Dr. Praveen Kumar and Dr. Vijendra Prabhu for their fruitful discussions.References
- S. M. Levchenko, A. Pliss, X. Peng, P. N. Prasad and J. Qu, Light: Sci. Appl., 2021, 10, 224 CrossRef CAS PubMed.
- K. Kainz, M. A. Bauer, F. Madeo and D. Carmona-Gutierrez, Microb. Cell, 2020, 7, 143–145 CrossRef PubMed.
- J. Talapko, M. Juzbašić, T. Matijević, E. Pustijanac, S. Bekić, I. Kotris and I. Škrlec, J. Fungi, 2021, 7, 79 CrossRef CAS PubMed.
- T. Vila, A. S. Sultan, D. Montelongo-Jauregui and M. A. Jabra-Rizk, J. Fungi, 2020, 6, 1–28 CrossRef PubMed.
- T. P. McCarty, C. M. White and P. G. Pappas, Infect. Dis. Clin. North Am., 2021, 35, 389–413 CrossRef PubMed.
- J. Geddes-McAlister and R. S. Shapiro, Ann. N. Y. Acad. Sci., 2019, 1435, 57–78 CrossRef PubMed.
- A. Prabhu, M. S. Giri Nandagopal, P. Peralam Yegneswaran, H. R. Singhal and N. K. Mani, Cellulose, 2020, 27, 7691–7701 CrossRef CAS.
- S. M. Mirmajlessi, M. Destefanis, R. A. Gottsberger, M. Mänd and E. Loit, Syst. Rev., 2015, 4, 9 CrossRef PubMed.
- Y. Hassan and L. T. L. Than, Annu. Res. Rev. Biol., 2020, 35, 33–44 CrossRef.
- T. Notomi, Y. Mori, N. Tomita and H. Kanda, J. Microbiol., 2015, 53, 1–5 CrossRef CAS PubMed.
- R. Augustine, A. Hasan, S. Das, R. Ahmed, Y. Mori, T. Notomi, B. D. Kevadiya and A. S. Thakor, Biology, 2020, 9, 182 CrossRef CAS PubMed.
- H. Huang, B. Phipps-todd, C. C. Leung and D. Elleithy, Different tolerance of loop-mediated isothermal amplification and polymerase chain reaction to inhibitors in chicken carcass rinse and feces for detecting Campylobacter jejuni, 2011, 3780.
- H. Zhang, Y. Xu, Z. Fohlerova, H. Chang, C. Iliescu and P. Neuzil, TrAC, Trends Anal. Chem., 2019, 113, 44–53 CrossRef CAS PubMed.
- P. Hardinge and J. A. H. Murray, BMC Biotechnol., 2019, 19, 1–15 CrossRef CAS PubMed.
- N. Garg, F. J. Ahmad and S. Kar, Curr. Res. Microb. Sci., 2022, 3, 100120 CAS.
- Y. Cao, L. Wang, L. Duan, J. Li, J. Ma, S. Xie, L. Shi and H. Li, Sci. Rep., 2017, 7, 13394 CrossRef PubMed.
- B. Y. Yang, X. L. Liu, Y. M. Wei, J. Q. Wang, X. Q. He, Y. Jin and Z. J. Wang, BMC Microbiol., 2014, 14, 2–8 CrossRef PubMed.
- X. Ding, K. Yin, Z. Li, V. Pandian, J. A. Smyth, Z. Helal and C. Liu, Sci. Rep., 2020, 10, 1–9 CrossRef PubMed.
- J. Maillard, K. Klehs, C. Rumble, E. Vauthey, M. Heilemann and A. Fürstenberg, Chem. Sci., 2021, 12, 1352–1362 RSC.
- A. P. Demchenko, Methods Appl. Fluoresc., 2020, 8, 22001 CrossRef CAS PubMed.
- J. Huang and K. Pu, Angew. Chem., Int. Ed., 2020, 59, 11717–11731 CrossRef CAS PubMed.
- M. Mariconti, M. Morel, D. Baigl and S. Rudiuk, Biomacromolecules, 2021, 22, 3431–3439 CrossRef CAS PubMed.
- A. Bergen, S. Rudiuk, M. Morel, T. Le Saux, H. Ihmels and D. Baigl, Nano Lett., 2016, 16, 773–780 CrossRef CAS PubMed.
- N. Moradi, Y. Zakrevskyy, A. Javadi, E. V. Aksenenko, V. B. Fainerman, N. Lomadze, S. Santer and R. Miller, Colloids Surf., A, 2016, 505, 186–192 CrossRef CAS.
- P. Evenou, J. Rossignol, G. Pembouong, A. Gothland, D. Colesnic, R. Barbeyron, S. Rudiuk, A.-G. Marcelin, M. Ménand and D. Baigl, Angew. Chem., Int. Ed., 2018, 57, 7753–7758 CrossRef CAS PubMed.
- A. Estévez-Torres and D. Baigl, Soft Matter, 2011, 7, 6746–6756 RSC.
- R. A. Gonçalves, K. Holmberg and B. Lindman, J. Mol. Liq., 2023, 121335 CrossRef.
- B. Pi-Boleda, S. Ramisetty, O. Illa, V. Branchadell, R. S. Dias and R. M. Ortuño, ACS Appl. Bio Mater., 2021, 4, 7034–7043 CrossRef CAS PubMed.
- A. Zinchenko, N. V. Berezhnoy, Q. Chen and L. Nordenskiöld, Biophys. J., 2018, 114, 2326–2335 CrossRef CAS PubMed.
- A. G. Wang, T. Dong, H. Mansour, G. Matamoros, A. L. Sanchez and F. Li, ACS Sens., 2018, 3, 205–210 CrossRef CAS PubMed.
- M. Serucnik, C. Podlipnik and B. Hribar-Lee, J. Phys. Chem. B, 2018, 122, 5381–5388 CrossRef CAS PubMed.
- B. Lindman, B. Medronho, L. Alves, M. Norgren and L. Nordenskiöld, Q. Rev. Biophys., 2021, 54, e3 CrossRef CAS PubMed.
- S. Rudiuk, K. Yoshikawa and D. Baigl, J. Colloid Interface Sci., 2012, 368, 372–377 CrossRef CAS PubMed.
- S. Rudiuk, A. Venancio-Marques and D. Baigl, Angew. Chem., 2012, 124, 12866–12870 CrossRef.
- A. Venancio-Marques, A. Bergen, C. Rossi-Gendron, S. Rudiuk and D. Baigl, ACS Nano, 2014, 8, 3654–3663 CrossRef CAS PubMed.
- X. Li, D. Sun, Y. Chen, K. Wang, Q. He and G. Wang, Biochem. Biophys. Res. Commun., 2018, 495, 2559–2565 CrossRef CAS PubMed.
- N. K. Mani, S. Rudiuk and D. Baigl, Chem. Commun., 2013, 49, 6858–6860 RSC.
- M. Cristofalo, C. A. Marrano, D. Salerno, R. Corti, V. Cassina, A. Mammola, M. Gherardi, B. Sclavi, M. C. Lagomarsino and F. Mantegazza, Biochim. Biophys. Acta, Gen. Subj., 2020, 1864, 129725 CrossRef CAS PubMed.
- V. V. Vasilevskaya, A. R. Khokhlov, Y. Matsuzawa and K. Yoshikawa, J. Chem. Phys., 1995, 102, 6595–6602 CrossRef CAS.
- T. Nishio, Y. Yoshikawa and K. Yoshikawa, PLoS One, 2021, 16, e0261736 CrossRef CAS PubMed.
- M. L. Ainalem, A. Bartles, J. Muck, R. S. Dias, A. M. Carnerup, D. Zink and T. Nylander, PLoS One, 2014, 9, e92692 CrossRef PubMed.
- M. López-López, P. López-Cornejo, V. I. Martín, F. J. Ostos, C. Checa-Rodríguez, R. Prados-Carvajal, J. A. Lebrón, P. Huertas and M. L. Moyá, J. Colloid Interface Sci., 2018, 521, 197–205 CrossRef PubMed.
- S. Gupta, N. Tiwari and M. Munde, Sci. Rep., 2019, 9, 5891 CrossRef PubMed.
- J. Carlstedt, D. Lundberg, R. S. Dias and B. Lindman, Langmuir, 2012, 28, 7976–7989 CrossRef CAS PubMed.
- B. Hao, K. Wang, Y. Zhou, C. Sui, L. Wang, R. Bai and Z. Yang, ACS Omega, 2020, 5, 1109–1119 CrossRef CAS PubMed.
- F. Muzzopappa, M. Hertzog and F. Erdel, Biophys. J., 2021, 120, 1288–1300 CrossRef CAS PubMed.
- S. L. Lai, C. H. Chen and K. L. Yang, Langmuir, 2011, 27, 5659–5664 CrossRef CAS PubMed.
- E. Grueso, C. Cerrillos, J. Hidalgo and P. Lopez-Cornejo, Langmuir, 2012, 28, 10968–10979 CrossRef CAS PubMed.
- T. L. Quyen, T. A. Ngo, D. D. Bang, M. Madsen and A. Wolff, Front. Microbiol., 2019, 10, 2234 CrossRef PubMed.
- A. Tárnok, Cytometry, Part A, 2008, 73, 477–479 CrossRef PubMed.
- D. Yang, J. Männik, S. T. Retterer and J. Männik, Mol. Microbiol., 2020, 113, 1022–1037 CrossRef CAS PubMed.
- P. J. Flory and A. Abe, Macromolecules, 1978, 11, 1119–1122 CrossRef CAS.
- A. Zinchenko, Q. Chen, N. V. Berezhnoy, S. Wang and L. Nordenskiöld, Soft Matter, 2020, 16, 4366–4372 RSC.
- C. Sun, X. Jiang, B. Li, S. Li and X. Z. Kong, ACS Sustainable Chem. Eng., 2021, 9, 5166–5178 CrossRef CAS.
- M. He and Z. Liu, Anal. Chem., 2013, 85, 11691–11694 CrossRef CAS PubMed.
- M. Alafeef, P. Moitra, K. Dighe and D. Pan, Nat. Protoc., 2021, 16, 3141–3162 CrossRef CAS PubMed.
- Y. Xia, J. Si and Z. Li, Biosens. Bioelectron., 2016, 77, 774–789 CrossRef CAS PubMed.
- W. V. Gonzales, A. T. Mobashsher and A. Abbosh, The progress of glucose monitoring—A review of invasive to minimally and non-invasive techniques, devices and sensors, 2019, vol. 19 Search PubMed.
- A. Vesty, K. Biswas, M. W. Taylor, K. Gear and R. G. Douglas, PLoS One, 2017, 12, 1–13 CrossRef PubMed.
- N. Kelkar, A. Prabhu, A. Prabhu, M. S. Giri Nandagopal and N. K. Mani, Microchem. J., 2022, 174, 107069 CrossRef CAS.
- R. Ray, A. Prabhu, D. Prasad, V. Kumar Garlapati, T. M. Aminabhavi, N. K. Mani and J. Simal-Gandara, Food Chem., 2022, 390, 133173 CrossRef CAS PubMed.
- R. Ray, C. Noronha, A. Prabhu and N. K. Mani, Food Anal. Methods, 2022, 15, 2664–2674 CrossRef.
- A. Hasandka, A. R. Singh, A. Prabhu, H. R. Singhal, M. S. G. Nandagopal and N. K. Mani, Anal. Bioanal. Chem., 2022, 414, 847–865 CrossRef CAS PubMed.
- A. Hasandka, A. Prabhu, A. Prabhu, H. R. Singhal, M. S. G. Nandagopal, R. Shenoy and N. K. Mani, Anal. Methods, 2021, 13, 3172–3180 RSC.
- H. R. Singhal, A. Prabhu, M. S. Giri Nandagopal, T. Dheivasigamani and N. K. Mani, Microchem. J., 2021, 165, 106126 CrossRef CAS.
- A. Prabhu, M. S. G. Nandagopal, P. Peralam Yegneswaran, V. Prabhu, U. Verma and N. K. Mani, RSC Adv., 2020, 10, 26853–26861 RSC.
- K. Minagawa, Y. Matsuzawa, K. Yoshikawa, A. R. Khokhlov and M. Doi, Biopolymers, 1994, 34, 555–558 CrossRef CAS.
- R. Ray, A. Goyal, A. Prabhu, S. Parekkh, S. Maddasani and N. K. Mani, Food Chem., 2023, 403, 134484 CrossRef CAS PubMed.
- N. K. Mani, S. S. Das, S. Dawn and S. Chakraborty, Electrophoresis, 2020, 41, 615–620 CrossRef CAS PubMed.
- Life Technologies, Trizol reagent: Experimental protocol for DNA isolation, Thermo Fish. Sci. Inc., 2016, 1–4.
- K. Nagamine, T. Hase and T. Notomi, Mol. Cell. Probes, 2002, 16, 223–229 CrossRef CAS PubMed.
- A. Prabhu, H. Singhal, M. S. Giri Nandagopal, R. Kulal, P. Peralam Yegneswaran and N. K. Mani, ACS Omega, 2021, 6, 12667–12675 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00102d |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2023 |