Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Early detection of the initial stages of LED light-triggered non-alcoholic fatty liver disease by wax physisorption kinetics-Fourier transform infrared imaging†
Yi-Ting
Chen‡
abc,
Pei-Yu
Huang‡
d,
Chee-Yin
Chai
abce,
Sebastian
Yu
 fg,
Yu-Lin
Hsieh
fg,
Yu-Lin
Hsieh
 hij,
Hao-Chao
Chang
k,
Chin-Wei
Kuo
d,
Yao-Chang
Lee
hij,
Hao-Chao
Chang
k,
Chin-Wei
Kuo
d,
Yao-Chang
Lee
 *dlm and
Hsin-Su
Yu
*dlm and
Hsin-Su
Yu
 *fno
*fno
aGraduate Institute of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
bDepartment of Pathology, Kaohsiung Medical University Hospital, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
cDepartment of Pathology, Faculty of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
dLife Science Group, National Synchrotron Radiation Research Center, Hsinchu 30076, Taiwan
eInstitute of Biomedical Sciences, National Sun Yat-Sen University, Kaohsiung 80424, Taiwan
fDepartment of Dermatology, Kaohsiung Medical University Hospital, Kaohsiung Medical University, Kaohsiung 80708, Taiwan. E-mail: yup.kmu@gmail.com
gDepartment of Dermatology, School of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
hDepartment of Anatomy, School of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
iSchool of Post-Baccalaureate Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
jDepartment of Medical Research, Kaohsiung Medical University Hospital, Kaohsiung 80708, Taiwan
kTaiwan Instrument Research Institute, National Applied Research Laboratories, Hsinchu 30205, Taiwan
lDepartment of Optics and Photonics, National Central University, Taoyuan 320317, Taiwan
mChemistry Department, National Tsing Hua University, Hsinchu 30013, Taiwan
nNational Institute of Environmental Health Sciences, National Health Research Institutes, Miaoli County 35053, Taiwan
oGraduate Institute of Clinical Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
First published on 22nd December 2022
Abstract
Light-emitting diodes (LEDs), particularly in the blue waveform range, are regarded as a major source of circadian rhythm dysregulation. A circadian rhythm dysregulation induced by blue LEDs is associated with non-alcoholic fatty liver disease (NAFLD). Hepatocellular accumulation of lipids is a key event in the early stages of NAFLD. Kupffer cells (KCs) have been reported to be lost in the early onset of NAFLD followed by an inflammatory reaction that alters the liver response to lipid overload. This study focused on the detection of the initial stages (subpathological stages) of LED light-triggered NAFLD. Mice were exposed to either blue or white LED irradiation for 44 weeks. Synchrotron radiation-based Fourier-transform infrared microspectroscopy (SR-FTIRM) and wax physisorption kinetic-Fourier transform infrared (WPK-FTIR) imaging were used to evaluate the ratio of lipid to protein and the glycosylation of glycoprotein, respectively. Immunohistopathological studies on KCs and circadian-related proteins were performed. Although liver biopsy showed normal pathology, an SR-FTIRM study revealed a high hepatic lipid-to-protein ratio after receiving LED illumination. The results of WPK-FTIR demonstrated that a high inflammation index was found in the high irradiance of the blue LED illumnation group. These groups showed a decrease in KC number and an increase in Bmal1 and Reverbα circadian protein expression. These findings provide explanations for the reduction of KCs without subsequent inflammation. A significant reduction of Per2 and Cry1 expression is correlated with the findings of WPK-FTIR imaging. WPK-FTIR is a sensitive method for detecting initiative stages of NAFLD induced by long-term blue LED illumination.
1. Introduction
Visible light can induce non-visual effects in mammals. Visible light entering the eye is sensed through the retina, which regulates the circadian rhythm via the hypothalamus–pituitary–adrenal axis.1 In the retina, the photopigment of visible light, melanopsin (OPN4), is expressed in photosensitive retinal ganglion cells, which are involved in the regulation of circadian rhythms.2 Melanopsin absorbs light in the short wavelength of the visible spectrum, with maximum sensitivity to blue light.3 The circadian rhythm is a biological process involved in 24 h behavior and physiology in living organisms. In mammals, the circadian timing system is composed of a network of clock-controlled genes (CCGs) and an auto-regulatory feedback loop within most cells and tissues throughout the body. Physiological circadian rhythm activity or stability can be regulated in the suprachiasmatic nucleus (SCN) of the hypothalamus by the effect of light exposure through the eyes.4 The interconnected transcriptional and post-transcriptional loops play vital roles in generating and maintaining a circadian rhythm, including brain and muscle ARNT-like 1/circadian locomotor output cycles kaput (Bmal1/CLOCK), the repressor complex Period (Per1/Per2), and Cryptochrome (Cry1/Cry2). The core transcription factors Bmal1 and CLOCK exist as a heterodimer and are first regulated by CCGs. Per1/Per2 and Cry1/Cry2, which are also the targets of Bmal1, play roles in the negative regulation of Bmal1 expression.5,6 Nuclear hormone receptors Rev-erbα (repressor) and Ror (activator) form an additional stabilizing loop with Bmal1 to fine-tune the precision regulation of the clock.4 In mammals, these networks of circadian clocks are arranged in a hierarchical structure with a “master” circadian pacemaker located in the SCN, and tissue-specific peripheral oscillators located throughout the rest of the body with the function of controlling the local aspects of physiology. The mammalian circadian clock coordinates the diverse physiological processes by regulating the autonomic nervous system, metabolism, and immune responses.7 Artificial lighting allows people to extend daytime activity into the night. In industrialized countries, 75% of the total workforce is estimated to have been involved in shift work and night work.8 Shift work is a complex lifestyle that to some degree includes exposure to circadian disruption, sleep disturbance, altered phase-angle of entrainment, and psychological stress.9 Light-emitting diodes (LEDs) are widely used in electronic devices and are regarded as a major source of circadian dysregulation in modern societies. Although the light emitted by most LEDs appears white, LEDs have peak emission in the blue range.10 The shift work associated with the disruption of the circadian rhythm11 has been linked to increased risks of cancers, diabetes, obesity, coronary heart disease, and stroke,5 as well as a high mortality rate associated with the dysregulation of innate immunity.9 The circadian clock circuity plays a significant role in the control of the innate immune system's function.12–15 In circadian disruption, macrophages play a pivotal role in inflammatory immune responses.12Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in industrialized countries.16 NAFLD is a spectrum of disorders, beginning as simple steatosis, which may progress into steatohepatitis and fibrosis, often resulting in cirrhosis and even hepatocellular cancer.17 Hepatocellular accumulation of lipids is a key event in the early stages of NAFLD, while progression to steatohepatitis is strongly associated with inflammation. Kupffer cells (KCs), known as hepatic resident macrophages, are localized within the lumen of the liver sinusoids. KCs are thought to play a key role in this progression.18 There is still only limited knowledge of the relationships between circadian dysregulation and KCs.
Glycosylation of proteins is one of the most critical roles of posttranslational modifications in cellular biology, cell differentiation, growth, malignant transformation, cell–matrix interactions, and disease progression.19 Aberrant protein glycosylation associated with the neo-synthesis and incomplete synthesis can cause elongated or branched protein-linked glycan residues of glycoprotein in cancer development.20 An alteration of the chain length of the glycan residues of glycoconjugates anchored in the cell membrane and tissue has been associated with inflammatory diseases.21 Synchrotron radiation-based Fourier-transform infrared microspectroscopy (SR-FTIRM) and wax physisorption kinetics-Fourier transform infrared (WPK-FTIR) imaging were conducted for liver tissue section and cell surface glycoprotein analysis22 and used to investigate the accumulation of lipids and glycogen in the liver tissue and to assess the inflammation status of liver tissue.
This study aims to explore the effects of chronic circadian dysregulation induced by LED illumination on the subpathological stages of NAFLD in mice. We found that SR-FTIRM and WPK-FTIR imaging revealed a higher ratio of lipid to protein (L/P) and inflammation indices in the hepatocytes of the blue LED illumination groups. These findings are correlated with the reduced KC number without subsequent inflammation and circadian protein expression in the liver. Noteworthily, the initial stage of NAFLD was submicroscopic. The population of lipid droplets should be lower and the diameter of lipid droplets should also be small in hepatocytes in the initial phase of NAFLD disease and they are too small to be seen with a light microscope. Therefore, WPK-FTIR imaging is a precise and sensitive method for the detection of the initial stages of NAFLD induced by LED illumination.
2. Materials and methods
2.1. Animals
Wild-type male C57BL/6 mice were purchased from the National Laboratory Animal Center, National Applied Research Laboratories, Taiwan. The mice were housed in a humidity (40–60%) and temperature (22–26 °C)-controlled room with a 12 h light and 12 h dark cycle. The animals were provided with food and water ad libitum, all possible efforts were made to minimize animal suffering and all procedures were performed in a coded and blinded manner and in accordance with the ethical guidelines related to laboratory animals of the Institute Animal Care and approved by the Animal Ethics Committee of Kaohsiung Medical University (#108130).2.2. Light illumination
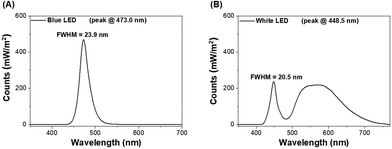
Two customized light-boxes with adjustable-intensity LEDs on the ceiling were utilized. LEDs of blue light (473.0 ± 12.0 nm, the major spectrum of electronic devices) and white light (the major spectrum of lightbulbs, consisting of around 17.4% blue light) were used in this study (Fig. 1). The lighting protocols involved the following factors: (1) the regular office days per week and (2) long-term night-shift work. Moreover, our pilot study showed that short-term LED lighting did not affect lipid and glycogen accumulation as well as the changes of the gut microbiota (unpublished data). Accordingly, the mice were exposed to blue LED illumination (3.6 J cm−2 or 7.2 J cm−2, each N = 3, total N = 6) or white LED illumination (3.6 J cm−2 or 7.2 J cm−2, each N = 3, total N = 6) at Zeitgeber time (ZT) 13.5–14 (equivalent to 09:30–10:00 a.m.) in an un-anesthetized state for 5 days per week, and long-term LED lighting illumination23 lasted for 44 weeks, to observe the alterations of liver metabolism and microbial community. The mice treated with the same protocol without LED illumination were defined as the control group (N = 3).2.3. Synchrotron-radiation-based Fourier-transform infrared microscopy (SR-FTIRM) of glycogen and lipids in mouse liver tissue
SR-FTIRM provides a method for detecting biological components such as lipids, proteins, carbohydrates, and their ratios.24 The FTIR spectra of liver tissue sections were acquired by accumulating 64 scans with a spectral resolution of 4 cm−1 at the end-station of SR-FTIRM TLS 14A1 of the National Synchrotron Radiation Research Center (NSRRC) in Taiwan. The end-station of SR-FTIRM includes an FTIR spectrometer (Nicolet 6700, Thermo Fisher Scientific, Madison, WI, USA) coupled with a confocal infrared microscope (Nicolet Continuum, Thermo Fisher Scientific) equipped with a liquid nitrogen-cooled mercury–cadmium–telluride detector. The baseline-corrected and normalized FTIR spectra of tissue section samples were obtained by using OMNICTM (v.8.5; Thermo Fisher Scientific). The SR-FTIR spectral images of the absorbance ratio of glycogen to protein (G/P) and lipids to protein (L/P) referred to the level of glycogen and lipids relative to the protein absorbance of each detection area; these were constructed by integrating the absorbances in the spectral range of 1180–970 cm−1, 2945–2900 cm−1, and 1585–1480 cm−1 for glycogen, lipids, and protein, respectively, in the detection area of 4500 × 1500 μm2 with a lateral resolution of 50 × 50 μm2.2.4. Wax physisorption kinetic-Fourier transform infrared (WPK-FTIR) imaging
In this study, WPK-FTIR imaging is utilized to measure the strength of physical adsorption between a non-polar wax reagent, playing the role of a glycan probe, and a glycan residue of glycoconjugates, including glycoproteins, anchoring in the cell membrane based on the dipole-induced dipole interaction.22 The amount of the remaining wax adsorbent adhering to the sample surface was acquired with FTIR imaging after the adsorption–desorption process, which was strongly correlated with the absorbance in the mid-IR spectral range of 3000–2800 cm−1 of the wax adsorbent. Therefore, a greater absorbance of the remaining wax adsorbent was observed, and a stronger interaction was expected between the wax adsorbent and glycan residue of glycoconjugates anchoring onto the sample surface. Moreover, there were four normal alkanes (n-alkane, n-CnH2n+2) with different carbon numbers (C.N.s) as the wax adsorbents, employed as glycan probes, for targeting the glycan residues of glycoconjugates with similar chain lengths based on the chemical similarity principle. The chain length alteration of the glycan residue of a glycoprotein can be correlated with the C.N. of the remaining n-alkane in the detection areas of interest in the liver tissue section surface22,25,26,73 under various doses of blue and white light LED illumination for mice.Formalin-fixed, paraffin-embedded liver tissue sections at 4 μm thickness were fixed on an IR low-E slide (Kevley Technologies, Chesterfield, OH, USA) and immersed in 40 mL xylene in a glass trough at 56.0 °C for 20 h to remove embedded paraffin within the tissue section samples. The first step of WPK was waxing of the mouse liver tissue section sample by soaking it in a 40 mL xylene-n-alkane solution consisting of 0.12 M n-alkane at 28.0 °C for 2 min and to remove xylene by heating the sample slide on a hot plate at 34.0 °C for 10 min. The second step of WPK was desorption of the wax from the waxed sample by soaking the waxed sample in 40 mL xylene at 28.0 °C for 1, 2.5, 5, and 7.5 s to desorb n-alkane from the waxed sample and then remove xylene by heating the sample on a hot plate at 34.0 °C for 2 min, respectively. The WPK-FTIR imaging result of the mouse liver tissue section of the control set exhibited a dramatic decrease of desorption from 1.0 to 2.5 s and from 5.0 to 7.5 s and a steady state of desorption from 2.5 to 5.0 s (Fig. 2). We proposed that there was a thermodynamic steady state for n-alkane desorption and resorption between 2.5 s and 5.0 s. In this study, the desorption time of 5.0 s was set for a sample in the WPK procedure.
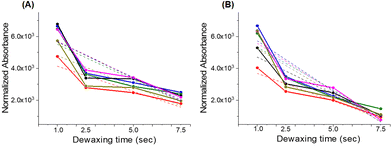 | ||
Fig. 2 The kinetic physisorption of C22 and C28 is presented for the liver tissue section of the control group. In the liver tissue section, six areas were selected and each area was 170 × 170 μm2 of a field of view as shown in Fig. 3A. The kinetic data of the remaining glycan probes (A) C22 and (B) C28 adhered to the liver tissue section surface and the linear regression curve corresponding to six areas ( area I, area I,  area II, area II,  area III, area III,  area IV, area IV,  area V, and area V, and  area VI, and dashed line corresponding to linear regression curve for each area). area VI, and dashed line corresponding to linear regression curve for each area). | ||
All the WPK procedures were completed by using a home-built computer-controlled system via the RS-232 interface for precisely controlling the time for wax sorption and desorption.22 The infrared absorbance of the remaining wax adhering to a tissue section surface was acquired with FTIR imaging to provide two-dimensional distribution of the remaining n-alkane wax adhering to the sample surface. The greater the amount of n-alkane wax adhering to the sample surface, the stronger the physical adsorption expected between n-alkane and the glycan residue of the glycoprotein.
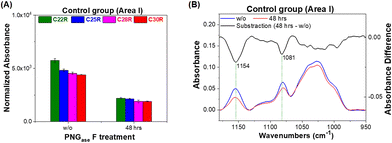
In this study, peptide: N-glycosidase F (PNGase F, purity ≥95%, Sigma-Aldrich, St Louis, MO, USA) was utilized to enzymatically cleave the N-link glycan including high mannose, hybrid, and complex glycan residues from glycoproteins on the surface of the mouse liver tissue section.27 Hence, PNGase F was used to digest tissue section samples for validating the capability of the wax adsorbent as a glycan probe for targeting the glycan residue of the glycoprotein anchoring onto the tissue section surface. The liver tissue section of the control group was incubated with PNGase F (100 IUB milliunits) at 45 °C for 48 h. However, PNGase F cannot remove an N-linked glycan residue from a glycoprotein with a fucose alpha 1,3-linked to the core region N-acetylglucosamine.28,29 The FTIR imaging system, including an FTIR spectrometer (INVENIO R, Bruker Optics, Ettlingen, Germany) equipped with an infrared microscope (Hyperion 3000, Bruker Optics) and an LN-cooled 64 × 64 pixels mercury–cadmium–telluride focal-plane-array amplified detector (Santa Barbara Focalplane, Goleta, CA, USA) was utilized to image a field of view of 170 × 170 μm2 for each of the areas of interest to acquire the absorbance of the remaining glycan probe adhering on liver tissue section samples.22
In this study, four n-alkanes were employed in the WPK procedures as glycan adsorbents, including n-C22H46 (C22), n-C25H52 (25), n-C28H58 (C28), and n-C30H62 (C30) (Sigma), for probing the chain length of the glycan residue of the glycoprotein on the liver tissue section surface. The amount of remaining n-alkane adhering to the tissue section surface can be used to correlate with the characteristic infrared absorbance of n-alkane, performed by integrating the absorption in the spectral range of 3000–2800 cm−1 with OPUS software (v. 8.1, Bruker Optics), as the input for constructing two-dimensional spectral images for every single pixel of remaining n-alkane in the detection area of 170 × 170 μm2.
2.5. Immunohistochemical staining for circadian proteins, scoring, and statistical analysis of liver tissue in LED illumination groups
Every case sample was 3 μm sections from formalin-fixed, paraffin-embedded liver tissue blocks. These sections were deparaffinized, rehydrated, and autoclaved at 121 °C for 10 min in a target retrieval solution, pH 6.0 (S2369; Dako, Glostrup, Denmark), to retrieve antigens. After 20 min at room temperature, 3% hydrogen peroxide for 5 min was used for blocking endogenous peroxidase in the sections at room temperature. After washing twice with Tris buffer, the sections were incubated with anti-F4/80 (D2S9R) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250; #70076; Cell Signaling Technology, Danvers, MA, USA), anti-Bmal-1 (1
250; #70076; Cell Signaling Technology, Danvers, MA, USA), anti-Bmal-1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400; NB100-2288; Novus Biologicals, Littleton, CO, USA), anti-Cryptochrome 1 (Cry1) (1
400; NB100-2288; Novus Biologicals, Littleton, CO, USA), anti-Cryptochrome 1 (Cry1) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; orb156463; Biorbyt, Cambridge, Cambridgeshire, UK), Period circadian protein 2 (Per2) (1
100; orb156463; Biorbyt, Cambridge, Cambridgeshire, UK), Period circadian protein 2 (Per2) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; bs-3927R; Bioss, Woburn, MA, USA), anti-Rev-erbα (1
100; bs-3927R; Bioss, Woburn, MA, USA), anti-Rev-erbα (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; orb6019; Biorbyt), and anti-TNF-α antibody (1
200; orb6019; Biorbyt), and anti-TNF-α antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25; SC-290189, Santa Cruz Biotechnology, Dallas, TX, USA) as the primary antibodies. After washing twice with Tris buffer, the sections were incubated with a secondary antibody conjugated with horseradish peroxidase for 30 min at room temperature. Finally, the slides were incubated in 3,3-diaminobenzidine (K5007; Dako) for 5 min followed by Mayer's haematoxylin counterstaining for 90 s, and mounted with Malinol.
25; SC-290189, Santa Cruz Biotechnology, Dallas, TX, USA) as the primary antibodies. After washing twice with Tris buffer, the sections were incubated with a secondary antibody conjugated with horseradish peroxidase for 30 min at room temperature. Finally, the slides were incubated in 3,3-diaminobenzidine (K5007; Dako) for 5 min followed by Mayer's haematoxylin counterstaining for 90 s, and mounted with Malinol.
A semiquantitative scoring system for immunopositive cells was evaluated by two independent pathologists (YTC and CYC). For the F4/80 stain, the number of immunoreactive cells was counted microscopically in three representative areas in the lower power field. For Bmal1, Cry1, Per2, and Rev-erbα stains, as well as for TNF-α, sections were scored based on the staining intensity score (0–3) multiplied percentage score (0–5).30 Percentages of immunoreactivity in hepatocytes or KCs were scored in the following manner: 0, 0%; 1, 1–9%; 2, 10–32%; 3, 33–65%; 4, 66–90%; and 5, 90–100%. All statistical analyses were performed with SPSS 24.0 software. Paired t-test was applied for correlation analysis. The test was 2-sided, and a p-value less than 0.05 was regarded as statistically significant.
3. Results and discussion
3.1. Histopathological examinations of liver tissues in different LED illumination groups
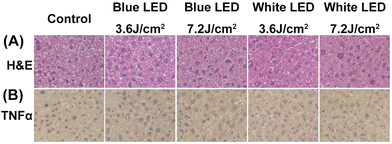
Haematoxylin and eosin stains of liver tissue revealed that the liver architectural framework is maintained and no obvious histological changes are seen in different LED illumination groups (Fig. 3A). The immunohistochemical expression of TNF-α protein is similar in both control and experimental groups (Fig. 3B).3.2. SR-FTIRM images of the absorbance ratio of lipid to protein (L/P) and glycogen to protein (G/P) after illumination with LED light
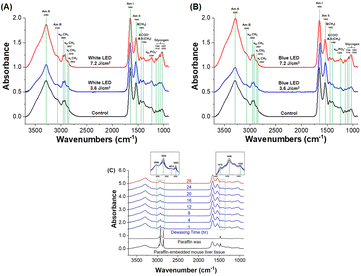
The de-paraffinization procedure of formalin-fixed, paraffin-embedded liver tissue section samples was the same as that of WPK-FTIR for the measurement of SR-FTIRM. The characteristic infrared absorption bands of SR-FTIRM spectra for the liver tissue section of mice irradiated at 3.6 J cm−2 and 7.2 J cm−2 by white and blue LED illumination were presented and assigned (Fig. 4A, B and Table 1); the absorbance of lipids, proteins, and glycogen was acquired by integrating infrared absorption in the spectral range 2945–2900 cm−1 (νas CH2 of lipids), 1585–1480 cm−1 (Am II of protein molecules), and 1180–970 cm−1 (glycosidic bond C–O, C–O–H of glycogen, and PO2− of DNA),31–33 respectively, and more detailed assignment of absorption bands is shown in Table 1. The infrared absorption of glycogen and lipids was increased after LED illumination, indicating that metabolic retardation occurred in mouse livers. Furthermore, the embedded paraffin within the liver tissue section samples was deparaffinized before acquiring FTIR images, and the spectral result showed that the remaining paraffin within the tissue section samples was nearly removed after 20 h of xylene immersion.34–38 The kinetic curve of the remaining paraffin within the tissue section showed a dramatic drop in the first hour of xylene immersion (ESI Fig. 1†) and the absorbance of lipids in the FTIR spectral images of 170 × 170 μm2 by integrating the spectral range of 3000–2800 cm−1 illustrated a nearly constant integration value after dewaxing for 20 h with xylene (Fig. 4C). The characteristic absorption bands of paraffin in the range of 2920 cm−1, 2850 cm−1, 1472 cm−1, and 1463 cm−1 were employed for monitoring the removal of paraffin within the tissue section with xylene treatment. The SR-FTIRM spectral images of the absorbance ratio of lipid to protein (L/P) and glycogen to protein (G/P) were established by using the characteristic absorbance of the lipid, protein, and glycogen mentioned above,39–41 respectively, for assessing the liver metabolic retardation after the mice received extra LED illumination. The spectral images of L/P showed a dramatic increase for the group that received 7.2 J cm−2 of blue LED illumination as compared with the control group (0.41 ± 0.08 vs. 0.24 ± 0.07, p = 0.013), which strongly suggests a lipid metabolic retardation in the liver tissue of mice after receiving extra LED illumination of blue and white light (Fig. 5). In addition, a higher G/P ratio also showed similar results to the L/P ratio, for example, more accumulated glycogen was found in the case receiving a dose of 7.2 J cm−2 of blue light. Moreover, the statistical results of SR-FTIRM for three sets of animal trials after receiving extra LED illumination of blue and white light revealed higher L/P and G/P ratios (ESI Fig. 2 and 3†). Based on these findings, we suggested that metabolic retardation occurred in the liver tissue of mice after receiving extra LED illumination.| Peak | Liver | Assignment | ||
|---|---|---|---|---|
| Control | WL | BL | ||
| Wavenumbers (cm−1) | ||||
| 1 | 1035 | 1034 | 1035 | δC–O–C (bending of carbohydrate) |
| 2 | 1081 | 1080 | 1081 | C–O–C (bending of carbohydrate), νs PO2− (PO2− symmetric stretching vibration of DNA) |
| 3 | 1154 | 1154 | 1155 | δC–O–H (bending of carbohydrate) |
| 4 | 1240 | 1237 | 1238 | ν as PO2− (PO2− antisymmetric stretching vibration of DNA) |
| 5 | 1399 | 1396 | 1396 | δ COO−δ CH3 (COO−, CH3 antisymmetric bending, lipids and proteins) |
| 6 | 1451 | 1453 | 1453 | δ CH2 (CH2 antisymmetric bending, lipids and proteins) |
| 7 | 1542 | 1540 | 1541 | Amide II (vibration motion coupled C–N stretching vibration and C–N–H bending vibration) |
| 8 | 1654 | 1651 | 1651 | Amide I (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration, proteins) O stretching vibration, proteins) |
| 9 | 2850 | 2852 | 2852 | ν s CH2 (CH2 symmetric stretching vibration, dominant contribution from lipids) |
| 10 | 2874 | 2876 | 2873 | ν s CH3 (CH3 symmetric stretching vibration, dominant contribution from proteins) |
| 11 | 2926 | 2927 | 2926 | ν asCH2 (CH2 antisymmetric stretching vibration, dominant contribution from lipids) |
| 12 | 2959 | 2960 | 2959 | ν asCH3 (CH3 antisymmetric stretching vibration, dominant contribution from proteins) |
| 13 | 3072 | 3066 | 3065 | Amide B (overtone of amide II) |
| 14 | 3293 | 3293 | 3286 | Amide A (N–H stretching vibration) |
3.3. Assessment of liver inflammation after extra LED illumination with WPK-FTIR imaging
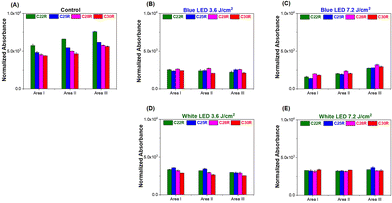
The additional glycosylation of glycoproteins, including IgG antibodies, is the characteristic of an anti-inflammatory process caused by the immune response, which stimulates the elongation of the glycan residue of the glycoprotein caused by galactosylation and/or sialylation.42 The KC glycoprotein receptor on the KC has also been reported to play a role in the clearance of galactose- and fructose-terminated glycoproteins from circulation.43 In this study, four glycan probes C22, C25, C28, and C30 were employed in the WPK-FTIR imaging to probe different chain-length glycans and further provide the correlation between the elongation degree of the glycan residue of the glycoprotein in the liver tissues by using the C.N. of the remaining glycan probes. The amount of the remaining glycan probes (C22R, C25R, C28R, and C30R) on the liver tissue section sample was measured in the order of C22R > C25R > C28R > C30R for the control group, and C28R > C30R > C25R ∼ C22R for the mouse group receiving a 7.2 J cm−2 blue LED illumination by comparing the absorbance in the spectral range of 3000–2800 cm−1 (ESI Fig. 3†). In addition, the order was C25R > C22R > C28R > C30R for the mouse group receiving a 3.6 J cm−2 LED white light dose, and a similar remaining amount for all absorbents was observed for the mice receiving a LED white light dose of 7.2 J cm−2 (Fig. 6A–E). Based on these findings, the glycan elongation was enhanced for all the mouse groups after receiving an extra dose of LED illumination, especially for the case receiving a blue light dose of 7.2 J cm−2. Besides, the remaining glycan probe was dramatically decreased for the control group after PNGase F treatment by about 61.4 ± 1.1%, 55.4 ± 1.3%, 57.3 ± 2.3%, and 56.4 ± 0.7% for C22R, C25R, C28R, and C30R, respectively, indicating that the n-alkane played a crucial role in physical binding of the glycan residue of the glycoprotein on the liver tissue section surface (Fig. 7A). Moreover, the characteristic absorption of the glycosidic bond C–O–C of the glycan residue of N-glycan and the C–N bond of asparagine(N)-linked glycoprotein decreased at 1154 cm−1 and 1081 cm−1, respectively, after PNGase F digestion treatment as shown in Fig. 7B.44 Based on the findings, we proposed that the N-linked glycoproteins were predominantly in the tissue, and the rest of the remaining N-linked glycoprotein should be alpha 1,3 fucosylated at the core N-acetylglucosamine (GlcNAc), O-linked glycoproteins and other glycoproteins in the liver tissue.According to these findings, when the dose of LED illumination that the mice received was higher, the remaining glycan probe increased with a greater C.N. observed on the mouse liver tissue sections as compared to that of the control group, indicating that the chain length of the glycan residue of the glycoprotein was elongated in the liver after receiving extra LED illumination. Consequently, the ratio of C28R/C22R was suggested to correlate with the population ratio of the long-chain to the short-chain glycan residues of the glycoprotein in the mouse livers, which could be utilized as an inflammation index to assess the liver inflammation in the very early stage of this animal trial mode after receiving an extra dose of LED illumination.
The WPK-FTIR spectral images presented the two-dimensional distribution of the remaining C28 and C22 adhering to the liver tissue section surface (Fig. 8). The result of the areas of interest in the liver tissue section showed a higher C28R/C22R ratio after the mice received an extra LED illumination as compared to the control group. The C28R/C22R ratio showed a positive correlation with the LED illumination dose received by the mice, especially that of the blue LED illumination, and higher G/P and L/P were also observed compared to those of the control group (Fig. 5). Therefore, the ratio of C28R/C22R could be a signpost for assessing the glycosylation level of glycoproteins distributed in the liver tissue sections of mice after receiving extra LED illumination. Previous studies showed that the galactosylation and sialylation of the fragment crystallizable (Fc) glycan of glycoproteins were validated owing to the anti-inflammatory activity of the immune response triggered by activated platelets in tissue.45 This revealed that a greater dose of LED illumination might enhance the inflammation of hepatocytes.
3.4. The number of KCs decreased after LED illumination
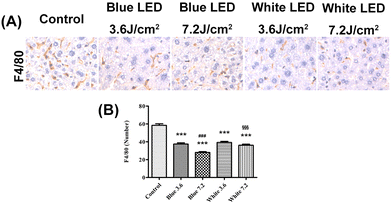
The number of KCs was evaluated in five different mouse groups, including the control group, and the white and blue light LED illumination at both 3.6 and 7.2 J cm−2, using the F4/80 antibody. The KCs were identified in sinusoid spaces, predominantly in zone 3 of liver tissue. The average number of KCs in the control group was 58.4 ± 4.8 in a low power field, 37.4 ± 3.7 (3.6 J cm−2), 28.0 ± 2.6 (7.2 J cm−2) in the group receiving blue LED illumination, and 39.2 ± 3.4 (3.6 J cm−2) and 36.1 ± 3.3 (7.2 J cm−2) in the group receiving white LED illumination. The number of KCs decreased in the mice exposed to 3.6 and 7.2 J cm−2 blue LED illumination (both p < 0.001) as compared with the control group, and the results differed between the two irradiances (p < 0.001). In the groups receiving white LED illumination, the number of KCs decreased in the 3.6 and 7.2 J cm−2 groups (both p < 0.001) compared with the control group; but there was no difference between the 3.6 and 7.2 J cm−2 white LED groups (p = 0.082). Under 7.2 J cm−2 illumination, the number of KCs significantly decreased in the blue LED group compared with the group receiving white LED illumination (p < 0.001) (Fig. 9A and B).3.5. The alteration of Bmal1, Cry1, Per2, and Rev-erbα proteins in LED illumination groups
In the immunohistochemical study, Bmal1 protein expression was noted in the cytoplasm of hepatocytes in the control group. In the control group, the Bmal1 immunoreactivity score of hepatocytes was 6.1 ± 1.7, while in the blue light LED illumination groups the scores were 7.9 ± 1.3 (3.6 J cm−2) and 10.2 ± 2.1 (7.2 J cm−2). The Bmal1 immunoreactivity scores of hepatocytes were 7.7 ± 2.1 (3.6 J cm−2) and 9.8 ± 2.1 (7.2 J cm−2) in the white light LED illumination groups. The Bmal1 expression increased in the liver parenchyma exposed to 3.6 and 7.2 J cm−2 of blue light LED illumination (p = 0.015 and 0.001, respectively), and the results differed between the two irradiances (p = 0.012). In the white light LED illumination group, Bmal1 expression also increased in the 7.2 J cm−2 group (p < 0.001) compared with the control one, but not in the 3.6 J cm−2 group (p = 0.064). There was no difference between groups receiving a different dose of white light LED illumination (p = 0.061). Furthermore, there was no significant difference between the groups receiving a dose of 7.2 J cm−2 of blue and white light LED illumination (p = 0.661) (Fig. 10A and B).On the other hand, Cry1 protein expression was identified in the hepatic sinusoids of the control group. The Cry1 immunoreactivity scores of hepatocytes were 13.3 ± 1.6 in the control group, and 10.4 ± 1.7 (3.6 J cm−2) and 9.1 ± 1.5 (7.2 J cm−2) in the blue light groups. In the white light groups, the Cry1 immunoreactivity scores of hepatocytes were 9.9 ± 1.7 (3.6 J cm−2) and 8.8 ± 1.0 (7.2 J cm−2). Cry1 expression significantly decreased in the liver parenchyma after 3.6 and 7.2 J cm−2 blue LED illumination (p = 0.002 and p < 0.001) as compared to the control group, but the results did not differ between the two irradiances (p = 0.090). In the white LED light group, Cry1 expression also decreased in the 3.6 and 7.2 J cm−2 groups (both p < 0.001) compared with the control, but the results exhibited no obvious difference between the illumination of the two white groups (p = 0.107). Moreover, there was no difference between the 7.2 J cm−2 blue or white illumination LED groups (p = 0.575) (Fig. 10A and B).
Per2 protein expression was identified in the cytoplasm of hepatocytes in the control group. The Per2 immunoreactivity scores of hepatocytes were 14.3 ± 1.3 in the control group, and 7.3 ± 3.3 (3.6 J cm−2) and 2.4 ± 3.1 (7.2 J cm−2) in the blue light groups. In the white light groups, the Per2 immunoreactivity scores of hepatocytes were 9.0 ± 1.9 (3.6 J cm−2) and 4.8 ± 4.2 (7.2 J cm−2). The expression of Per2 protein decreased significantly after 3.6 and 7.2 J cm−2 blue LED illumination (both p < 0.001) as compared to the control group; and the two irradiance groups responded differently (p = 0.014). In the white LED light group, Per2 expression also decreased in the 3.6 and 7.2 J cm−2 groups (both p < 0.001) compared with the control, and the two white LED irradiance groups responded differently (p = 0.017). However, there was no difference between the 7.2 J cm−2 blue illumination LED group and the 7.2 J cm−2 white one (p = 0.328) (Fig. 10A and B).
Rev-erbα protein expression was noted in the nuclei of hepatic sinusoids. The Rev-erbα immunoreactivity scores of KCs were 0 in the control group and the blue and white 3.6 J cm−2 groups. The Rev-erbα immunoreactivity scores of hepatocytes were 8.9 ± 3.2 in blue light and 8.4 ± 2.4 in white light at 7.2 J cm−2. In the 7.2 J cm−2 LED groups, Rev-erbα expression was significantly increased as compared with the control group (both p < 0.001), but no difference was found between the blue and white illumination groups (p = 0.740; Fig. 10A and B).
3.6. Discussion
Modern lifestyles, chronic jet lag, and shift work can lead to an irregular circadian rhythm which may induce an increased risk of health issues. LEDs are widely used in electronic devices and are regarded as a major risk factor for circadian dysfunction in modern societies.10 Dysregulation of the circadian rhythm in humans is known to be associated with metabolic diseases, such as obesity, diabetes, cardiovascular disease, and NAFLD.46 In this study, we found that chronic circadian dysregulation induced by LED illumination resulted in a decrease in the number of KCs and elongated glycan residue chains of glycoproteins in hepatocytes, which play significant roles in the initiation of NAFLD.Previous studies have indicated that chronic disruption of the circadian rhythm causes an inflammatory response and induces metabolic diseases.9,47 It is well known that macrophages play essential functions in the initiation of inflammation responses. Macrophages can be classified into bone marrow-derived macrophages and tissue-resident macrophages, which are derived from yolk-sac and/or fetal liver monocytes.48,49 Increasing evidence has revealed that macrophages play key roles in lipid metabolism and the pathogenesis of lipid-related diseases.50 In the pathogenesis of NAFLD, the accumulation of lipids is a key event in the early stages of NAFLD, while progression to non-alcoholic steatohepatitis is triggered by inflammation.18 KCs are specialized resident macrophages of the liver in the detection of invading pathogens and are located alongside sinusoid endothelial cells. KCs are known to play important roles in maintaining the functions of cholesterol, bilirubin, and iron metabolism. Additionally, KCs mediate phagocytosis, anti-inflammatory effects, and liver regeneration, which play dual roles in both intrahepatic and extrahepatic systems.51–54 KCs can regulate hepatic lipid metabolism and immune homeostasis and are the major effectors in the pathogenesis of NAFLD.50,55–58 KCs are critical in initiating liver damage and inflammation during experiments with NAFLD. The number of KCs was decreased in early disease onset with an increase in TNF-α production,59 followed by an infiltration of monocyte-derived KCs, which alter the liver response to lipid overload.60–62 In NAFLD, KCs can be activated by various factors, such as gut-derived endotoxins, free fatty acids, cholesterol, metabolites, and damaged hepatocytes.63
In this study, mice were exposed 5 days a week to blue LED (3.6 J cm−2 or 7.2 J cm−2) or white LED (3.6 J cm−2 or 7.2 cm−2) illumination at ZT 13.5–14 for 44 weeks. Immunohistochemical examinations of liver tissues revealed no inflammatory reaction as well as no TNF-α release in all LED illumination groups (Fig. 2). To gain a better understanding of dysregulations in hepatocytes, SR-FTIRM and WPK-FTIR imaging were used. These methods can identify the functional groups and chemical bonds that are present in biological tissues.64 SR-FTIRM was conducted to assess the mid-infrared absorbance ratios of lipid to protein and glycogen to protein within the area of interest in the liver sections. After receiving extra LED illumination, the infrared absorbance ratios L/P and G/P were dramatically greater than those of the control group, especially for the groups receiving the blue LED illumination, indicating that the metabolism of lipids and glycogen was retarded. WPK-FTIR imaging was conducted for analyzing the cell surface glycoprotein and is utilized to investigate the alteration of the chain length of the glycan residues of glycoproteins in the inflammatory hepatocytes. Alteration of the chain length of glycan residues owing to additional glycosylation of glycoproteins has been associated with inflammatory diseases.21 An inflammation index (C28R/C22R) of a liver tissue section of mice after receiving extra LED illumination (greater impact after blue LED illumination than that after white one) was found compared with the control groups.
In this study, the relationships between circadian dysregulation with the findings of immunohistopathological examinations, SR-FTIRM, and WPK-FTIR imaging were also investigated. We uncovered that the number of KCs in the LED illumination groups was significantly decreased, especially in the 7.2 J cm−2 blue LED illumination group without subsequent inflammation. Alterations of hepatic circadian protein expression via LED illumination play significant roles in the regulation of liver physiologic functions. In normal animal physiology, diurnal oscillation is involved in major pathways of liver homeostasis and is especially associated with the Bmal1 gene, including immune responses, fatty acid and glucose metabolism, and cell cycle by proteomics.65,66 Bmal1 may regulate macrophages in the oxidation pathway to limit the production of pro-inflammatory cytokines.67 We found that an increase in Bmal1 expression was demonstrated in hepatocytes after receiving extra LED illumination, especially in the 7.2 J cm−2 blue and white LED illumination groups. Overexpression of Bmal1 could suppress pro-inflammatory cytokine production in macrophages and play a role in the avoidance of subsequent inflammatory reactions. Furthermore, we noticed overexpression of Rev-erbα in the 7.2 J cm−2 blue and white LED illumination groups, but not in the 3.6 J cm−2 illumination group. Previous studies demonstrated that Rev-erbα regulates the inflammatory infiltration of macrophages through the suppression of CCL2 expression. CCL2 is an important chemokine that binds to CCR2 on monocytes/macrophages to stimulate their migration and initiate inflammation.68,69 It is a reasonable inference that the Rev-erbα overexpression found in our study might play a suppressive role in recruiting circulating monocytes to the liver after KC depletion. Accordingly, an increase in Bmal1 expression and overexpression of Rev-erbα would explain this study's finding of depletion of KCs without subsequent inflammation after LED illumination.
Circadian protein Cry1 is an inhibitor of gluconeogenesis in the liver, and as the target of the lysosome, it is degraded by liver macroautophagy. Autophagic degradation of Cry1 causes repression of gluconeogenesis and increases glucose levels.70 In addition, Per-2 is a negative regulator of lipogenesis in hepatocytes, and downregulation of Per-2 restores lipogenic gene expression and reverses the reduced TG levels in hepatocytes.71 We found significantly decreased Per2 and Cry1 proteins in the liver after LED illumination, especially in the 7.2 J cm−2 blue and white LED illumination groups. These findings echoed the results identified by SR-FTIRM. Per2 expresses a protective effect on liver injury and fibrosis during cholestasis. Decreased Per2 levels might exacerbate the cholestatic liver injury and fibrosis.72 Our result of the extreme reduction of Per2 expression in 7.2 J cm−2 blue LED illumination mice coincides with the findings of inflammation from WPK-FTIR.
Our work demonstrated that long-term LED illumination can decrease the number of KCs and induce dysregulation of circadian rhythm in the liver. We unfolded three significant discoveries. First, we found that decreased KC numbers failed to induce subsequent inflammatory reactions in the liver. Second, we identified a significant increase in Bmal1 and Rev-erbα protein expression, while there was a decrease in Cry1 and Per2 protein expression in the liver. The increased expression of Bmal1 and Rev-erbα provides possible directions for interpretation regarding the normal histopathological findings without obvious inflammatory reactions in the LED-illuminated liver. Third, by using SR-FTIRM and WPK-FTIR imaging technologies higher ratios of L/P and C28R/C22R (inflammation index) were both noticed in the liver of the blue light illumination groups, as compared to the control group and groups that received white LED illumination. These findings significantly correlated with a decrease in Per2 and Cry1 expression in the blue LED illumination groups. Since KCs and circadian proteins play significant roles in the development of NAFLD, our findings provide scientific evidence that chronic circadian dysregulation induced by LED illumination plays a predisposing role in inducing NAFLD. Furthermore, WPK-FTIR imaging is a sensitive method for the early detection of the initial stages of LED light-triggered NAFLD.
4. Conclusions
WPK-FTIR is a sensitive method for detecting the initial stages of NAFLD induced by long-term blue LED illumination. KCs and circadian proteins play significant roles in the pathogenesis of blue LED-induced NAFLD.Author contributions
Conceptualization, HSY and YCL; methodology, YTC, PYH, YCL and CWK; resource of the LED light box, HCC; software, PYH and YCL; formal analysis, YTC; investigation, PYH, SY and YLH; data curation, SY and YLH; writing—original draft, CYC and YTC; writing—review and editing, HSY and YCL; visualization, CWK; supervision, CYC and HSY; project administration, HSY; and funding acquisition, HSY. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Distinguished Chair Professor Ding-Shinn Chen of Internal Medicine, National Taiwan University College of Medicine, for his guidance and comments, and Mr Chao-Hung Cheng of the National Institute of Environmental Health Sciences, National Health Research Institutes, for his assistance with animal experiments. This research was funded by the Ministry of Science and Technology, Taiwan, grant number MOST-111-2320-B-037-010-MY3 and MOST-111-2314-B-037-001-, 002- and 003-.References
- C. E. Koch, B. Leinweber, B. C. Drengberg, C. Blaum and H. Oster, Neurobiol. Stress, 2017, 6, 57–67 CrossRef PubMed.
- M. Spitschan, Curr. Opin. Behav. Sci., 2019, 30, 67–72 CrossRef PubMed.
- H. J. Bailes and R. J. Lucas, Proc. Biol. Sci., 2013, 280, 20122987 Search PubMed.
- M. Dudek and Q. J. Meng, Biochem. J., 2014, 463, 1–8 CrossRef PubMed.
- J. Richards and M. L. Gumz, Am. J. Physiol.: Regul., Integr. Comp. Physiol., 2013, 304, R1053–R1064 CrossRef PubMed.
- H. Ukai and H. R. Ueda, Annu. Rev. Physiol., 2010, 72, 579–603 CrossRef PubMed.
- N. Cermakian, T. Lange, D. Golombek, D. Sarkar, A. Nakao, S. Shibata and G. Mazzoccoli, Chronobiol. Int., 2013, 30, 870–888 CrossRef PubMed.
- Y. Touitou, A. Reinberg and D. Touitou, Life Sci., 2017, 173, 94–106 CrossRef PubMed.
- O. Castanon-Cervantes, M. Wu, J. C. Ehlen, K. Paul, K. L. Gamble, R. L. Johnson, R. C. Besing, M. Menaker, A. T. Gewirtz and A. J. Davidson, J. Immunol., 2010, 185, 5796–5805 CrossRef PubMed.
- G. Tosini, I. Ferguson and K. Tsubota, Mol. Vision, 2016, 22, 61–72 Search PubMed.
- R. G. Stevens, Chronobiol. Int., 2016, 33, 589–594 CrossRef PubMed.
- M. Keller, J. Mazuch, U. Abraham, G. D. Eom, E. D. Herzog, H. D. Volk, A. Kramer and B. Maier, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 21407–21412 CrossRef PubMed.
- T. Bollinger, A. Leutz, A. Leliavski, L. Skrum, J. Kovac, L. Bonacina, C. Benedict, T. Lange, J. Westermann, H. Oster and W. Solbach, PLoS One, 2011, 6, e29801 CrossRef CAS PubMed.
- A. C. Silver, A. Arjona, W. E. Walker and E. Fikrig, Immunity, 2012, 36, 251–261 CrossRef CAS.
- A. M. Curtis, M. M. Bellet, P. Sassone-Corsi and L. A. O'Neill, Immunity, 2014, 40, 178–186 CrossRef CAS.
- M. Sayiner, A. Koenig, L. Henry and Z. M. Younossi, Clin. Liver Dis., 2016, 20, 205–214 CrossRef.
- J. A. Marrero, R. J. Fontana, G. L. Su, H. S. Conjeevaram, D. M. Emick and A. S. Lok, Hepatology, 2002, 36, 1349–1354 CrossRef.
- Z. Wenfeng, W. Yakun, M. Di, G. Jianping, W. Chuanxin and H. Chun, Ann. Hepatol., 2014, 13, 489–495 CrossRef.
- R. Peracaula, S. Barrabés, A. Sarrats, P. M. Rudd and R. de Llorens, Dis. Markers, 2008, 25, 207–218 CrossRef CAS PubMed.
- R. Kannagi, J. Yin, K. Miyazaki and M. Izawa, Biochim. Biophys. Acta, 2008, 1780, 525–531 CrossRef CAS PubMed.
- X. Verhelst, A. M. Dias, J. F. Colombel, S. Vermeire, H. Van Vlierberghe, N. Callewaert and S. S. Pinho, Gastroenterology, 2020, 158, 95–110 CrossRef CAS.
- C. H. Lee, C. Y. Hsu, P. Y. Huang, C. I. Chen, Y. C. Lee and H. S. Yu, Int. J. Mol. Sci., 2016, 17, 427 CrossRef PubMed.
- S. Zhang, Y. Wang, Z. Wang, H. Wang, C. Xue, Q. Li, W. Guan and J. Yuan, Occup. Environ. Med., 2020, 77, 333–339 CrossRef PubMed.
- P. Yu, Br. J. Nutr., 2004, 92, 869–885 CrossRef PubMed.
- M. M. Hsu, P. Y. Huang, Y. C. Lee, Y. C. Fang, M. W. Chan and C. I. Lee, Int. J. Mol. Sci., 2014, 15, 17963–17973 CrossRef PubMed.
- L. F. Chiu, P. Y. Huang, W. F. Chiang, T. Y. Wong, S. H. Lin, Y. C. Lee and D. B. Shieh, Anal. Bioanal. Chem., 2013, 405, 1995–2007 CrossRef PubMed.
- N. Shakiba, C. A. White, Y. Y. Lipsitz, A. Yachie-Kinoshita, P. D. Tonge, S. M. I. Hussein, M. C. Puri, J. Elbaz, J. Morrissey-Scoot, M. Li, J. Munoz, M. Benevento, I. M. Rogers, J. H. Hanna, A. J. R. Heck, B. Wollscheid, A. Nagy and P. W. Zandstra, Nat. Commun., 2015, 6, 7329 CrossRef PubMed.
- A. L. Tarentino, R. B. Trimble and T. H. Plummer, Jr., Methods Cell Biol., 1989, 32, 111–139 CrossRef PubMed.
- F. Maley, R. B. Trimble, A. L. Tarentino and T. H. Plummer, Jr., Anal. Biochem., 1989, 180, 195–204 CrossRef PubMed.
- G. Wiggins and M. Legge, J. Reprod. Infertil., 2016, 17, 192–198 Search PubMed.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sule-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Nat. Protoc., 2014, 9, 1771–1791 CrossRef CAS PubMed.
- A. Sala, D. J. Anderson, P. M. Brennan, H. J. Butler, J. M. Cameron, M. D. Jenkinson, C. Rinaldi, A. G. Theakstone and M. J. Baker, Cancer Lett., 2020, 477, 122–130 CrossRef CAS PubMed.
- E. Staniszewska, K. Malek and M. Baranska, Spectrochim. Acta, Part A, 2014, 118, 981–986 CrossRef CAS PubMed.
- E. O. Faolain, M. B. Hunter, J. M. Byrne, P. Kelehan, H. A. Lambkin, H. J. Byrne and F. M. Lyng, J. Histochem. Cytochem., 2005, 53, 121–129 CrossRef CAS PubMed.
- R. Gaifulina, D. J. Caruana, D. Oukrif, N. J. Guppy, S. Culley, R. Brown, I. Bell, M. Rodriguez-Justo, K. Lau and G. M. H. Thomas, Analyst, 2020, 145, 1499–1510 RSC.
- J. Chwiej, A. Skoczen, K. Janeczko, J. Kutorasinska, K. Matusiak, H. Figiel, P. Dumas, C. Sandt and Z. Setkowicz, Analyst, 2015, 140, 2190–2204 RSC.
- M. J. Hackett, J. A. McQuillan, F. El-Assaad, J. B. Aitken, A. Levina, D. D. Cohen, R. Siegele, E. A. Carter, G. E. Grau, N. H. Hunt and P. A. Lay, Analyst, 2011, 136, 2941–2952 RSC.
- M. J. Hackett, N. J. Sylvain, H. Hou, S. Caine, M. Alaverdashvili, M. J. Pushie and M. E. Kelly, Anal. Chem., 2016, 88, 10949–10956 CrossRef CAS PubMed.
- C. L. Effio and J. Hubbuch, Biotechnol. J., 2015, 10, 715–727 CrossRef CAS.
- B. Szalontai, Y. Nishiyama, Z. Gombos and N. Murata, Biochim. Biophys. Acta, 2000, 1509, 409–419 CrossRef CAS PubMed.
- O. Bozkurt, S. Haman Bayari, M. Severcan, C. Krafft, J. Popp and F. Severcan, J. Biomed. Opt., 2012, 17(7), 076023 CrossRef PubMed.
- D. M. Oswald, M. B. Jones and B. A. Cobb, Glycobiology, 2020, 30, 346–359 CrossRef CAS PubMed.
- A. J. Fadden, O. J. Holt and K. Drickamer, Glycobiology, 2003, 13, 529–537 CrossRef CAS PubMed.
- N. Islam, H. Wang, F. Maqbool and V. Ferro, Molecules, 2019, 24(7), 1271–1288 CrossRef CAS PubMed.
- J. D. Pagan, M. Kitaoka and R. M. Anthony, Cell, 2018, 172(3), 564–577 CrossRef CAS PubMed.
- J. Bass and J. S. Takahashi, Science, 2010, 330, 1349–1354 CrossRef CAS PubMed.
- Z. Yang, H. Kim, A. Ali, Z. Zheng and K. Zhang, Liver Res., 2017, 1, 156–162 CrossRef PubMed.
- F. Ginhoux and M. Guilliams, Immunity, 2016, 44, 439–449 CrossRef CAS PubMed.
- C. Schulz, E. Gomez Perdiguero, L. Chorro, H. Szabo-Rogers, N. Cagnard, K. Kierdorf, M. Prinz, B. Wu, S. E. Jacobsen, J. W. Pollard, J. Frampton, K. J. Liu and F. Geissmann, Science, 2012, 336(6077), 86–90 CrossRef CAS PubMed.
- A. Remmerie and C. L. Scott, Cell. Immunol., 2018, 330, 27–42 CrossRef CAS PubMed.
- D. Hirayama, T. Iida and H. Nakase, Int. J. Mol. Sci., 2017, 19(1), 92–106 CrossRef PubMed.
- C. C. Qin, Y. N. Liu, Y. Hu, Y. Yang and Z. Chen, World J. Gastroenterol., 2017, 23, 3043–3052 CrossRef CAS PubMed.
- Y. Kakinuma, T. Kimura and Y. Watanabe, Can. J. Gastroenterol. Hepatol., 2017, 2017(2896809), 1–10 Search PubMed.
- C. Mehrfeld, C. Eckert, H. Julich-Haertel, S. Urban, M. Kornek, F. Lammert and V. Lukacs-Kornek, J. Hepatol., 2017, 66, S166 CrossRef.
- J. H. Lefkowitch, J. H. Haythe and N. Regent, Mod. Pathol., 2002, 15, 699–704 CrossRef PubMed.
- J. W. Park, G. Jeong, S. J. Kim, M. K. Kim and S. M. Park, J. Gastroenterol. Hepatol., 2007, 22, 491–497 CrossRef PubMed.
- C. L. Scott, F. Zheng, P. De Baetselier, L. Martens, Y. Saeys, S. De Prijck, S. Lippens, C. Abels, S. Schoonooghe, G. Raes, N. Devoogdt, B. N. Lambrecht, A. Beschin and M. Guilliams, Nat. Commun., 2016, 7, 10321 CrossRef PubMed.
- C. L. Scott and M. Guilliams, J. Hepatol., 2018, 69, 1197–1199 CrossRef PubMed.
- A. C. Tosello-Trampont, S. G. Landes, V. Nguyen, T. I. Novobrantseva and Y. S. Hahn, J. Biol. Chem., 2012, 287, 40161–40172 CrossRef PubMed.
- J. Bonnardel, W. T’Jonck, D. Gaublomme, R. Browaeys, C. L. Scott, L. Martens, B. Vanneste, S. De Prijck, S. A. Nedospasov, A. Kremer, E. Van Hamme, P. Borghgraef, W. Toussaint, P. De Bleser, I. Mannaerts, A. Beschin, L. A. van Grunsven, B. N. Lambrecht, T. Taghon, S. Lippens, D. Elewaut, Y. Saeys and M. Guilliams, Immunity, 2019, 51(4), 638–654 CrossRef PubMed.
- D. T. Reid, J. L. Reyes, B. A. McDonald, T. Vo, R. A. Reimer and B. Eksteen, PLoS One, 2016, 11, e0159524 CrossRef CAS PubMed.
- S. Tran, I. Baba, L. Poupel, S. Dussaud, M. Moreau, A. Gélineau, G. Marcelin, E. Magréau-Davy, M. Ouhachi, P. Lesnik, A. Boissonnas, W. Le Goff, B. E. Clausen, L. Yvan-Charvet, F. Sennlaub, T. Huby and E. L. Gautier, Immunity, 2020, 53, 627–640.e625 CrossRef CAS PubMed.
- K. Kazankov, S. M. D. Jørgensen, K. L. Thomsen, H. J. Møller, H. Vilstrup, J. George, D. Schuppan and H. Grønbæk, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 145–159 CrossRef CAS PubMed.
- I. u. Rehman, Z. Movasaghi and S. Rehman, Vibrational Spectroscopy for Tissue Analysis, CRC Press, 2013 Search PubMed.
- Y. Wang, L. Song, M. Liu, R. Ge, Q. Zhou, W. Liu, R. Li, J. Qie, B. Zhen, Y. Wang, F. He, J. Qin and C. Ding, Nat. Commun., 2018, 9, 1553 CrossRef PubMed.
- D. Zhou, Y. Wang, L. Chen, L. Jia, J. Yuan, M. Sun, W. Zhang, P. Wang, J. Zuo, Z. Xu and J. Luan, Oncotarget, 2016, 7, 8625–8639 CrossRef PubMed.
- J. O. Early, D. Menon, C. A. Wyse, M. P. Cervantes-Silva, Z. Zaslona, R. G. Carroll, E. M. Palsson-McDermott, S. Angiari, D. G. Ryan, S. E. Corcoran, G. Timmons, S. S. Geiger, D. J. Fitzpatrick, D. O'Connell, R. J. Xavier, K. Hokamp, L. A. J. O'Neill and A. M. Curtis, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E8460–e8468 CrossRef CAS PubMed.
- A. Rot and U. H. von Andrian, Annu. Rev. Immunol., 2004, 22, 891–928 CrossRef CAS PubMed.
- S. Sato, T. Sakurai, J. Ogasawara, M. Takahashi, T. Izawa, K. Imaizumi, N. Taniguchi, H. Ohno and T. Kizaki, J. Immunol., 2014, 192, 407–417 CrossRef PubMed.
- M. Toledo, A. Batista-Gonzalez, E. Merheb, M. L. Aoun, E. Tarabra, D. Feng, J. Sarparanta, P. Merlo, F. Botrè, G. J. Schwartz, J. E. Pessin and R. Singh, Cell. Metab., 2018, 28, 268–281 CrossRef PubMed.
- C. F. Chou, X. Zhu, Y. Y. Lin, K. L. Gamble, W. T. Garvey and C. Y. Chen, J. Lipid Res., 2015, 56, 227–240 CrossRef PubMed.
- P. Chen, X. Kakan, S. Wang, W. Dong, A. Jia, C. Cai and J. Zhang, Exp. Toxicol. Pathol., 2013, 65, 427–432 CrossRef PubMed.
- Y. Y. Chen, P. Y. Huang, J. Y. Wang, Y. C. Lee and C. Y. Chai, Sci. Rep., 2022, 12, 17168–17168 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an01546c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |