Advancements in microfluidics for skin cosmetic screening
Nianfang
Hu
a,
Kerun
Cheng
b,
Shuhan
Zhang
b,
Shan
Liu
c,
Lijun
Wang
d,
Xiaoxin
Du
e,
Yong
Li†
*a and
Chenzhong
Li†
 *f
*f
aDepartment of Dermatology, West China Hospital, Sichuan University, China. E-mail: 676354940@qq.com
bWest China School of Medicine, Sichuan University, China
cSichuan Provincial Key Laboratory for Human Disease Gene Study, Department of Medical Genetics, Sichuan Academy of Medical Sciences & Sichuan Provincial People's Hospital, University of Electronic Science and Technology of China, Chengdu 610072, China
dDepartment of Ophthalmology, The Third People's Hospital of Chengdu, The Affiliated Hospital of Southwest Jiaotong University, Chengdu 610031, China
eOffice of Scientific Research & Development, University of Electronic Science and Technology, Chengdu, China
fBiomedical Engineering, School of Medicine, The Chinese University of Hong Kong, Shenzhen 518172, China. E-mail: chenzhongbiosensor@gmail.com
First published on 24th February 2023
Abstract
With the global penetration of skin care awareness and upgrading of personal care awareness, the use rate of cosmetics and personal skin care products has been increasing worldwide. It is particularly important to monitor the quality and safety of skin cosmetics. In accordance with the requirements of the 7th Amendment of the European Cosmetics Directive 1223/2009, in vitro test methods have been developed to replace animal experiments, such as the 2D test, 3D test, microfluidic skin chip test, etc. The microfluidic skin chip overcomes the shortcomings of the 2D test and the 3D test that lack the complexity of human skin through fine control of the human skin microenvironment and induction of relevant mechanical stimulation. High similarity to real human skin through simulation of the vascular system and immune response. Therefore, the microfluidic skin chip is considered as a valuable and effective tool for the in vitro screening of cosmetics. In this paper, we reviewed the detection methods and technologies of common chemical substances, toxic elements, active substances and adverse reactions in vitro in quality monitoring of cosmetics. The most advantageous microfluidic skin chip technology is also introduced. The material and technology progress of skin chips used in cosmetic screening is reviewed and discussed. Then the application of microfluidic design in cosmetic screening in vitro is summarized.
1. Introduction
The purpose of cosmetics is to beautify the skin or protect or change a person's appearance.1 In the early primitive society, animal oils and fatty skin care products were used to make the skin smooth and shiny. Later, skin care products entered the era of mineral oil and natural ingredients,2 including cleaning, moisturizing and UV protection ingredients. With the progress of science and technology and the diversification of consumer demand, modern cosmetics have more complex ingredients and diverse functions. Therefore, the detection of the safety and effectiveness of cosmetic ingredients has become necessary.3Cosmetic detection includes chemical analysis, toxic element detection, biochemical detection of active substances and in vitro adverse reaction detection. The chemical properties (stability, pH, antioxidant, etc.), the content of toxic elements and active substances and the possible adverse reactions of the substances contained in cosmetics are tested to comprehensively evaluate the function and safety of cosmetics. Among them, in vitro adverse reaction detection is based on the EU Directive 2010/63/EU,4 REACH5 (Registration, Evaluation, Authorization and Restriction of Chemicals) and other regulations for the protection of experimental animals. These regulations suggest that biochemical methods should be used instead of animal experiments. Researchers have developed a series of standardized animal in vitro alternative testing methods to achieve this goal, including the 2D test, the 3D test, and the skin chip test (i.e. microfluidic technology). A two-dimensional (2D) cell culture was established by co-culturing various cell types6 and a three-dimensional (3D) skin model was established by simulating the three layers of epidermis, dermis and subcutaneous adipose tissue, as well as the tight and gap connection between different cell types.7
The microfluidic technology chip, which is established by more realistically simulating the 3D microenvironment of real skin (such as skin appendages and blood vessels) and manipulating the physical and biochemical parameters therein, is also known as the skin on a chip.8 Among them, the skin chip testing technology provides an effective testing and screening method through the application of microfluidic technology and the establishment of a layered 3D environment and other special structures, and has a good development and application prospect. In the future, through the seamless integration of microfluidic technology, cell biology and tissue engineering technology, innovative micro-engineered cell culture systems will become possible,9–11 which also allows a small amount of precise control of the cell microenvironment,12,13 providing a great impetus for more accurate simulation of the real skin environment to replace animal experiments in vivo.
It should also be noted that biosensors have been associated with the strategic integration of the Internet of Things (IoT), the fifth generation (5G) communication, artificial intelligence (AI) and machine learning (ML), and gradually integrated into social life, which is conducive to the development of instant detection (POCT),14,15 and has a good prospect in the field of cosmetic detection. Biosensor technology can not only be used to detect cosmetics independently, but also be widely used in 2D testing, 3D testing, skin chip testing and routine chemical, toxic element and active substance testing. Biosensor technology plays an important role in the above three in vitro substitution tests. It can convert tiny biological binding reactions into observable electrical signals, which is very helpful for visual analysis of experimental results.
In this paper, we summarized the basic structure of skin related to cosmetics and the role of the corresponding cosmetics, and reviewed the detection methods and technologies of common chemical substances, toxic elements, active substances and in vitro adverse reactions in quality monitoring of cosmetics. The most advantageous microfluidic skin chip technology is introduced. The material and technology progress of skin chips used in cosmetic screening is reviewed and discussed. Then the paper summarizes the practical application of skin chips in in vitro screening of cosmetics in recent years, including the construction of disease models, toxicological evaluation, permeability evaluation, screening of active ingredients in cosmetics, and the construction of aging skin models. A multi-organ chip system with a good application prospect in cosmetic screening is also introduced. Finally, this review points out the challenges that the current microfluidic skin chip model still faces, and points out the future development direction.
2. Skin structures and the role of cosmetics
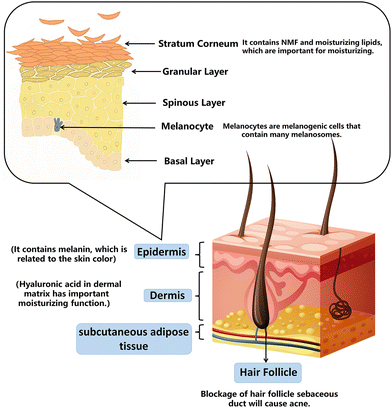
The dermis and epidermis compose the body's skin. The four layers of the epidermis—the base layer (stratum germinatum), the squamous layer (stratum spinosum), the granular layer (stratum granulosum), and the cornified layer (stratum corneum; SC)—are formed by keratinocyte differentiation and cornification, which occurs from deep to shallow.2.1. Skin barrier and moisturizing
Skin plays an essential role in maintaining normal physiological functions and water levels as a barrier. The basic concept of a skin barrier was developed after introducing the “brick and mortar” model.16 The skin barrier is mainly located in the stratum corneum (SC), composed of keratinocytes (“bricks”), surrounded by intercellular lipids (“mortar”), and linked by keratinocyte desmosomes (CD) (“rivets”). The maintenance of skin moisturizing ability is attributed to the SC. A combination of tiny water-soluble molecules makes up the natural moisturizing factor (NMF) in the SC. Skin becomes brittle and dry when the NMF is removed because it decreases the molecular fluidity of keratin filaments and SC lipids.17 The moisturizing lipids in the SC are composed of squalene, wax, fat, and triglycerides. Moisturizing lipids lubricate the skin, reduce water evaporation on the surface, and adjust the pH. Hyaluronic acid in the dermal matrix has essential moisturizing functions; it contains many hydrophilic groups that form hydrogen bonds with water and bind large amounts of water. A small amount of hyaluronic acid locks in a substantial amount of water necessary to regulate skin metabolism and mediate anti-wrinkle and beautifying effects.18The principle of moisturizing products on the market is based on the structural design of the skin barrier. Moisturizing creams add water, NMF, hyaluronic acid, and lipids to achieve moisturization.19 By measuring water losses in the skin after removing the moisturizer residue from the surface, the NMF temporarily increases hydration by being absorbed into the epidermis.202.2. Skin aging and anti-aging
Aging is characterized by thin, smooth, pale, dry skin with low elasticity and fine wrinkles.21Both intrinsic and extrinsic variables have an impact on skin aging. Intrinsic aging is slow and inevitable and shows significant differences across individuals and races and even parts of the body in the same person.22 Extrinsic aging is caused by external environmental factors such as air pollution, sunlight exposure, and smoking.23 Long-term sunlight exposure is the most critical factor leading to aging, also called photo-aging. As early as 1969, it was proposed that sunlight leads to skin aging.24 Photo-aging causes the skin to become rough, wrinkled, yellow, and mottled, and lose elasticity. Under UV irradiation, the expression of collagen in keratinocytes decreases, weakening the connection between the dermis and epidermis and causing thinning and wrinkles.25 Photo-aging also causes elastin molecules to become susceptible to decomposition by enzymes, accelerating its degradation and causing loss of elasticity (Kligman, 1969).24 Using sunscreen every day is an effective anti-aging method.2.3. Skin and whitening skincare products
Skin color represents health and beauty, and its social and psychological significance often exceeds its biological function.26 In Asian cultures, most women desire light skin associated with beauty and light skin can increase self-confidence. In particular, in China, India, and Japan, there is a substantial demand for whitening cosmetics, and the investment in whitening agents increases yearly.27Normal human skin color depends on the expression of several biological pigments, including melanin, red oxyhemoglobin, blue oxyhemoglobin, and carotene. Skin color is determined by the proportions of these pigments, their distribution levels, skin thickness, and other factors. Of these, the amount of melanin and its distribution are most critical.28 This is because melanin-producing cells are in the epidermis, the outermost layer. When skin is irradiated with UV light, tyrosinase (the key enzyme of melanin production) is activated to initiate melanin synthesis. Normal melanin can protect the skin and resist UV radiation. However, excessive melanin secretion can lead to skin anomalies such as freckles, solar freckles (age spots), and chloasma.29
Melanin metabolism is divided into synthesis, transfer to keratinocytes, and final transport to the SC. Most commercially-available whiteners act by inhibiting, blocking, or disrupting melanin production and transportation.30
2.4. Acne and anti-acne cosmetics
Acne is a chronic inflammatory disease of skin and hair follicles characterized by papules, pustules, cysts, nodules, disfigured scars, and poor quality of life.31 According to the Global Burden of Condition Study 2010, acne vulgaris is the eighth most prevalent disease worldwide.32With an overall frequency of 9.38% across all ages, it is also the second most prevalent skin disease.33 The pathogenesis of acne is complex and includes seborrhea, keratosis of sebaceous ducts, colonization of bacilli, and inflammation.34 The common pathogenic principle of acne is that sebum secretion increases, the proliferation of keratinocytes increases, and shedding decreases, resulting in the combination of sebum and keratinocytes, causing skin keratosis, sebaceous duct blockage in hair follicles, and finally, the formation of microcomedones.34,35 The proportions of Propionibacterium acnes and Staphylococcus epidermidis are maladjusted, causing infection of sebaceous units and resulting in acne.36 It is worth noting that bacterial resistance and potential side effects must be considered. Most anti-acne cosmetics on the market are added with anti-inflammatory, oil control, skin barrier repairing, and exfoliating substances.2.5. Summary
We can understand each structure of the skin and its corresponding function by examining a simplified image (Fig. 1). The functions and skin structure related to each skincare product are summarized in Table 1.| Purpose | Cosmetics | Related skin structure/related mechanism | The function of cosmetics |
|---|---|---|---|
| Moisture | Moisturizing cream | NMF and lipids in the stratum corneum; hyaluronic acid in the dermal matrix | Moisturizing |
| Anti-aging | Sunscreen, anti-aging products | Collagen decreased and free radicals increased | Maintain skin moisture, slow down the degradation rate of elastin and remove free radicals |
| Whitening | Whitening products which are added with vitamin C, arbutin, nicotinamide, tranexamic acid, kojic acid, glutathione, etc. | Melanin accumulation | Inhibiting, blocking or affecting melanin production and transportation |
| Anti-acne | Anti-acne agents (salicylic acid etc.) and anti-inflammatory agents | Blockage of hair follicle sebaceous ducts and sebaceous unit infection | Clean oil on the skin surface, relieve follicle blockage, control oil and treat bacterial inflammation |
3. Cosmetic assessment (cell culture and in vitro culture)
According to the EC Cosmetics Regulation introduced in 2009,1 animal testing of cosmetic products will be banned entirely in Europe. The REACH regulation requires the adoption of substitutes for animal tests for safety assessments of cosmetics.5 The industry was compelled to adopt in vitro testing programs for these reasons. Animal skin corrosion and irritation models, severe eye damage and irritation, skin allergy, mutagenicity, and genotoxicity have led to the relative development of substitutes. However, animal testing is still used to determine acute systemic toxicity, repeated dosage toxicity, reproductive toxicity, and developmental toxicity.3 Currently, no replacement tests can identify mild eye irritation.37In vitro experiments include 2D, 3D, and skin-on-a-chip tests (Table 2).| Human health endpoint | In vitro model | Characteristics | OECD No. | |
|---|---|---|---|---|
| 2D | Skin irritation | SkinEthic™ RHE | - The containment properties of the RhE model prevent the passage of a material around the stratum corneum to the viable tissue | OECD 439 |
| TER tset | - Skin clipper from young, approximately 22 day-old, male or female rats | OECD 430 | ||
| - The skin impedance is measured as TER using a low-voltage, alternating current Wheatstone bridge | ||||
| Skin sensitization | KeratinoSens™ | - Makes use of an immortalised adherent cell line derived from human keratinocytes | OECD 442D | |
| - Quantitative measurement of luciferase gene induction as an indicator of the activity of Nrf2 transcription | ||||
| Phototoxicity | 3T3 NRU | - Uses normal BALB/c 3T3 mouse fibroblasts to measure the concentration-dependent reduction in neutral red uptake by the cells after exposure to a test material either in the presence or absence of UVA light | OECD 432 | |
| Skin absorption | — | - Split thickness skin (200–400 μm thick) prepared with a dermatome | OECD 428 | |
| - The test system includes the donor chamber, the skin surface rinsing, the skin preparation and the receptor fluid/chamber | ||||
| 3D | Skin irritation | epiCS® | - Comprised of non-transformed, human-derived epidermal keratinocytes, which have been a highly differentiated model of the human epidermis | OECD 431 |
| - Consists of organized basal, spinous and granular layers, and a multi-layered stratum corneum containing intercellular lamellar lipid layers. | ||||
| Eye irritation | EpiOcular™ | - Three-dimensional RhCE tissue constructs that are produced using either primary human epidermal keratinocytes human-derived cells should be used to reconstruct the cornea-like epithelium | OECD 492 | |
| - The EpiOcular™ EIT validation database contained 113 chemicals in total | ||||
| HCE | - Human immortalized corneal epithelial cells which should be composed of progressively stratified but not cornified cells. | |||
| Skin-on-a-chip | Skin sensitization and hepatotoxicity | Skin–nerve hybrid model | - The medium flows along the channel by gravity when a microfluidic channel is used to tilt the chip | — |
| Pumpless SOC | - The keratinocytes were cultured on the top layer of a vertical microfluidic chip at the air–liquid interface to prepare a skin–nerve hybrid model | — |
3.1. 2D tests
The traditional model is the 2D model, in which keratinocytes are co-cultured with immune cells and dermal fibroblasts.38 Because of its easier preparation and mature technology, most test methods used for skin corrosion and skin sensitivity in Regulation (EC) No. 440/2008 are of this type. The in vitro 3T3 NRU phototoxicity test (OECD TG 432)39 is listed as a required in vitro method to evaluate phototoxicity in Regulation 440/2008. This technique measures the concentration-dependent decrease in neutral red absorption by cells following exposure to the test substance using regular BALB/c 3T3 small murine fibroblasts (with or without UVA light). Four crucial biological events must occur in order to detect a skin allergy: (i) molecular trigger events, (ii) altered keratinocyte gene expression related to particular cell signaling pathways and inflammatory responses, (iii) activation of dendritic cells through the expression of particular cell surface markers (cytokines and chemokines), and (iv) T cell proliferation.40,41 The direct peptide reactivity assay42 and the KeratinoSens™ assay are two techniques for measuring photosensitivity. The method known as KeratinoSens™ uses selective plasmid transfection to measure the luciferase gene's induction in an immortalized adherent human keratinocyte cell line (HaCaT cell line). This is a measurement of the antioxidant/electrophoresis response element using Keap1-Nrf2-A.433.2. 3D tests
Although 2D models are simple to create, they are unable to replicate in vivo interactions between cells and the matrix. Extracellular matrix materials are utilized to create 3D models in order to more closely mimic the actual human microenvironment.7 The three levels of epidermis, dermis, and subcutaneous adipose tissue, as well as the creation of tight and gap junctions between various cell types, are necessary for the 3D model.7 Drug diffusion is the main distinction between 2D and 3D cosmetic detection. Similar to the barrier function of human skin, the SC structure produced in the 3D model can slow the drug diffusion rate since the drug frequently needs to diffuse across numerous layers of cells before reaching the final target.44Three-dimensional models have been taken seriously by regulations in the European Community and are used in cosmetic safety testing, especially in eye irritation testing. The recombinant HCE™ model (SkinEthic Laboratorie, Nice, France) (OECD TG 492)45 consists of an inert permeable polycarbonate insert in immortalized corneal epithelial cells for toxicity assessment of corneal irritants, while the EpiOcular™ model (MatTek Corporation, USA)46 replicates the corneal epithelium using stratified but unkeratinized cells that are generated in synthetic cell tissue and placed in porous membrane inserts that allow nutrients to enter the cells.
3.3. Skin-on-a-chip tests
There are some drawbacks of 3D representations, such as the absence of skin appendages and vascular system. There are a number of criteria that must be completed before testing can begin. These involve collecting cellular components at precise sites for in-depth biological investigation and sampling luminal contents to examine medication adsorption, distribution, metabolism, elimination, and toxicity. The skin-on-a-chip design was developed,47 which involves cultivating skin tissue in a microfluidic system48 and 3D bioprinting technology49 as shown in Fig. 2. To mimic the operation of the 3D microenvironment of real skin, physical and biochemical parameters can be manipulated, such as medium flow, mechanical stresses, and biochemical gradients.48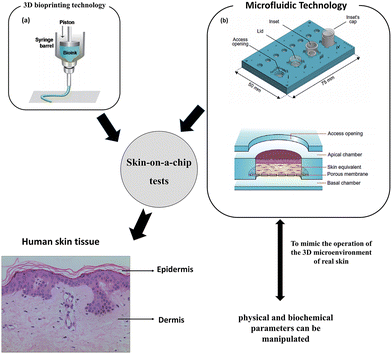 | ||
| Fig. 2 Skin-on-a-chip tests. Skin-on-a-chip models mimicking native human skin structures using microfluidic and 3D bioprinting technology. Images (a and b) are reproduced with permission from ref. 50 and 51, respectively. | ||
3.4. Biosensors
When it comes to the detection of cosmetics, in addition to the above methods, biosensors must be mentioned. Biosensors play an important role in cosmetic-related detection. They can not only carry out antioxidant testing, toxic element testing, and active substance testing of cosmetics, but also play an important role in cosmetics’ health and safety testing.Described as “an independent analytical device that blends biological components with physical and chemical devices to detect analytes of biological interest”,8 a biosensor is a device that is sensitive to biological substances and converts their concentration into electrical signals for detection. A wide range of chemical, physical, or biological interactions can be converted into electrical signals using sensors used in biosensors.52 Generally, biosensors can be divided into enzyme biosensors, cell sensors, immune sensors, microbial sensors, etc. according to the biological components used.53 A variety of practices have shown that these biosensors are also effective means of safety detection of cosmetics.54
3.5. Oxidation resistance test
Electrochemical biosensors are the most representative of all biosensors. Because of their high sensitivity, high throughput and low cost, they have received extensive attention. The detection principle is based on antigens, antibodies, enzymes, nucleic acids, aptamers and other biological recognition elements. Capturing the antioxidant substances to be measured will cause changes in the current, impedance, potential or conductivity of the sensor surface.55 For example, cell lines (such as Caco-2, HepG2 and IPEC-J2), biosensors56 and antioxidant enzyme activity detection (catalase, superoxide dismutase)57 can be used to detect antioxidant activity based on cells and enzymes.3.6. Toxic element detection
Biosensors are highly sensitive to the detection of heavy metals in the routine detection of cosmetics. Taking the detection of mercury as an example, enzyme biosensors, optical fiber biosensors, cell sensors, immunosensors and some new sensors can be used for detection. An enzyme biosensor for heavy metal detection uses the inhibition of heavy metal ions on enzymes and convert them into electrical signals, thereby determining the components. For example, a biosensor using fixed glucose oxidase detects the decomposition of hydrogen peroxide by an amperometric method, and determines the inhibition effect of Hg2+, Ag+, and Cu2+ plasma on enzymes, respectively, so as to detect heavy metal ions.58An optical fiber biosensor monitors the heavy metal ions based on the inhibition of urease activity. Heavy metal ions can reduce the biocatalytic activity, reduce the amount of urea hydrolysis and cause pH changes. They are detected by the optical fiber biosensor at a wavelength of 615 nm.59
Cell sensors are also used in the detection of heavy metal ions in cosmetics. A kind of biosensor uses heart bottom cells as biosensors to detect the harm of heavy metal ions. Hg2+, Pb2+, Cu2+ and Zn2+ can cause cardiomyocytes to show changes in the frequency, amplitude and duration within 15 minutes.60
The test strip based on microbial biosensors for qualitative detection of soluble and insoluble mercury pollution in cosmetics is one of the new sensors that have emerged at the same time. Considering that the biosensor's cells are capable of automatically converting insoluble Hg2Cl2 and Hg(NH2)Cl into soluble Hg(II) ions, when the test paper turns red, it indicates whether the overall mercury pollution in cosmetics exceeds 1 mg kg−1. No specific tools are required for the entire detection process.61 As a result, this paper offers a straightforward, affordable detection approach.
3.7. Active substance detection
Biosensors can also detect the content of active substances such as ferulic acid added in cosmetics. Ferulic acid in skin care has a good effect of boosting whitening and relieving inflammation. Enzyme biosensors are mostly used to determine its composition, and the enzyme is installed on the biosensitive membrane using immobilized technology. It can react to provide usable information material if the sample includes the correct enzyme substrate, showing that the electrode has modifications that can be translated into electrical signals. The existence and amount of chemicals can be estimated from this alteration.62 For instance, the electrochemical detection of ferulic acid is carried out using a novel biosensor called CNF GNP ty/SPE that is based on nanomaterials and tyrosinase. This approach is popular because it is precise, easy, and affordable. It can be used to conduct routine analyses for the quality control of samples for cosmetics, medications, and other products.543.8. Microbiological indicators of cosmetic testing
Microbial contamination of cosmetics is a key issue of social concern. Contaminated microorganisms may produce some metabolites and stimulate skin inflammation or allergic reactions. Therefore, it is very important to regulate the effective supervision and management of cosmetics’ quality. The possibility of using biosensors to find microbes in cosmetics is also promising. Taking Staphylococcus aureus as an example, the traditional method is the conventional culture method. This method has high reliability and low cost, but it has low sensitivity and a long detection cycle. Electrochemical biosensors and spectrum analysis sensors have recently been used in new detection techniques as a result of the ongoing advancements in science and technology.55Electrochemical biosensors are the most representative of all biosensors. Due to their high sensitivity, high throughput, and low cost, they have received a great deal of attention. Their detection principle is based on biological identification elements such as antigens/antibodies, enzymes, nucleic acids, aptamers, etc. Capturing Staphylococcus aureus for testing will cause changes in parameters such as current, impedance, potential or conductivity on the surface of the sensor. Quantitative analysis of the concentration of Staphylococcus aureus can be performed by monitoring the changes of these chemical signals.63 For example, a multiple electrochemical biosensor based on ultra-sensitive peptides is used to detect Staphylococcus aureus. This biosensor is made up of a number of carbon electrodes that have been modified with gold nanoparticles and fixed with magnetic nanoparticles attached to particular peptides using the streptavidin–biotin interaction. Within a minute, the proteolytic activity of two bacteria's protease on particular peptides can be observed. This platform for biosensor arrays has attained exceptional sensitivity.64 This approach offers hope for the simultaneous, quick, low-cost detection of several microorganisms.
Because of their high sensitivity, excellent specificity, and quick detection speed, optical biosensors have gradually replaced the relay chemical detection approach for the detection of Staphylococcus aureus.63 For example, a surface plasmon resonance (SPR) sensor with a micro-contact imprinting sensor chip is used to detect Staphylococcus aureus. The principle is to recognize Staphylococcus aureus through micro-contact imprinting and optical sensor technology.65
3.9. Cosmetic safety test
Antibiotics have bactericidal and bacteriostatic effects. Antibiotics in cosmetics mainly include metronidazole, chloramphenicol, ofloxacin, etc., which can inhibit skin microorganisms, enhance the ability of skin to resist bacterial infection, and achieve the effect of skin protection on the surface.66 The long-term use of cosmetics with antibiotics can cause adverse reactions such as contact dermatitis, such as erythema, edema, scaling, exudation and burning. The long-term use of antibiotics will also lead to the increase of bacterial antibiotic resistance, which will reduce the efficacy and delay the treatment.67 In the past decade, biosensors have been widely used to detect antibiotic residues because of their high sensitivity, rapid response, easy miniaturization and low price. Antibiotics have been suggested to be detected using nanostructured electrochemical platforms made of several materials, including carbon nanoparticles, metal nanoparticles, magnetic nanoparticles, metal–organic frameworks, and quantum dots.68 The electrochemical biosensor that uses receptors and enzymes can use a variety of enzymes and receptors that have been nano-materially tagged to detect different kinds of antibiotics. For example, Kling et al. designed an experiment using a highly sensitive biomolecular sensor system to simultaneously detect tetracycline and streptococcin, two commonly used antibiotics.69Although different types of biosensors have their own advantages, their application has greatly improved the detection specificity, sensitivity and anti-interference ability, and further shortened the detection time. However, there are still some problems to be improved: the accuracy of sensor recognition is not accurate enough and the biometric elements of some sensors are mainly antibodies, which have poor stability, easy deactivation and a long preparation cycle.55,68 Moreover, biosensors are not suitable for the detection of all substances in cosmetics, such as preservatives, hence, high performance liquid chromatography is mainly used for their detection instead of biosensors.70 The principles and detection substances of several biosensors mainly used to detect cosmetic substances are summarized as follows (Table 3).
| Biosensor | Mechanism | Tested substances |
|---|---|---|
| Enzyme biosensor | 1. An enzyme biosensor for heavy metal detection uses the inhibition of heavy metal ions on enzymes to affect the activity of enzymes and change the concentration of substrates or products | 1. Heavy metal ions |
| 2. Additives in cosmetics, such as ferulic acid | ||
| 2. The main principle of an enzyme biosensor for the detection of substances other than heavy metal ions is to use immobilized technology to install the enzyme on the bio-sensitive membrane. If the sample contains the corresponding enzyme substrate, it can react to produce acceptable information material, indicating that the electrode has changes that can be converted into electrical signals. According to this change, the existence and quantity of substances can be determined | 3. Staphylococcus aureus and other microorganisms | |
| 4. Antibiotics | ||
| Optical biosensor | 1. The inhibition of heavy metal ions on urease is monitored. Heavy metal ions can reduce the biocatalytic activity, reduce the amount of urea hydrolysis and cause pH changes. It is detected by an optical fiber biosensor at 615 nm wavelength | 1. Heavy metal ions |
| 2. The principle of detecting Staphylococcus aureus and other microorganisms is to recognize Staphylococcus aureus through micro-contact imprinting and optical sensor technology | 2. Staphylococcus aureus and other microorganisms |
4. Microfluidic design for in vitro screening of cosmetics
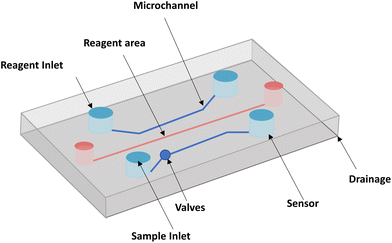
Microfluidic chips typically include reagent inlets, sample inlets, valves, microchannels, sensor assemblies, and drainage systems, as shown in Fig. 3. With the development of microfluidics allowing dynamic culture and precise control of the internal cellular microenvironment, a more comprehensive mimic of human skin has been achieved. With the test of time, microfluidic platforms have also proven to be an effective tool for in vitro screening of cosmetics.71 The following sections summarize the materials required for the microfluidic design of in vitro cosmetic screening and related technological advances.4.1 Fabrication materials
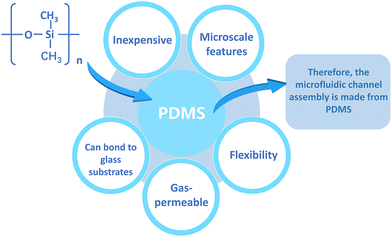
Skin-on-a-chip models are created by combining skin culture models with microfluidics. Complex tissue-like structures can be created using microfluidic chips. There are multiple methods to construct microfluidic devices and different materials are utilized to fabricate them. Various materials include silicon-based elastomers such as polydimethylsiloxane (PDMS), glass, and thermoplastic polymers such as polystyrene (PS), polycarbonate (PC), and polymethyl methacrylate (PMMA).72 Among these materials, PDMS is very popular because it is relatively inexpensive and possesses the ability to map microscale features, in addition, it is flexible, gas permeable, and able to bond to glass substrates.73,74 These advantages are summarized in Fig. 4. Therefore, PDMS is often used to construct microfluidic channel assemblies.75,76However, the hydrophobic and porous nature of PDMS can cause adsorption of numerous medicinal drugs and other tiny molecules.74 Furthermore, the mass production and commercialization of PDMS-based devices have been a challenge due to the high cost of the current manufacturing strategies. In contrast, thermoplastic polymers are not permeable and therefore do not adsorb small molecules and can be sculpted using a variety of microfabrication techniques.77 Thermoplastics are also receiving increasing attention because they offer several advantages over PDMS: they are cheaper, have less absorption and evaporation, and have higher chemical resistivity.78 The use of thermoplastics can be more easily transitioned from academia to industry, as they can be fabricated through rapid prototyping and industrial-scale manufacturing techniques. The integration of membranes on thermoplastics can be performed using various techniques such as thermal bonding, ultrasonic welding, laser welding, solvent bonding and surface functionalization.76 Alternatively, these materials can be reversibly sealed using clamps or magnets, thus imposing minimal requirements on membrane materials.79 Sun et al.80 reported the creation of a gravity-driven, microfluidic-based full-thickness human skin-on-a-chip platform that incorporates an endothelialized, perfusable microvascular network for simulating HSV infection, host immune response, and antiviral drug efficacy. The vascularized microchip skin then becomes free of polydimethylsiloxane (PDMS), a widely used material in microfluidic systems, but since it has a well-known tendency to absorb small molecules and interfere with drug delivery, glass was used instead.
4.2. Advances in microfluidics
An organ-on-a-chip (OoC) is a cell culture system that integrates tissue engineering and microfluidics and aims to build bionic human functional units in vitro. The skin-on-a-chip model for cosmetic testing needs to be a more comprehensive simulation of human skin, which would include the major layers of human skin (dermis and epidermis) and blood vessels, nerves and appendages. Simulation of mechanical signals such as cyclic tensile and shear stresses should also be considered to build a more realistic dynamic model. The most important technology is the integration of sensors in the skin-on-a-chip to monitor skin functions in real time, which is the key to the effective use of microfluidics. The following table summarizes the relevant developments of skin chips in recent years.The skin protects against pathogens, bacteria, fungi, and viruses through the physical barrier of the epidermis and the innate and adaptive immune system, which includes immune cells and biomolecules in the skin.85 The implementation of skin-on-a-chip models that can correctly mimic the immune system will not only allow the evaluation of possible adverse reactions triggered by cosmetic products and assess their molecular toxicity, but will also help understand their cellular mechanisms, simulate skin diseases and investigate the possible therapeutic approaches.
The methods of simulating immune responses in the skin in microfluidic devices are divided into two types: adding cytokines to induce immune responses or directly doping immune cells in the microarray.86 These 3D skin models that integrate immune components consist of different cell types, so it is important to select the appropriate immunocompetent 3D skin model according to the targeting mechanism of the drug under test.87
Notably, the microbiome is often forgotten in skin-on-a-chip models and there is still a lack of reports on integrated microbiota.88 It is well known that human skin is chronically exposed to air and thus colonized by numerous microorganisms, and the host and symbiotic microbiota rely on immune cell and molecular network interactions to establish and maintain healthy skin homeostasis.89 Integrating microbiota in skin-on-a-chip devices helps to fully replicate human skin models and construct more realistic human skin equivalents. The addition of other microorganisms from the skin and sebaceous glands, blood flow or other mechanical requirements will more closely represent human skin, facilitating permeability studies or simulation of different pathologies caused by microbiota alterations.
At the same time, human skin microbial communities are richly diverse, and their composition is influenced by age, body site, and gender. The abundance and composition of the skin flora vary greatly from site to site and are closely related to the distribution of skin sebaceous glands and skin humidity.90 The integration of microbiota in skin-on-a-chip devices allows the study of personalized treatment of pathologies caused by microbiome dysfunction by adding specific microbiota groups of affected patients to the skin-on-a-chip model.89
One of the decisive features of the microfluidic device is the perfusion of the culture medium. It can not only remove the waste produced by the cells and help the culture medium to update the nutrient content, but also, more importantly, can endow the cells with shear stress. Shear stress refers to the force parallel to the object caused by particles flowing or sliding through the object. Shear stress is common in the human body. All our organs are exposed to a certain degree of shear stress, and our skin is no exception. Applying shear stress to skin tissue can improve barrier function, improve epidermal differentiation, and increase the density of fibroblasts in skin tissue.91 Now there are many methods for the perfusion of culture media, and these media can be divided into pump-driven or gravity-driven solutions. Microfluidic devices usually use peristaltic pumps or injection pumps to generate fluid flow. Both methods have their advantages and disadvantages. The pumping equipment can be used for a long time and can provide programmable fine-tuning flow control.92 The gravity-driven method is simpler to install, owing to which the culture medium is recycled through the swing platform to avoid the pipeline and complex setting requirements.93 However, the use of pipelines for pumping equipment will increase the risk of cross contamination, and it is very time-consuming to set up; the gravity-driven method lacks precise control of the flow rate, and the advantages of fresh medium infusion and waste removal are not significant, and it is also easy to lead to the change of hydrostatic pressure with time.83,94 Therefore, it is of great significance to continuously improve the relevant performance of the perfusion equipment of the microfluidic equipment, to ensure that the shear stress is continuously applied to the base layer and top layer, and improve the fine control of the fluid environment to improve the authenticity of the skin model.
5. Microfluidic skin model and its application in cosmetics
5.1 Microfluidic skin disease models
The study of disease and the identification of drug and cosmetic targets require the availability of comprehensive model systems that can reproducibly generalize developmental and physiological processes. As the skin-on-a-chip technology continues to mature, an increasing number of skin microfluidic models have emerged to assist researchers in the in vitro screening of drugs and cosmetics. Currently, the most relevant models are focused on the field of skin inflammation, which has stimulated more research due to the complex etiology and large patient population. Secondly, some relatively novel models of skin diseases in recent years, such as new advances in skin wound healing models, are presented in this section for your reference.Recently, Gupta et al.106 provided a microcapillary shear stress-influenced microchip skin wound healing model of fibroblast-derived wound healing. This work describes the use of microfluidic devices to simulate wounds on fibroblast monolayers via trypsin flow and PDMS barriers. In this study, a microfluidic chip that provides 3D cell culture conditions simulating a 3D extracellular matrix (ECM)-like environment and allows real-time monitoring of cells is reported. A dual-chip design was prepared: one with a central inlet for trypsin-containing medium flow and the second with a removable column for wound creation to monitor cell migration. Wounds created using the removable PDMS barrier were reproducible and simple. Design 1 resulted in the creation of wounds with high variability and poor reproducibility, while Design 2 created wounds with high precision and reproducibility. This experiment overcomes the lack of reproducibility of traditional wound healing assay techniques by preparing a simple and effective wound healing assay using a microfluidic chip. This dynamic wound healing assay will provide greater convenience for conducting research related to drug discovery, cellular pathway studies, and wound healing mechanisms. As the number of channels increases, a larger number of cells can be integrated into the chip to study the dynamic and complex skin microenvironment.
5.2 Microfluidic design for in vitro cosmetic screening applications
Interspecific differences lead to the deviation of animal skin from human skin in physiology and immunity, as well as the lack of a physiologically related tissue microenvironment. Traditional animal models and two-dimensional models in vitro cannot accurately describe the toxic effects and predict the actual in vivo reactions.87 In order to address these gaps, the past few years have witnessed the rapid development of several complex three-dimensional (3D) models and skin-on-a-chip platforms. The SoC platform is expected to replicate the physiological structure and functions of native human skin on a relatively low-cost and high-throughput platform, which can better represent skin in vitro and replace the ethically complex animal experiments. The following describes the application of skin chips in cosmetic toxicology, the transdermal absorption effect and the screening of active ingredients in cosmetics, and research and development of anti-aging cosmetics.Before putting new cosmetic ingredients on the market, it is necessary to evaluate their safety, including the skin sensitization hazard and efficacy. Exposure to skin sensitizers can induce specific immune reactions, and repeated local exposure may even cause allergic contact dermatitis. Skin sensitization is a complex immune process. In view of the need for information on skin sensitization in the safety assessment of EU cosmetics regulations, it has become the subject of many tests. 3D organ chip models with normal immune functions have been explored to produce accurate models for evaluating the reaction or toxicity of a cell monolayer.107 Wuefer et al.108 developed a miniature model of human skin in a microfluidic platform composed of the epidermis, dermis and endothelium. Each layer is separated by a transparent porous membrane to allow inter-layer communication and simulate skin biology (Fig. 5a). They showed that their SOC model could be used to build an in vitro skin disease model or test the toxicity of cosmetics. In addition, Mori et al.91 proposed a 3D chip skin micro-device, whose vascular channels are coated with endothelial cells (Fig. 5b). This micro-device includes a skin equivalent fixed on a culture device connected to an external pump and tube. This model can be used for the development of skin therapy and cosmetics. Légues et al.109 also developed an innovative 3D full-size bioprinted human skin model, which contains immune cells, for screening cosmetic ingredients aimed at reducing or preventing inflammation. Lee et al.110 reported a mixed human skin model capable of performing a detailed toxicological evaluation of drug and cosmetic compounds (Fig. 5c). This model successfully simulates the skin–nerve mixed model and the skin–liver model. This model can detect the changes of neuron activity in combination with calcium imaging technology to conduct real-time quantitative skin sensitization analysis, and can also evaluate the potential hepatotoxicity of chemicals applied to the skin in the model. This 3D hybrid skin chip will provide a useful human skin model for the cosmetics industry for toxicological screening.
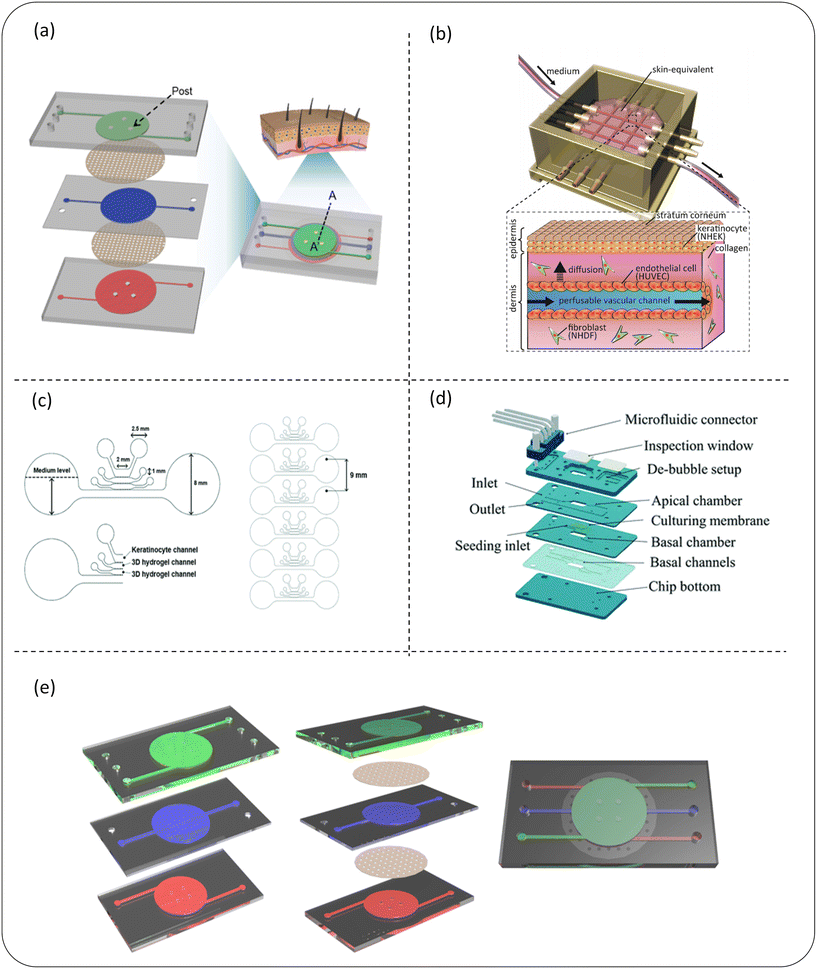 | ||
| Fig. 5 Schematic diagram of a microfluidic device for skin sensitization (a–c) or skin irritation (d and e) detection of cosmetics. (a) The 3D skin chip device of the chip epidermis system composed of three PDMS layers and two PET porous membranes can be used to build an in vitro skin disease model or test the toxicity of cosmetics; (b) the schematic diagram of the skin chip integrated with the perfusion vascular channel which can be used to test the toxicity of cosmetics; (c) a microfluidic chip design of a mixed skin model can not only quantitatively analyze skin sensitization, but can also evaluate the potential hepatotoxicity of chemicals applied to skin in this model; (d) it is composed of four main PMMA layers and a microfluidic connector, which can be used for skin irritation evaluation of cosmetic ingredients in vitro; (e) a skin model (keratinocyte – green, fibroblast – blue and endothelial cell – red) on a chip composed of three layers representing the composition of the epidermis, dermis and endothelium; images (a–e) are reproduced from ref. 91, 108, 110, 112 and 113. | ||
Skin irritation refers to the reversible damage to the skin caused by the expansion and increase in the permeability of endothelial cells after contact with a substance or mixture.111 Alternatives to animal testing methods used for skin irritation assessment, such as the reconstructed human skin model, do not fully represent the physiological response caused by skin irritants. In recent years, skin chips have shown the potential to test the irritation of cosmetics. Zhang et al.112 developed a microfluidic chip based on polymethylmethacrylate (PMMA) (Fig. 5d). The functional chip skin was used to test the stimulation of 10 known toxins and non-toxins. Because the complexity of skin irritation cascade reactions cannot be predicted with certainty only by cytotoxicity measurement, several other irritation reactions, such as inflammatory cytokine release, have also been preliminarily performed to assess the impact of exposure to some of these chemicals. These results show that this organotypic chip is a potential alternative to stimulation detection and can support scalable testing as a physiologically and clinically relevant in vitro epidermal model. The upper surface of this high-fidelity chip provides a potential substitute for the evaluation of skin irritation of cosmetic ingredients in vitro. Jeon et al.113 invented a skin model on a chip consisting of three layers representing the composition of the epidermis, dermis and endothelium (Fig. 5e). After further development, this chip could simulate the physiological response of skin irritation, so it is specially used for skin irritation tests. It solves the problem of the current RhE model's inability to simulate physiological skin irritation. This model evaluated chemical-induced edema by observing the tight connection of endothelial cells, and was further improved and specifically used to evaluate physiological skin irritation, and its prediction ability was enhanced by the two-parameter model. Based on the in vivo data, the sensitivity and accuracy of the classification ability of 20 test substances using the dual-parameter chip model is higher than that of the reconstructed human skin model. This experiment provides a dual-parameter chip model with great potential, opens up a new method for the skin irritation test in vitro, and is expected to become a new paradigm for animal testing to replace the irritation test of cosmetic ingredients.
Historically, the Franz cell has been a widely used in vitro method for assessing molecular permeability, which has the advantages of (i) less tissue treatment, (ii) no need for continuous sample collection and (iii) low drug volume required for analysis. However, this in vitro model not only requires moral consent, but the skin will metabolise some chemicals during the percutaneous absorption process.114 The innovative platform provided by skin chips is crucial to overcome the limitations of diffusion cell technology.115 The main advantage of these devices is that they simulate the microcirculation of the skin through a dynamic arrangement, which is easy to use and small in size (less tissue, cell, test substance and carrier requirements).
Varga-Medveczky et al.116 developed a new concept of a microfluidic device to test the penetration of drugs through the skin by improving the first generation microfluidic diffusion chamber (MDC) designed and fabricated by Lukács et al. (Fig. 6a). The hydrophilic model drug caffeine cream was placed in the microchip donor space, and the peripheral perfusion fluid (PPF) passed through the system at a rate of 4 μL min−1. The results showed similar transport kinetics and good reproducibility when comparing the transport of caffeine in the artificial skin equivalent with the human skin sample. The transdermal permeation curves of two P-glycoprotein substrates, erythromycin and quinidine, were studied in the presence and absence of a P-gp inhibitor (PSC-833). It has been demonstrated that the MDC system can be used for the study of the interaction between the skin chip microfluidic system and the transdermal drug delivery and transport protein at the dermal barrier. In conclusion, MDC can be used for in vitro/in vivo transdermal drug or cosmetic delivery. Chen et al.117 reported a dermal fibroblast sphere (DFS) microfluidic (MF) chip model, which is used to grow a large number of dermal fibroblast sphere (DFS) arrays in bionic hydrogels under near-physiological flow conditions, and is capable of screening skin care active ingredients (CI) (Fig. 6b). Although spheroid-based skin models are considered promising models for screening AI candidates for skin care products, they are currently not widely used in skin care research.24,25 However, the microfluidic (MF) model designed by Chen et al. based on dermal spheroids retains its inherent function in the production and organization of the dermal ECM and shows the capability of easy fabrication, amplification and time-saving AI screening. In the proof-of-concept design, DFS on-a-chip devices can screen 12 different AI or AI combinations on a single chip. This model may be extended to high-throughput screening of AI used in skin care products. This work shows a method of screening AI for time, cost and labor efficiency in research laboratories and the cosmetic industry.
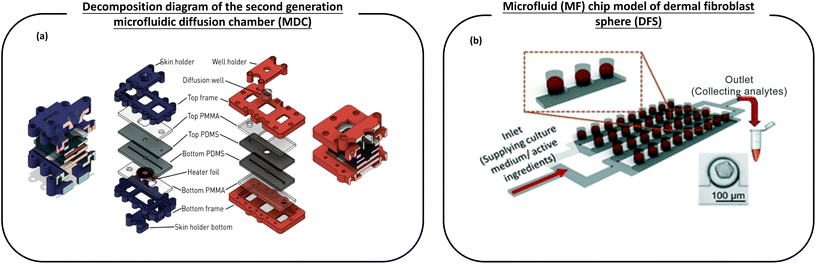 | ||
| Fig. 6 The schematic diagram of the microfluidic device used for the transdermal absorption effect and screening of active ingredients in cosmetics. (a) Left (blue): cross section and layer-by-layer view of the first generation temperature control equipment with a skin sample rack. Right (red): the decomposition diagram of the second generation microfluidic diffusion chamber (MDC) with a diffusion well support improved by Varga-Medveczky, which is used to test the penetration of drugs through the skin; (b) a microfluidic (MF) chip model of dermal fibroblast spheroids (DFS), which is used to grow a large number of dermal fibroblast spheroid (DFS) arrays in bionic hydrogels under conditions close to physiological flow, and has the ability to screen active ingredients (AI) of skin care products. Images (a and b) are reproduced with from ref. 116, 117. | ||
Jeong et al.120 used a flexible chip skin (FSOC) and a mechanical stimulation actuation system (MSAS). Mechanical compression stimulation reflecting circadian rhythms is applied to 3D skin to generate an ageing skin model (Fig. 7a). The ageing markers were examined in a 28-day comparative culture experiment to demonstrate the progression of the ageing process in the dermis and epidermis. The experimental results showed that the equivalent shrinkage of the full-thickness skin and the thickness of the epidermis decreased after mechanical stimulation, and β-galactosidase gene expression increased, indicating that the ageing effect of the model is successful. It is expected that the new aging mechanism, which can be used for screening and efficacy testing of new anti-aging substances, will be revealed using this new skin chip aging model. This is of great importance for the development of new drugs for the treatment of skin diseases and for the development of anti-aging cosmetics.
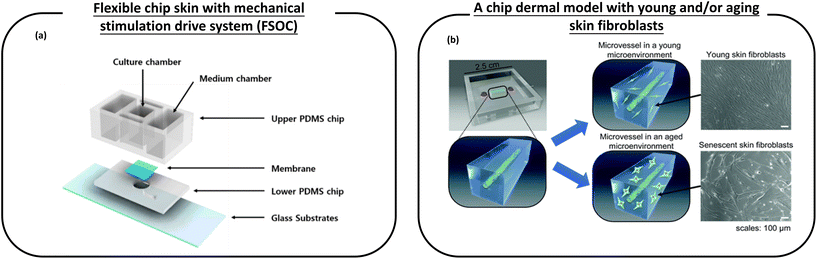 | ||
| Fig. 7 Schematic diagram of a microfluidic device used for anti-aging cosmetic research. (a) A flexible chip skin (FSOC) with a mechanical stimulation drive system can be used for screening and efficacy testing of new anti-aging substances; (b) a chip dermal model, which has young and/or aging skin fibroblasts, thus allowing the study of changes caused by aging dermal fibroblasts on microvessels, provides a platform for the development of anti-aging drugs. Images (a and b) are reproduced from ref. 120 and 121. | ||
Pauty et al.121 have created a skin model on a chip that makes it possible to study the changes caused by ageing skin fibroblasts on microvessels (Fig. 7b). This model consists of an in vitro 3D model of blood vessels embedded in a collagen type I ECM. Young and/or ageing skin fibroblasts are attached to the ECM. This simple setup allows monitoring only the effects of ageing fibroblasts, which are the main participants in skin ageing, and is unaffected by other skin cells (such as neurons or immune cells). The experiments showed that SASPs are involved in angiogenic activation and they tend to show excess traction stress associated with morphological changes in the ECM that ultimately lead to angiogenesis. Therefore, this experiment provides a platform for the development of anti-aging drugs.
At the same time, we note that Hausmann et al.122 have proposed a new idea with emphasis on the individualised construction of a skin ageing model. The expression of ageing is heterogeneous due to internal and external factors. At present, the development of a skin ageing model ignores the highly individualised ageing process, which is beneficial for the general conclusion of elderly patients. Therefore, Hausmann et al. proposed a strategy to optimise drug treatment for elderly patients. It is necessary to take into account the heterogeneity of patients to unlock their full potential in order to narrow the gap between the model and its internal counterpart. This means that the multi-organ combined chip (which was introduced in section 2.3) is the development direction of the future skin ageing model, which will be conducive to the development of anti-aging cosmetics that are better suited to personal characteristics.
5.3 Multi-organ joint chip
Adverse skin drug reactions are mediated by at least two different mechanisms, both of which involve systemic interactions between the liver and immune and dermal tissues. The existing in vitro skin models cannot comprehensively summarize these complex multi-cellular interactions to predict the skin sensitization potential of drugs. At the same time, considering the heterogeneity of the human body and the integrity of the internal system of the body, a human–computer chip formed by connecting the SoC platform to multiple OoC devices has emerged to evaluate the system-wide impact.123 This combination of multiple OoC devices will provide more authoritative and comprehensive evidence for toxicological tests of cosmetic screening.It is noteworthy that Chong et al.124 have recently reported a novel in vitro drug screening platform, which includes a microfluidic multicellular co-culture array (MCA). It simulates different mechanisms of action using a series of simple cell assays, and integrates the obtained readings with machine learning algorithms to predict the skin sensitization potential of systemic drugs. MCA is composed of two PDMS layers and four cell culture chambers connected by diffusion microchannels. Cell culture is carried out using hepatocyte spheroids (HHS) derived from human cell lines HepaRG, U937 myelocytes, HaCaT (keratinocytes) and human dermal fibroblasts. This platform can realize the crosstalk between hepatocytes that produce drug metabolites, antigen presenting cells (APC) that detect the immunogenicity of drug metabolites, and keratinocytes and dermal fibroblasts. Single drug screening using MCA can generate five readings at the same time. These readings are integrated using a support vector machine (SVM) and principal component analysis (PCA) to classify and visualize drugs as skin sensitizers or non-skin sensitizers. The hierarchical 4-fold cross validation (CV) on SVM proves that MCA has 100% sensitivity, 87.5% accuracy and 75% specificity to predict the skin sensitization potential of drugs. This platform simplifies complex potential mechanisms into several simple cell models, including liver-mediated drug biological activation models and downstream immune and skin models, and combines the multi-indicator phenotypic readings obtained from these cell models with machine learning algorithms. At the same time, it also shows that the MCA system has the potential of development and can be used as a high-throughput drug and cosmetic screening platform, especially for skin sensitization and toxicity after further validation research.
Hausmann et al.122 proposed a strategy to optimize drug treatment for elderly patients. In order to narrow the gap between the model and its internal counterpart, it is necessary to consider the heterogeneity of patients to release their full potential. The perfusion and combination of individual tissues on the chip create an interconnected organ system to access system data in vitro. If the most relevant organs and diseases are provided on the chip, these models will provide unprecedented insights into the systematic fate of drugs in vitro. The patients on the chip, especially the elderly chip patients, will more accurately predict the pharmacokinetics and pharmacodynamics of the elderly. In particular, the combined effects of changes in the skin barrier function and vascular diseases and the effects of decreased liver and kidney functions are predictable in the future.
6. Challenges, conclusions and prospects
So far, great progress has been made in the use of SoC equipment to develop bionic artificial skin and for cosmetic screening. The development of artificial skin tissue continues to improve comprehensively. These achievements, combined with the latest progress in tissue engineering, provide high-throughput, automated and realistic skin models for cosmetic and pharmaceutical applications. It is foreseeable that with the continuous improvement of more and more skin-on-chip devices combined with microfluidic technology, it is expected to provide more diversified skin disease models for drug research and cosmetics screening. The skin-on-a-chip model will also become a promising platform for in vitro evaluation of cosmetic toxicology, the transdermal absorption effect and the screening of active ingredients in cosmetics, and research and development of anti-aging cosmetics. However, the skin-on-a-chip model still faces many challenges. No device can completely provide a truly complete method that can replace animal testing. In the field of cosmetic research and development and screening, researchers believe that the following challenges still exist.6.1 The technology of complete replication of skin appendages and the microenvironment
Microfluidic technology has made continuous progress, which has solved many limitations of traditional skin tissue engineering, such as vascularization, adding immune components, etc. However, skin is a complex structure composed of the epidermis, dermis, subcutaneous tissue and skin appendages (hair, hair follicles, sweat glands, sebaceous glands and fingernails). The current SoC model still cannot be fully replicated and integrated. More importantly, the skin surface has an important micro-ecosystem, and there is still a lack of reports and research related to skin chips. Skin microbiome is an ecosystem composed of various microorganisms and the microenvironment of the host skin and skin surface. Skin microbiota consists of bacteria, viruses, fungi, mites and other microorganisms that are planted on the skin, with bacteria being the most prevalent microorganism. When endogenous and exogenous factors lead to the imbalance of micro-ecology, the resident bacteria multiply in large numbers, leading to the occurrence of skin diseases, such as acne caused by Propionibacterium acnes or Seborrheic dermatitis caused by Malassezia. Therefore, how to completely replicate skin appendages and integrate the skin microenvironment is an important issue that calls for a more complete platform for the construction of an in vitro screening model of cosmetics. By more closely imitating human tissue, the SoC model can limit the number of drugs or cosmetics that have successfully passed pre-clinical tests but failed in clinical trials, and reduce the risk of cosmetics that have failed in animals but may be effective for humans.6.2 Perfection of disfiguring disease models (such as pigmented diseases)
At present, the number of skin disease models of SoC devices has increased, but the model of disfiguring disease is still blank. Disruptive diseases, such as pigmented diseases, are caused by genetic and environmental factors and a large number of patients suffer from such diseases worldwide. Although most pigmented skin diseases do not pose a major threat to health, they hinder beauty, thus causing mental pressure on patients and affecting their work, study and life. Therefore, searching for more effective cosmetic active ingredients for the treatment of the corresponding diseases will help find useful drug therapeutic-assisted alternative therapies for this group of patients. The construction of a disfiguring disease model requires the mechanism of related diseases to be simulated on the basis of the original skin chip. However, the mechanism of disfiguring diseases is complex, so how to build relevant models on skin chips in the future is still a field that needs to be explored continuously.6.3 The future direction of improving SoC devices should focus on the following areas: providing a personalized skin chip platform
More and more researchers are putting forward the concept of personalization, which is due to the era background of precision medicine. This means that pluripotent stem cells (iPSCs) that can differentiate into almost all types of skin cells will be used more and more in the construction of skin chips in the future. IPSCs from specific patients will be used in the chip design that simulates the health and disease characteristics of patients, which is of great significance for the pre-clinical evaluation (including therapeutic drugs and immunotherapy) that characterizes the immune mechanism behind the results of a wide range of diseases.6.4. Improving repeatability and efficiency through technology and test standardization
Building a 3D skin model is time-consuming and laborious. With the complete replication of skin appendages and the microenvironment, the skin chip structure will become more complex. The increase of technological and biological differences is inevitable. Therefore, the future standardization of chip design, materials and manufacturing technology is an inevitable trend. For example, with the help of bioprinting technology, the scale and quality of skin models are kept constant, technical differences are reduced, and the repeatability and efficiency of 3D skin models are improved. This will greatly simplify the cross-laboratory validation of these devices. At the same time, the future skin-on-a-chip platform is crucial for real-time, sensitive and accurate monitoring of biological parameters. According to the specific background and research objectives, the selection and integration of biosensors should also have standardization restrictions.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (Grant No. 81904207), the Natural Science Foundation of Sichuan Provincial Department of Science and Technology (2022NSFSC1446) and the Popularized Application Project of Sichuan Provincial Health Commission (No. chuan-gan-yan2023-214).References
- EC. REGULATION (EC) No 1223/2009 on cosmetic products, Off. J. Eur. Union, 2009, L342, 59–209 Search PubMed
.
- L. C. Parish and J. T. Crissey, Clin. Dermatol., 1988, 6, 1–4 CrossRef CAS PubMed
.
- F. Pistollato, F. Madia, R. Corvi, S. Munn, E. Grignard, A. Paini, A. Worth, A. Bal-Price, P. Prieto, S. Casati, E. Berggren, S. K. Bopp and V. Zuang, Arch. Toxicol., 2021, 95, 1867–1897 CrossRef CAS PubMed
.
-
E. Parliament, EU Directive 2010/63/EU on the protection of animals used for scientific purposes, 2010 Search PubMed
.
-
E. Parliament, Regulation (EC) No 1907/2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), 2020 Search PubMed
.
- S. H. Mathes, H. Ruffner and U. Graf-Hausner, Adv. Drug Delivery Rev., 2014, 69–70, 81–102 CrossRef CAS PubMed
.
- S. Choudhury and A. Das, Cytotherapy, 2021, 23, 1–9 CrossRef CAS PubMed
.
- A. Hasan, M. Nurunnabi, M. Morshed, A. Paul, A. Polini, T. Kuila, M. Al Hariri, Y. K. Lee and A. A. Jaffa, BioMed Res. Int., 2014, 2014, 307519 Search PubMed
.
- S. N. Bhatia and D. E. Ingber, Nat. Biotechnol., 2014, 32, 760–772 CrossRef CAS PubMed
.
- D. Huh, G. A. Hamilton and D. E. Ingber, Trends Cell Biol., 2011, 21, 745–754 CrossRef CAS PubMed
.
- S. Ahadian, R. Civitarese, D. Bannerman, M. H. Mohammadi, R. Lu, E. Wang, L. Davenport-Huyer, B. Lai, B. Zhang, Y. Zhao, S. Mandla, A. Korolj and M. Radisic, Adv. Healthcare Mater., 2018, 7, 1700506 CrossRef PubMed
.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed
.
- M. Astolfi, B. Peant, M. A. Lateef, N. Rousset, J. Kendall-Dupont, E. Carmona, F. Monet, F. Saad, D. Provencher, A. M. Mes-Masson and T. Gervais, Lab Chip, 2016, 16, 312–325 RSC
.
- V. Chaudhary, V. Khanna, H. T. Ahmed Awan, K. Singh, M. Khalid, Y. K. Mishra, S. Bhansali, C.-Z. Li and A. Kaushik, Biosens. Bioelectron., 2023, 220, 114847 CrossRef CAS PubMed
.
- P. Manickam, S. A. Mariappan, S. M. Murugesan, S. Hansda, A. Kaushik, R. Shinde and S. P. Thipperudraswamy, Biosensors, 2022, 12, 562 CrossRef CAS PubMed
.
- P. M. Elias and J. S. Wakefield, Clin. Rev. Allergy Immunol., 2011, 41, 282–295 CrossRef CAS PubMed
.
- M. Gunnarsson, E. H. Mojumdar, D. Topgaard and E. Sparr, J. Colloid Interface Sci., 2021, 604, 480–491 CrossRef CAS PubMed
.
- M. Hara-Chikuma and A. S. Verkman, Biol. Cell, 2005, 97, 479–486 CrossRef CAS PubMed
.
- T. Prasch, G. Knübel, K. Schmidt-Fonk, S. Ortanderl, S. Nieveler and T. Förster, Int. J. Cosmet. Sci., 2000, 22, 371–383 CrossRef CAS PubMed
.
- M. Loden, Acta Derm.-Venereol., 1992, 72, 327–330 CAS
.
- J. Krutmann, A. Bouloc, G. Sore, B. A. Bernard and T. Passeron, J. Dermatol. Sci., 2017, 85, 152–161 CrossRef PubMed
.
- D. J. Tobin, J. Tissue Viability, 2017, 26, 37–46 CrossRef PubMed
.
- A. C. Mora Huertas, C. E. H. Schmelzer, W. Hoehenwarter, F. Heyroth and A. Heinz, Biochimie, 2016, 128–129, 163–173 CrossRef CAS PubMed
.
- A. M. Kligman, J. Am. Med. Assoc., 1969, 210, 2377–2380 CrossRef CAS
.
- T. Quan and G. J. Fisher, Gerontology, 2015, 61, 427–434 CrossRef CAS PubMed
.
- N. Arjinpathana and P. Asawanonda, J. Dermatol. Treat., 2012, 23, 97–102 CrossRef CAS PubMed
.
- T. Pillaiyar, M. Manickam and V. Namasivayam, J. Enzyme Inhib. Med. Chem., 2017, 32, 403–425 CrossRef CAS PubMed
.
- B. Desmedt, P. Courselle, J. O. De Beer, V. Rogiers, M. Grosber, E. Deconinck and K. De Paepe, J. Eur. Acad. Dermatol. Venereol., 2016, 30, 943–950 CrossRef CAS PubMed
.
- N. Unver, P. Freyschmidt-Paul, S. Horster, H. Wenck, F. Stab, T. Blatt and H. P. Elsasser, Br. J. Dermatol., 2006, 155, 119–128 CrossRef CAS PubMed
.
- F. Liu, L. Qu, H. Li, J. He, L. Wang, Y. Fang, X. Yan, Q. Yang, B. Peng, W. Wu, L. Jin and D. Sun, Pharmaceutics, 2022, 14, 2308 CrossRef CAS PubMed
.
- W. P. Bowe, J. J. Leyden, C. E. Crerand, D. B. Sarwer and D. J. Margolis, J. Am. Acad. Dermatol., 2007, 57, 222–230 CrossRef PubMed
.
- T. Vos, A. D. Flaxman, M. Naghavi, R. Lozano, C. Michaud, M. Ezzati, K. Shibuya, J. A. Salomon, S. Abdalla, V. Aboyans, J. Abraham, I. Ackerman, R. Aggarwal, S. Y. Ahn, M. K. Ali, M. Alvarado, H. R. Anderson, L. M. Anderson, K. G. Andrews, C. Atkinson, L. M. Baddour, A. N. Bahalim, S. Barker-Collo, L. H. Barrero, D. H. Bartels, M. G. Basanez, A. Baxter, M. L. Bell, E. J. Benjamin, D. Bennett, E. Bernabe, K. Bhalla, B. Bhandari, B. Bikbov, A. Bin Abdulhak, G. Birbeck, J. A. Black, H. Blencowe, J. D. Blore, F. Blyth, I. Bolliger, A. Bonaventure, S. Boufous, R. Bourne, M. Boussinesq, T. Braithwaite, C. Brayne, L. Bridgett, S. Brooker, P. Brooks, T. S. Brugha, C. Bryan-Hancock, C. Bucello, R. Buchbinder, G. Buckle, C. M. Budke, M. Burch, P. Burney, R. Burstein, B. Calabria, B. Campbell, C. E. Canter, H. Carabin, J. Carapetis, L. Carmona, C. Cella, F. Charlson, H. Chen, A. T. Cheng, D. Chou, S. S. Chugh, L. E. Coffeng, S. D. Colan, S. Colquhoun, K. E. Colson, J. Condon, M. D. Connor, L. T. Cooper, M. Corriere, M. Cortinovis, K. C. de Vaccaro, W. Couser, B. C. Cowie, M. H. Criqui, M. Cross, K. C. Dabhadkar, M. Dahiya, N. Dahodwala, J. Damsere-Derry, G. Danaei, A. Davis, D. De Leo, L. Degenhardt, R. Dellavalle, A. Delossantos, J. Denenberg, S. Derrett, D. C. Des Jarlais, S. D. Dharmaratne, M. Dherani, C. Diaz-Torne, H. Dolk, E. R. Dorsey, T. Driscoll, H. Duber, B. Ebel, K. Edmond, A. Elbaz, S. E. Ali, H. Erskine, P. J. Erwin, P. Espindola, S. E. Ewoigbokhan, F. Farzadfar, V. Feigin, D. T. Felson, A. Ferrari, C. P. Ferri, E. M. Fevre, M. M. Finucane, S. Flaxman, L. Flood, K. Foreman, M. H. Forouzanfar, F. G. Fowkes, R. Franklin, M. Fransen, M. K. Freeman, B. J. Gabbe, S. E. Gabriel, E. Gakidou, H. A. Ganatra, B. Garcia, F. Gaspari, R. F. Gillum, G. Gmel, R. Gosselin, R. Grainger, J. Groeger, F. Guillemin, D. Gunnell, R. Gupta, J. Haagsma, H. Hagan, Y. A. Halasa, W. Hall, D. Haring, J. M. Haro, J. E. Harrison, R. Havmoeller, R. J. Hay, H. Higashi, C. Hill, B. Hoen, H. Hoffman, P. J. Hotez, D. Hoy, J. J. Huang, S. E. Ibeanusi, K. H. Jacobsen, S. L. James, D. Jarvis, R. Jasrasaria, S. Jayaraman, N. Johns, J. B. Jonas, G. Karthikeyan, N. Kassebaum, N. Kawakami, A. Keren, J. P. Khoo, C. H. King, L. M. Knowlton, O. Kobusingye, A. Koranteng, R. Krishnamurthi, R. Lalloo, L. L. Laslett, T. Lathlean, J. L. Leasher, Y. Y. Lee, J. Leigh, S. S. Lim, E. Limb, J. K. Lin, M. Lipnick, S. E. Lipshultz, W. Liu, M. Loane, S. L. Ohno, R. Lyons, J. Ma, J. Mabweijano, M. F. MacIntyre, R. Malekzadeh, L. Mallinger, S. Manivannan, W. Marcenes, L. March, D. J. Margolis, G. B. Marks, R. Marks, A. Matsumori, R. Matzopoulos, B. M. Mayosi, J. H. McAnulty, M. M. McDermott, N. McGill, J. McGrath, M. E. Medina-Mora, M. Meltzer, G. A. Mensah, T. R. Merriman, A. C. Meyer, V. Miglioli, M. Miller, T. R. Miller, P. B. Mitchell, A. O. Mocumbi, T. E. Moffitt, A. A. Mokdad, L. Monasta, M. Montico, M. Moradi-Lakeh, A. Moran, L. Morawska, R. Mori, M. E. Murdoch, M. K. Mwaniki, K. Naidoo, M. N. Nair, L. Naldi, K. M. Narayan, P. K. Nelson, R. G. Nelson, M. C. Nevitt, C. R. Newton, S. Nolte, P. Norman, R. Norman, M. O'Donnell, S. O'Hanlon, C. Olives, S. B. Omer, K. Ortblad, R. Osborne, D. Ozgediz, A. Page, B. Pahari, J. D. Pandian, A. P. Rivero, S. B. Patten, N. Pearce, R. P. Padilla, F. Perez-Ruiz, N. Perico, K. Pesudovs, D. Phillips, M. R. Phillips, K. Pierce, S. Pion, G. V. Polanczyk, S. Polinder, C. A. Pope, S. Popova, E. Porrini, F. Pourmalek, M. Prince, R. L. Pullan, K. D. Ramaiah, D. Ranganathan, H. Razavi, M. Regan, J. T. Rehm, D. B. Rein, G. Remuzzi, K. Richardson, F. P. Rivara, T. Roberts, C. Robinson, F. R. De Leon, L. Ronfani, R. Room, L. C. Rosenfeld, L. Rushton, R. L. Sacco, S. Saha, U. Sampson, L. Sanchez-Riera, E. Sanman, D. C. Schwebel, J. G. Scott, M. Segui-Gomez, S. Shahraz, D. S. Shepard, H. Shin, R. Shivakoti, D. Singh, G. M. Singh, J. A. Singh, J. Singleton, D. A. Sleet, K. Sliwa, E. Smith, J. L. Smith, N. J. Stapelberg, A. Steer, T. Steiner, W. A. Stolk, L. J. Stovner, C. Sudfeld, S. Syed, G. Tamburlini, M. Tavakkoli, H. R. Taylor, J. A. Taylor, W. J. Taylor, B. Thomas, W. M. Thomson, G. D. Thurston, I. M. Tleyjeh, M. Tonelli, J. A. Towbin, T. Truelsen, M. K. Tsilimbaris, C. Ubeda, E. A. Undurraga, M. J. van der Werf, J. van Os, M. S. Vavilala, N. Venketasubramanian, M. Wang, W. Wang, K. Watt, D. J. Weatherall, M. A. Weinstock, R. Weintraub, M. G. Weisskopf, M. M. Weissman, R. A. White, H. Whiteford, S. T. Wiersma, J. D. Wilkinson, H. C. Williams, S. R. Williams, E. Witt, F. Wolfe, A. D. Woolf, S. Wulf, P. H. Yeh, A. K. Zaidi, Z. J. Zheng, D. Zonies, A. D. Lopez, C. J. Murray, M. A. AlMazroa and Z. A. Memish, Lancet, 2012, 380, 2163–2196 CrossRef PubMed
.
- A. H. S. Heng and F. T. Chew, Sci. Rep., 2020, 10, 5754 CrossRef CAS PubMed
.
- T.-X. Cong, D. Hao, X. Wen, X.-H. Li, G. He and X. Jiang, Arch. Dermatol. Res., 2019, 311, 337–349 CrossRef CAS PubMed
.
- W. J. Cunliffe, D. B. Holland, S. M. Clark and G. I. Stables, Br. J. Dermatol., 2000, 142, 1084–1091 CrossRef CAS PubMed
.
- S. Nishijima, I. Kurokawa, N. Katoh and K. Watanabe, J. Dermatol., 2000, 27, 318–323 CrossRef CAS PubMed
.
-
OECD, Test No. 405: Acute Eye Irritation/Corrosion, 2021 Search PubMed
.
- H. Abaci, Z. Guo, Y. Doucet, J. Jacków and A. Christiano, Exp. Biol. Med., 2017, 242, 1657–1668 CrossRef CAS PubMed
.
- OECD, Test No. 432: In Vitro 3T3 NRU Phototoxicity Test, 2004 Search PubMed.
- A. Almeida, B. Sarmento and F. Rodrigues, Int. J. Pharm., 2017, 519, 178–185 CrossRef CAS PubMed
.
- D. S. Macmillan, S. J. Canipa, M. L. Chilton, R. V. Williams and C. G. Barber, Regul. Toxicol. Pharmacol., 2016, 76, 30–38 CrossRef CAS PubMed
.
- G. F. Gerberick, J. D. Vassallo, R. E. Bailey, J. G. Chaney, S. W. Morrall and J. P. Lepoittevin, Toxicol. Sci., 2004, 81, 332–343 CrossRef CAS PubMed
.
-
OECD, Test No. 442D: In Vitro Skin Sensitisation, 2018 Search PubMed
.
- A. Polini, L. Prodanov, N. S. Bhise, V. Manoharan, M. R. Dokmeci and A. Khademhosseini, Expert Opin. Drug Discovery, 2014, 9, 335–352 CrossRef CAS PubMed
.
-
OECD, Test No. 492: Reconstructed human Cornea-like Epithelium (RhCE) test method for identifying chemicals not requiring classification and labelling for eye irritation or serious eye damage, 2019 Search PubMed
.
- Y. Kaluzhny, H. Kandarova, Y. Handa, J. DeLuca, T. Truong, A. Hunter, P. Kearney, L. d'Argembeau-Thornton and M. Klausner, ATLA, Altern. Lab. Anim., 2015, 43, 101–127 CrossRef PubMed
.
- V. van Duinen, S. J. Trietsch, J. Joore, P. Vulto and T. Hankemeier, Curr. Opin. Biotechnol, 2015, 35, 118–126 CrossRef CAS PubMed
.
- A. Hasan, A. Paul, N. E. Vrana, X. Zhao, A. Memic, Y. S. Hwang, M. R. Dokmeci and A. Khademhosseini, Biomaterials, 2014, 35, 7308–7325 CrossRef CAS PubMed
.
- V. Lee, G. Singh, J. P. Trasatti, C. Bjornsson, X. Xu, T. N. Tran, S.-S. Yoo, G. Dai and P. Karande, Tissue Eng., Part C, 2014, 20, 473–484 CrossRef CAS PubMed
.
- F. Pati, J. Gantelius and H. A. Svahn, Angew. Chem., Int. Ed., 2016, 55, 4650–4665 CrossRef CAS PubMed
.
- G. Sriram, M. Alberti, Y. Dancik, B. Wu, R. Wu, Z. Feng, S. Ramasamy, P. L. Bigliardi, M. Bigliardi-Qi and Z. Wang, Mater. Today, 2017, 21, 326–340 CrossRef
.
- M. Y. Mulla, L. Torsi and K. Manoli, Methods Enzymol., 2020, 642, 403–433 Search PubMed
.
- H. A. Alhadrami, Biotechnol. Appl. Biochem., 2018, 65, 497–508 CrossRef CAS PubMed
.
- A. V. Bounegru and C. Apetrei, Sensors (Basel), 2020, 20, 6724 CrossRef CAS PubMed
.
- A. Joshi and K.-H. Kim, Biosens. Bioelectron., 2020, 153, 112046 CrossRef CAS PubMed
.
- H. Wan, D. Liu, X. Yu, H. Sun and Y. Li, Food Chem., 2015, 175, 601–608 CrossRef CAS PubMed
.
- M. N. Alam, N. J. Bristi and M. Rafiquzzaman, Saudi Pharm. J., 2013, 21, 143–152 CrossRef PubMed
.
- M. R. Guascito, E. Filippo, C. Malitesta, D. Manno, A. Serra and A. Turco, Biosens. Bioelectron., 2008, 24, 1063–1069 CrossRef CAS PubMed
.
- B. Kuswandi, Anal. Bioanal. Chem., 2003, 376, 1104–1110 CrossRef CAS PubMed
.
- Q. Liu, H. Cai, Y. Xu, L. Xiao, M. Yang and P. Wang, Biosens. Bioelectron., 2007, 22, 3224–3229 CrossRef CAS PubMed
.
- M. Guo, J. Wang, R. Du, Y. Liu, J. Chi, X. He, K. Huang, Y. Luo and W. Xu, Biosens. Bioelectron., 2020, 150, 111899 CrossRef CAS PubMed
.
- S. Bhavaniramya, R. Vanajothi, S. Vishnupriya, K. Premkumar, M. S. Al-Aboody, R. Vijayakumar and D. Baskaran, Curr. Pharm. Des., 2019, 25, 2661–2676 CrossRef CAS PubMed
.
- M. Rubab, H. M. Shahbaz, A. N. Olaimat and D.-H. Oh, Biosens. Bioelectron., 2018, 105, 49–57 CrossRef CAS PubMed
.
- S. Eissa and M. Zourob, Mikrochim. Acta, 2020, 187, 486 CrossRef CAS PubMed
.
- N. Idil, M. Bakhshpour, I. Perçin and B. Mattiasson, Biosensors, 2021, 11, 140 CrossRef CAS PubMed
.
- H. Xu and H. Li, Am. J. Clin. Dermatol., 2019, 20, 335–344 CrossRef PubMed
.
- M. Frieri, K. Kumar and A. Boutin, J. Infect. Public Health, 2017, 10, 369–378 CrossRef PubMed
.
- L. Lan, Y. Yao, J. Ping and Y. Ying, Biosens. Bioelectron., 2017, 91, 504–514 CrossRef CAS PubMed
.
- A. Kling, C. Chatelle, L. Armbrecht, E. Qelibari, J. Kieninger, C. Dincer, W. Weber and G. Urban, Anal. Chem., 2016, 88, 10036–10043 CrossRef CAS PubMed
.
- B. Dréno, T. Zuberbier, C. Gelmetti, G. Gontijo and M. Marinovich, J. Eur. Acad. Dermatol. Venereol., 2019, 33(Suppl 7), 15–24 CrossRef PubMed
.
- K. Kim, J. Kim, H. Kim and G. Y. Sung, Int. J. Mol. Sci., 2021, 22, 2160 CrossRef CAS PubMed
.
- B. K. Nahak, A. Mishra, S. Preetam and A. Tiwari, ACS Appl. Bio Mater., 2022, 5, 3576–3607 CrossRef CAS PubMed
.
- R. Martinez-Duarte, Electrophoresis, 2012, 33, 3110–3132 CrossRef CAS PubMed
.
- T. A. Moore, P. Brodersen and E. W. K. Young, Anal. Chem., 2017, 89, 11391–11398 CrossRef CAS PubMed
.
- J. J. Kim, F. Ellett, C. N. Thomas, F. Jalali, R. R. Anderson, D. Irimia and A. B. Raff, Lab Chip, 2019, 19, 3094–3103 RSC
.
- J. Kim, K. Kim and G. Y. Sung, Int. J. Mol. Sci., 2020, 21, 8475 CrossRef CAS PubMed
.
- E. Sano, C. Mori, N. Matsuoka, Y. Ozaki, K. Yagi, A. Wada, K. Tashima, S. Yamasaki, K. Tanabe, K. Yano and Y.-S. Torisawa, Micromachines, 2019, 10, 793 CrossRef PubMed
.
- A. E. Ongaro, D. Di Giuseppe, A. Kermanizadeh, A. Miguelez Crespo, A. Mencattini, L. Ghibelli, V. Mancini, K. L. Wlodarczyk, D. P. Hand, E. Martinelli, V. Stone, N. Howarth, V. La Carrubba, V. Pensabene and M. Kersaudy-Kerhoas, Anal. Chem., 2020, 92, 6693–6701 CrossRef CAS PubMed
.
- S. Schneider, D. Gruner, A. Richter and P. Loskill, Lab Chip, 2021, 21, 1866–1885 RSC
.
- S. Sun, L. Jin, Y. Zheng and J. Zhu, Nat. Commun., 2022, 13, 5481 CrossRef CAS PubMed
.
- I. Risueño, L. Valencia, J. L. Jorcano and D. Velasco, APL Bioeng., 2021, 5, 030901 CrossRef PubMed
.
- E. Dellambra, T. Odorisio, D. D'Arcangelo, C. M. Failla and A. Facchiano, ALTEX, 2019, 36, 177–202 Search PubMed
.
- E. Sutterby, P. Thurgood, S. Baratchi, K. Khoshmanesh and E. Pirogova, Small, 2020, 16, e2002515 CrossRef PubMed
.
- C. Gao, C. Lu, H. Qiao, Y. Zhang, H. Liu, Z. Jian, Z. Guo and Y. Liu, Biomater. Sci., 2022, 10, 4724–4739 RSC
.
- A. V. Nguyen and A. M. Soulika, Int. J. Mol. Sci., 2019, 20, 1811 CrossRef CAS PubMed
.
- S. Moon, D. H. Kim and J. U. Shin, Yonsei Med. J., 2021, 62, 969–980 CrossRef CAS PubMed
.
- E. Filaire, R. Nachat-Kappes, C. Laporte, M.-F. Harmand, M. Simon and C. Poinsot, Int. J. Cosmet. Sci., 2022, 44, 604–613 CrossRef CAS PubMed
.
- E. Fernandez-Carro, M. Angenent, T. Gracia-Cazaña, Y. Gilaberte, C. Alcaine and J. Ciriza, Pharmaceutics, 2022, 14, 1417 CrossRef CAS PubMed
.
- A. L. Byrd, Y. Belkaid and J. A. Segre, Nat. Rev. Microbiol., 2018, 16, 143–155 CrossRef CAS PubMed
.
- Y. Belkaid and J. A. Segre, Science, 2014, 346, 954–959 CrossRef CAS PubMed
.
- N. Mori, Y. Morimoto and S. Takeuchi, Biomaterials, 2017, 116, 48–56 CrossRef CAS PubMed
.
- F. Forouzandeh, A. Alfadhel, A. Arevalo and D. A. Borkholder, Sens. Actuators, A, 2021, 326, 112602 CrossRef CAS PubMed
.
- M. Busek, A. Aizenshtadt, T. Koch, A. Frank, L. Delon, M. A. Martinez, A. Golovin, C. Dumas, J. Stokowiec, S. Gruenzner, E. Melum and S. Krauss, Lab Chip, 2023, 23, 591–608 RSC
.
- H. E. Abaci, K. Gledhill, Z. Guo, A. M. Christiano and M. L. Shuler, Lab Chip, 2015, 15, 882–888 RSC
.
- B. W. Graf and S. A. Boppart, Methods Mol. Biol., 2010, 591, 211–227 CrossRef PubMed
.
- Y. B. Arık, M. W. van der Helm, M. Odijk, L. I. Segerink, R. Passier, A. van den Berg and A. D. van der Meer, Biomicrofluidics, 2018, 12, 042218 CrossRef PubMed
.
- N. Renous, M. D. Kiri, R. A. Barnea, R. Rauti, Y. Leichtmann-Bardoogo and B. M. Maoz, Lab Chip, 2021, 22, 71–79 RSC
.
- K. Takeuchi, N. Takama, K. Sharma, O. Paul, P. Ruther, T. Suga and B. Kim, Drug Delivery Transl. Res., 2022, 12, 435–443 CrossRef CAS PubMed
.
- P. Zoio and A. Oliva, Pharmaceutics, 2022, 14, 682 CrossRef CAS PubMed
.
- L. Bay and H. C. Ring, APMIS, 2022, 130, 706–718 CrossRef PubMed
.
- M. D. Arpa, İ. M. Seçen, Ü. C. Erim, A. Hoş and N. Üstündağ Okur, Pharm. Dev. Technol., 2022, 27, 268–281 CrossRef CAS PubMed
.
- Q. Quan, D. Weng, X. Li, Q. An, Y. Yang, B. Yu, Y. Ma and J. Wang, Front. Bioeng. Biotechnol., 2022, 10, 939629 CrossRef PubMed
.
- B. S. Kwak, S.-P. Jin, S. J. Kim, E. J. Kim, J. H. Chung and J. H. Sung, Biotechnol. Bioeng., 2020, 117, 1853–1863 CrossRef CAS PubMed
.
- X. Ren, A. E. Getschman, S. Hwang, B. F. Volkman, T. Klonisch, D. Levin, M. Zhao, S. Santos, S. Liu, J. Cheng and F. Lin, Lab Chip, 2021, 21, 1527–1539 RSC
.
- G. Shabestani Monfared, P. Ertl and M. Rothbauer, Pharmaceutics, 2021, 13, 793 CrossRef PubMed
.
- S. Gupta, L. Patel, K. Mitra and A. Bit, Micromachines, 2022, 13, 305 CrossRef PubMed
.
- S. Maharjan, B. Cecen and Y. S. Zhang, Small Methods, 2020, 4, 2000235 CrossRef CAS PubMed
.
- M. Wufuer, G. Lee, W. Hur, B. Jeon, B. J. Kim, T. H. Choi and S. Lee, Sci. Rep., 2016, 6, 37471 CrossRef CAS PubMed
.
- M. Légues, C. Milet and N. Forraz, IFSCC Mag.
- J. S. Lee, J. Kim, B. Cui, S. K. Kim, S.-A. Cho, S. An and S.-W. Cho, Lab Chip, 2022, 22, 343–353 RSC
.
- X. Ma, H. Wang, Y. Song and Y. Pan, J. Cosmet. Dermatol., 2021, 20, 195–203 CrossRef PubMed
.
- J. Zhang, Z. Chen, Y. Zhang, X. Wang, J. Ouyang, J. Zhu, Y. Yan, X. Sun, F. Wang, X. Li, H. Ye, S. Sun, Q. Yu, J. Sun, J. Ge, Q. Li, Q. Han, Y. Pu and Z. Gu, Lab Chip, 2021, 21, 3804–3818 RSC
.
- B. Jeon, G. Lee, M. Wufuer, Y. Huang, Y. Choi, S. Kim and T. H. Choi, Toxicol In Vitro, 2020, 68, 104955 CrossRef CAS PubMed
.
- N. Jusoh, J. Ko and N. L. Jeon, APL Bioeng., 2019, 3, 036101 CrossRef PubMed
.
- A. Bal-Öztürk, B. Miccoli, M. Avci-Adali, F. Mogtader, F. Sharifi, B. Çeçen, G. Yaşayan, D. Braeken and E. Alarcin, Curr. Pharm. Des., 2018, 24, 5437–5457 CrossRef PubMed
.
- Z. Varga-Medveczky, D. Kocsis, M. B. Naszlady, K. Fónagy and F. Erdő, Pharmaceutics, 2021, 13, 1852 CrossRef CAS PubMed
.
- Z. Chen, S. Kheiri, A. Gevorkian, E. W. K. Young, V. Andre, T. Deisenroth and E. Kumacheva, Lab Chip, 2021, 21, 3952–3962 RSC
.
- C. C. Zouboulis and E. Makrantonaki, Clin. Dermatol., 2011, 29, 3–14 CrossRef PubMed
.
- A. Kazanci, M. Kurus and A. Atasever, Sking Res. Technol., 2017, 23, 48–60 CrossRef CAS PubMed
.
- S. Jeong, J. Kim, H. M. Jeon, K. Kim and G. Y. Sung, Int. J. Mol. Sci., 2021, 22, 12788 CrossRef CAS PubMed
.
- J. Pauty, S. Nakano, R. Usuba, T. Nakajima, Y. Johmura, S. Omori, N. Sakamoto, A. Kikuchi, M. Nakanishi and Y. T. Matsunaga, Biomater. Sci., 2021, 9, 199–211 RSC
.
- C. Hausmann, A. Vogt, M. Kerscher, K. Ghoreschi, M. Schäfer-Korting and C. Zoschke, Drug Discovery Today, 2020, 25, 851–861 CrossRef CAS PubMed
.
- N. Ashammakhi, M. A. Darabi, B. Çelebi-Saltik, R. Tutar, M. C. Hartel, J. Lee, S. Hussein, M. J. Goudie, M. B. Cornelius, M. R. Dokmeci and A. Khademhosseini, Small Methods, 2020, 4, 1900589 CrossRef CAS PubMed
.
- L. H. Chong, T. Ching, H. J. Farm, G. Grenci, K.-H. Chiam and Y.-C. Toh, Lab Chip, 2022, 22, 1890–1904 RSC
.
Footnote |
| † These authors contributed equally to this work and should be considered co-corresponding authors. |
| This journal is © The Royal Society of Chemistry 2023 |