Open Access Article
Open Access ArticleNovel fluorescence-based method for rapid quantification of live bacteria in river water and treated wastewater†
Manna
Wang
a,
Mohamed
Ateia
 *bc,
Yuta
Hatano
a,
Kazuhiko
Miyanaga
d and
Chihiro
Yoshimura
*a
*bc,
Yuta
Hatano
a,
Kazuhiko
Miyanaga
d and
Chihiro
Yoshimura
*a
aDepartment of Civil and Environmental Engineering, Tokyo Institute of Technology, Tokyo 152-8552, Japan. E-mail: yoshimura.c.aa@m.titech.ac.jp
bDepartment of Chemistry, Northwestern University, Evanston, Illinois 60208, USA. E-mail: Ibrahim.mohamed@epa.gov
cUnited States Environmental Protection Agency, Center for Environmental Solutions & Emergency Response, Cincinnati, OH, USA
dDepartment of Life Science and Technology, Tokyo Institute of Technology, Yokohama 226-8501, Japan
First published on 3rd November 2021
Abstract
Monitoring bacteria is essential for ensuring microbial safety of water sources, including river water and treated wastewater. The plate count method is common for monitoring bacterial abundance, although it cannot detect all live bacteria such as viable but non-culturable bacteria, causing underestimation of microbial risks. Live/Dead BacLight kit, involving fluorochromes SYTO 9 and propidium iodide (PI), provides an alternative to assess bacterial viability using flow cytometry or microscopy. However, its application is limited due to the high cost of flow cytometry and the inapplicability of microscopy to most environmental waters. Thus, this study introduces the combination of BacLight kit and fluorescence spectroscopy for quantifying live bacteria in river water and treated wastewater. Mixtures of live and dead Escherichia coli (E. coli) with various ratios and total cell concentrations were stained with SYTO 9 and PI and measured by fluorescence spectroscopy. The fluorescence emission peak area of SYTO 9 in the range of 500–510 nm at the excitation wavelength of 470 nm correlates linearly with the viable cell counts (R2 > 0.99, p < 0.0001) with only slight variations in the complex water matrix. The tested method can quantify the live E. coli from 3.67 × 104 to 2.70 × 107 cells per mL. This method is simple, sensitive and reliable for quantifying live bacteria in environmental water, which can be later integrated into real-time monitoring systems.
Environmental significanceThe microbiological examination of water is an essential task to ensure the water safety, and monitoring bacteria is one of the most important steps in this task. Heterotrophic plate count can indicate the efficiency of water treatment or quality of natural water. However, the traditional culture-dependent method fails in detecting all viable bacteria and is time-consuming. Therefore, developing an effective and reliable method for monitoring bacterial viability is needed. |
1. Introduction
Quantifying bacteria is an essential task to ensure the microbial safety of river water and treated wastewater. The traditional plate count method for bacterial quantification is time-consuming and can only detect culturable cells under the set conditions (e.g., media type, temperature, incubation time).1 However, a large number of bacteria in water, such as viable but non-culturable (VBNC) bacteria, cannot be cultured under laboratory conditions, resulting in underestimating the actual bacterial population and thus their biological risk.2,3 Besides their potential to reactivate and regrow, VBNC bacteria were found to retain a certain level of plasmid gene transfer efficiency4 as well as their resistance to antibiotics.5 Therefore, it is crucial to quantify the total viable cells, consisting of culturable and VBNC bacteria, especially in environmental water samples such as river water and treated wastewater.Among various bacterial detection methods (Table S1†), Live/Dead BacLight kit has been widely used in bacterial viability tests.6,7 The kit typically contains fluorescent nucleic acid-binding stains SYTO 9 and propidium iodide (PI). Due to their different ability to permeate cell membranes,8 when used alone, SYTO 9 and PI can label total cells green and dead cells red, respectively. Qualitative or quantitative evaluation of bacterial viability has been conducted through fluorescence detection using flow cytometry and fluorescence microscopy.9–14 However, the high facility cost and requirement of technicians restrict the application of flow cytometry.6,15 And fluorescence microscopy is time-consuming, especially when analyzing a large number of samples16 and not applicable in complex water matrices, which is shown by the preliminary work of this study (Fig. S1†). Therefore, a rapid and effective method for fluorescence detection and bacterial quantification is required.
Several studies have followed the BacLight kit instructions to establish a standard curve of red to green fluorescence ratio against the proportion of live cells in a sample in saline solution6,17–19 (Table S2†). However, the fluorescence-based bacterial quantification using BacLight in complex water matrices (e.g., river water) has not been explored except in two studies, which tested river water using flow cytometry20 and growth media using a fluorescence optrode.21 Since minimizing viable microbial loads in river water and treated water is dependent on accurate quantification of viable bacteria, evaluation of methods such as BacLight kit using fluorescence spectroscopy is needed.
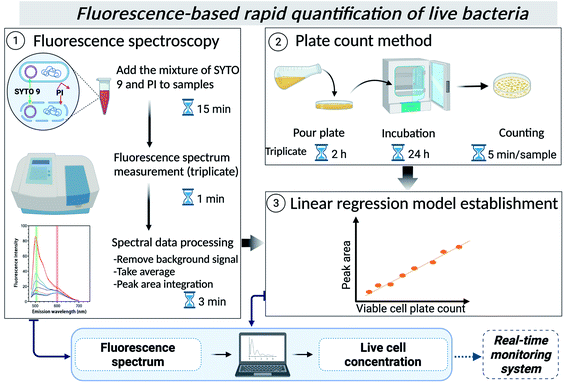
In this study, therefore, we developed and verified a fluorescence spectroscopy protocol for quantifying viable bacteria in environmental water using the Live/Dead BacLight kit and a reference bacterial strain for the first time (Scheme 1). A simple linear relationship between fluorescence peak intensity and cell plate counts was built for several types of waters, showing the utility of the method in quantifying live bacteria in other types of environmental waters. This method would allow rapid quantification of live bacteria, including VBNC cells and can be automated to be later integrated into real-time monitoring systems for on-site applications.
2. Methods
2.1 River water and treated wastewater
The river water sample was taken from the downstream section of the Tama River (Tokyo, Japan) on March 4th, 2021, which receives some treated domestic wastewater. Treated wastewater was sampled from the Nomi River (Tokyo, Japan) on May 12th, 2021, as 100% of the discharge in this river comes from domestic wastewater treatment plants. Each water sample was filtered with a 0.22 μm membrane to ensure the removal of bacterial cells, and this was confirmed by the negative culture on Luria–Bertani (LB) agar plates. The dissolved organic carbon measured using the TOC-CHP analyzer (Shimadzu V-series, Japan) is 2.198 and 9.725 mg C per L in Tama River and Nomi River water samples, respectively.2.2 Preparation of mixtures of live and dead E. coli
E. coli (NBRC3301) (NITE Biological Resource Center, Japan) was inoculated into LB medium (L3022, Sigma-Aldrich) and cultured overnight at 37 °C in a shaker incubator. Live and dead E. coli were prepared following the instructions of the BacLight viability kit with some modifications (Fig. S2†). The prepared live and dead E. coli suspensions were diluted in 0.85% NaCl solution (SA), Tama River water (TM), Nomi River water (NM), the mixture of TM and SA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (TM + SA), and the mixture of NM and SA (1
1) (TM + SA), and the mixture of NM and SA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (NM + SA), to achieve concentrations of ∼105, ∼106, and ∼107 cells per mL. Then, live and dead cells in each water were mixed in ratios of 0
1) (NM + SA), to achieve concentrations of ∼105, ∼106, and ∼107 cells per mL. Then, live and dead cells in each water were mixed in ratios of 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 20
100, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80, 50
80, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 80
50, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, and 100
20, and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0. We expected no formation of VBNC cells during this process because of the absence of environmental stress to E. coli. Finally, the mixtures were analyzed by the plate count method and fluorescence viability test using fluorescence spectroscopy. The saline solution, glassware and centrifuge tubes were autoclaved prior to use.
0. We expected no formation of VBNC cells during this process because of the absence of environmental stress to E. coli. Finally, the mixtures were analyzed by the plate count method and fluorescence viability test using fluorescence spectroscopy. The saline solution, glassware and centrifuge tubes were autoclaved prior to use.
2.3 Bacterial quantification by the plate count method and using the Live/Dead BacLight kit
Each sample was diluted at an appropriate concentration with sterilized 0.85% NaCl solution and pour plated in triplicate using LB agar (L2897, Sigma-Aldrich) as the growth medium. The detection limit is 1 CFU mL−1 for undiluted samples and 10 CFU mL−1 for diluted samples. Live/Dead BacLight bacterial viability kit (L7012, Invitrogen™) was used to quantify total live cells. SYTO 9 can penetrate the membrane of viable cells while PI cannot penetrate it, and PI has a higher affinity to nucleic acids than SYTO 9.22 So, when both dyes (i.e., SYTO 9 and PI) are applied, viable cells show a strong green fluorescence signal while dead cells show red fluorescence. 3 μL of the mixture of SYTO 9 (3.34 mM in dimethyl sulfoxide) and PI (20 mM in dimethyl sulfoxide) was added for each mL of the sample. The final concentrations of SYTO 9 and PI were 10.02 μM and 60 μM. After mixing, the stained samples were incubated at room temperature in the dark for 15 minutes. Subsequently, the fluorescence emission spectrum of each sample was measured in triplicate in the range of 480–700 nm by fixing excitation wavelength at 470 nm using a fluorescence spectrophotometer (RF-5300 PC, Shimadzu, Japan). The fluorescence spectrum of each sample was measured without a prior washing step to prevent cell loss.21,23 Each water matrix without spiking of E. coli was also stained and measured by fluorescence spectroscopy to obtain the background spectrum of each water matrix. Finally, to preprocess the obtained spectra of bacterial samples, the background spectrum of each water matrix was subtracted from each spectrum, and the average spectrum of the results in triplicate was taken for regression model calculation.2.4 Application of the regression model and determination of the detection limit
Preprocessed spectra were used to calculate the integrated fluorescence intensity of SYTO 9 and PI peaks in GraphPad Prism (version 9.1.2). The fluorescence peaks in all acquired spectra were between 500 and 510 nm for SYTO 9, and 595 and 605 nm for PI. In addition, the ratio and the adjusted ratio of peak area between SYTO 9 and PI were also calculated according to the kit instructions and a previous study.24 The calculated peak areas of SYTO 9 and peak ratios between SYTO 9 and PI were used to make scatter plots against viable cell plate count. Then, each scatter plot was analyzed by regression analysis in GraphPad Prism (version 9.1.2) or Microsoft Excel. In the regression analysis, viable cell plate count (CFU mL−1) and the peak area or the peak ratio were considered independent and dependent variables, respectively. The final regression analysis omitted data points with a viable cell plate count less than 104 CFU mL−1. The goodness of the regression model was quantified by the coefficient of determination (R2) and p-value.Based on the experimental results and calculated models, the limit of detection (LOD) of viable cell concentration in each type of water was determined. Here, we define experimental LOD and mathematical LOD. The experimentally defined LOD of the viable cell was the smallest value of viable cell plate count used for linear model calculation. The mathematically defined LOD of the viable cell was obtained by assuming the smallest value of the dependent variable, assuring a positive value of the cell concentration.
3. Results and discussion
3.1 Fluorescence spectra and intensity peaks
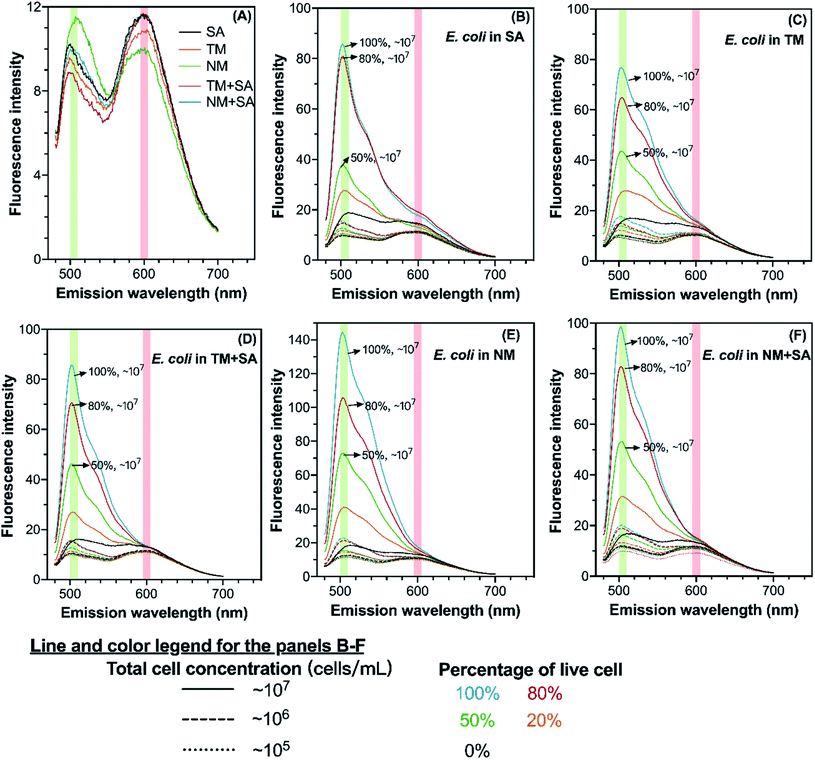
The spectra of all stained samples in different water matrices exhibit peaks at the emission wavelength at 505 nm and 600 nm corresponding to SYTO 9 and PI, respectively (Fig. 1). The BacLight kit recommends using the regions of 510–540 nm and 620–650 nm for calculating peak areas of SYTO 9 and PI, respectively. Meanwhile, previous studies determined the integrated fluorescence intensity in regions of 505–515 nm and 509–529 nm for SYTO 9, and 600–610 nm and 609–629 nm for PI.21,24 Though the reason for such variations in the position of the fluorescence peaks across studies is unclear, it could be the difference in the equipment used for fluorescence measurement or the difference in the water matrix (Table S2†). In addition, peaks of SYTO 9 and PI were also observed in stained samples without E. coli spiking (Fig. 1(A)). The appearance of the peaks in cell-free water was probably due to the existence of extracellular DNA or RNA, which can also bind to SYTO 9 and PI.25,26 Therefore, the background fluorescence signal was removed from all samples by subtracting the corresponding background spectrum from sample spectra (preprocessed spectra seen in Fig. S3†). The difference in the preprocessed spectra among the water matrices basically revealed that E. coli cells took up different amounts of SYTO 9 and PI, which could be attributed to three factors. First, the number of active cells is not the same in those water matrices, inferring the different capacity of SYTO 9 and PI uptake due to the varying amount of nucleic acid. Second, the cell membrane permeability, which determines the ability of intaking SYTO 9 and blocking PI, depends on various factors such as pH and nutrient scarcity. For example, nutrient scarcity can alter the cell wall and reduce dye permeability.20 Third, the binding of SYTO 9 or PI with nucleic acids is influenced by the water matrix such as the presence of metal ions and urea.273.2 Regression model calculation and LOD determination
Simple linear regression analysis showed that the SYTO 9 peak area across all four wavelength ranges was linearly correlated with viable cell plate count for all five types of water (R2 > 0.90) (Fig. S4†). In the comparison of several wavelength ranges, fluorescence integration across the wavelength range 500–510 nm resulted in the best linear fits with the highest R2 values (R2 > 0.95 for saline solution and R2 > 0.98 for the others), though other wavelength ranges also gave rather high R2 values over 0.93 (Fig. S4†). Yet, Ou et al.6 reported that the integrated SYTO 9 peak intensity (509–529 nm) did not have a linear relationship with cell counts, which could be due to the intrinsically complex interactions of fluorochromes or between fluorochromes and nucleic acids.Meanwhile, we found that the relationship of the peak ratio of SYTO 9 to PI and the percentage of live cells could not be represented by simple linearity, as several total cell concentrations (∼105, ∼106, or ∼107 cells per mL) were involved in the investigation. Nonlinear relationships between the peak ratio or adjusted ratio and logarithmic cell plate count were observed in all types of water (Fig. S5†). Nonlinear models using the peak regions suggested by the kit instruction gave higher R2 values than those using other peak regions. This result was contradictory to the BacLight kit instruction and previous studies,21 which present a linear relationship between the peak ratio and percentage of live bacteria when the total cell concentration was fixed. Utilizing the full fluorescence spectra for area calculation or complex model construction (e.g., principal component regression and support vector regression) could involve more spectral information such as the slight shift of peak position and relative peak intensity, and the relative intensity of the two dyes6,28 (Table S2†). Yet, our investigation on five types of water suggested that complex nonlinear models did not outperform simple linear models, and the information from the SYTO 9 peak in the fluorescence emission spectrum was sufficient for model calculation. Therefore, we conducted the data analysis using simple linear regression based on the proposed SYTO 9 peak region (500–510 nm), where the emission peak appears in this study.
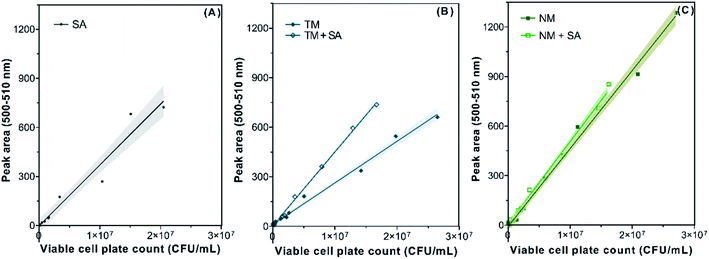
Compiling the data points for relatively low viable cell plate count values (Fig. S4†), we proceeded with more careful data analysis by removing those points with cell counts less than 104 CFU mL−1. Recalculated linear regression models (Fig. 2 and Table 1) showed slightly improved goodness of linear fit (R2 > 0.95 for saline solution and R2 > 0.99 for the others) compared with those models built on unfiltered data values (Fig. S4†). In determining the LOD of the fluorescence-based method, we found the experimentally and mathematically determined LOD values were very close and less than 105 cells per mL except in Nomi River water (Table 1). The LOD values determined in our experimental system were one to two orders lower than previous investigations.6,21
| Water type | Linear regression modela,b | R 2 | Experimentally defined LOD of the viable cellc (cells per mL) | Mathematically defined LOD of the viable celld (cells per mL) |
|---|---|---|---|---|
| a Here, y is the SYTO 9 peak area in the 500–510 nm range, and x is the viable cell plate count (CFU mL−1). b The p-value of all the linear regression models was less than 0.0001. c The experimentally defined LOD of the viable cell was the smallest value of viable cell plate count used for linear model calculation. d The mathematically defined LOD of the viable cell was obtained by assuming the smallest value of y. When the y-intercept was a positive value, the smallest y assured that the product of the slope and x was one. When the y-intercept was a negative value, the smallest y was zero. | ||||
| 0.85% saline solution | y = 3.72 × 10−5x − 1.67 | 0.96 | 3.67 × 104 | 4.49 × 104 |
| Tama River water | y = 2.52 × 10−5x + 10.93 | 0.99 | 5.07 × 104 | 3.97 × 104 |
| The mixture of 0.85% saline solution and Tama River water | y = 4.46 × 10−5x + 3.95 | 1.00 | 5.37 × 104 | 2.24 × 104 |
| Nomi River water | y = 4.73 × 10−5x − 9.64 | 0.99 | 1.80 × 105 | 2.04 × 105 |
| The mixture of 0.85% saline solution and Nomi River water | y = 5.00 × 10−5x + 7.14 | 0.99 | 3.63 × 104 | 2.00 × 104 |
Once the linear model is established for a specific water matrix, the presented method could quantify viable cells, including VBNC bacteria, within 30 minutes, including staining, sample measurement, and data analysis. This process is relatively fast compared to the plate count method (time consumption > 24 h), though the LOD is not as low as the plate count method (Table S1†). Given the disadvantages of flow cytometry and microscopy mentioned above, the presented method using fluorescence spectroscopy is a promising tool for fluorescence analysis with the BacLight kit. Besides the application to quantify live bacteria in environmental water demonstrated in this study, the method could have a broader range of applications, such as in laboratory-scale disinfection experiments and antimicrobial susceptibility tests. The limitations of this study are that only a single species, E. coli, was tested and that the LOD was much higher than the plate count method. For instance, the governmental regulations for bacteria in environmental water and treated wastewater are commonly lower than 104 CFU mL−1, for which the presented method cannot suffice in terms of measurement accuracy. Improvement of the LOD is out of the scope of the current study that aimed to develop a rapid and simple method for quantifying live bacteria. However, this limitation might be overcome in future work for example by adjusting the dye volume, adding a step of washing the samples, and using a more sensitive instrument for fluorescence measurement. Therefore, future work can focus on other species or microbial communities and the improvement of the LOD.
4. Conclusions
For the first time, this study demonstrated the use of fluorescence spectroscopy to determine the concentration of viable E. coli in saline solution, river water, and treated wastewater. The peak area of SYTO 9 (500–510 nm) is linearly dependent on the viable cell plate counts (R2 > 0.99, p < 0.0001), although the parameters of the regression models slightly varied with the water matrix. Though the LOD of the proposed method was not lower than 104 cells per mL, the presented results suggested the potential of fluorescence spectroscopy and the BacLight kit for quantifying live bacterial cells in environmental water. Future work can be extended to more bacterial species or bacterial communities. In addition, it is worth investigating ways to lower the LOD.Disclaimer
The research presented was not performed or funded by EPA and was not subject to EPA's quality system requirements. The views expressed in this article are those of the author(s) and do not necessarily represent the views or the policies of the U.S. Environmental Protection Agency.Author contributions
Manna Wang: conceptualization, methodology, investigation, data analysis, visualization, writing – original draft; Mohamed Ateia: methodology, visualization, writing – review & editing; Yuta Hatano: investigation, writing – review & editing; Kazuhiko Miyanaga: methodology, writing – review & editing; Chihiro Yoshimura: supervision, project administration, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This research was supported by the Japan Society for the Promotion of Science (JSPS KAKENHI) grant numbers 21H01462 and 21J13596. The authors are also grateful to the Center for Biological Resources and Informatics, Tokyo Institute of Technology, for providing access to the fluorescence microscope and technical support on its operation. M. A. is grateful for financial support by the Collaborative Water-Energy Research Center (CoWERC), supported by the Binational Industrial Research and Development Foundation under Energy Center grant EC-15. Figures were created using “http://BioRender.com”.References
- B. Papić, et al., New approaches on quantification of Campylobacter jejuni in poultry samples: The use of digital PCR and real-time PCR against the ISO standard plate count method, Front. Microbiol., 2017, 8, 331 CrossRef PubMed.
- M. Wang, M. Ateia, D. Awfa and C. Yoshimura, Regrowth of bacteria after light-based disinfection — What we know and where we go from here, Chemosphere, 2021, 268, 128850 CrossRef CAS PubMed.
- M. J. Allen, S. C. Edberg and D. J. Reasoner, Heterotrophic plate count bacteria - What is their significance in drinking water?, Int. J. Food Microbiol., 2004, 92, 265–274 CrossRef PubMed.
- D. E. Salcedo and S. Kim, Fate of tetracycline resistance in synthetic livestock carcass leachate for two years, J. Environ. Manage., 2017, 187, 220–228 CrossRef CAS PubMed.
- M. Amarasiri, D. Sano and S. Suzuki, Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered, Crit. Rev. Environ. Sci. Technol., 2019, 1–44 Search PubMed.
- F. Ou, C. McGoverin, S. Swift and F. Vanholsbeeck, Near real-time enumeration of live and dead bacteria using a fibre-based spectroscopic device, Sci. Rep., 2019, 9, 1–10 CAS.
- Y. Deng, L. Wang, Y. Chen and Y. Long, Optimization of staining with SYTO 9/propidium iodide: Interplay, kinetics and impact on Brevibacillus brevis, Biotechniques, 2020, 69, 89–99 CrossRef PubMed.
- L. Boulos, M. Prévost, B. Barbeau, J. Coallier and R. Desjardins, LIVE/DEAD(®) BacLight(TM): Application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water, J. Microbiol. Methods, 1999, 37, 77–86 CrossRef CAS PubMed.
- M. S. Gião, S. A. Wilks, N. F. Azevedo, M. J. Vieira and C. W. Keevil, Validation of SYTO 9/propidium iodide uptake for rapid detection of viable but noncultivable Legionella pneumophila, Microb. Ecol., 2009, 58, 56–62 CrossRef PubMed.
- J. Song, et al., Highly flexible, core-shell heterostructured, and visible-light-driven titania-based nanofibrous membranes for antibiotic removal and E. coil inactivation, Chem. Eng. J., 2020, 379, 122269 CrossRef CAS.
- R. Xiao, et al., Elucidating sulfate radical-mediated disinfection profiles and mechanisms of Escherichia coli and Enterococcus faecalis in municipal wastewater, Water Res., 2020, 173, 115552 CrossRef CAS PubMed.
- S. Das, et al., Ag@SnO2@ZnO core-shell nanocomposites assisted solar-photocatalysis downregulates multidrug resistance in Bacillus sp.: A catalytic approach to impede antibiotic resistance, Appl. Catal., B, 2019, 259, 118065 CrossRef CAS.
- S. Kim, et al., Bacterial inactivation in water, DNA strand breaking, and membrane damage induced by ultraviolet-assisted titanium dioxide photocatalysis, Water Res., 2013, 47, 4403–4411 CrossRef CAS PubMed.
- S. Chaiyanan, S. Chaiyanan, A. Huq, T. Maugel and R. R. Colwell, Viability of the nonculturable Vibrio cholerae O1 and O139, Syst. Appl. Microbiol., 2001, 24, 331–341 CrossRef CAS PubMed.
- Y. Wang, F. Hammes, K. De Roy, W. Verstraete and N. Boon, Past, present and future applications of flow cytometry in aquatic microbiology, Trends Biotechnol., 2010, 28, 416–424 CrossRef CAS PubMed.
- C. Jin, M. M. F. Mesquita, J. L. Deglint, M. B. Emelko and A. Wong, Quantification of cyanobacterial cells via a novel imaging-driven technique with an integrated fluorescence signature, Sci. Rep., 2018, 8, 1–12 Search PubMed.
- S. Y. Park and C. G. Kim, A comparative study of three different viability tests for chemically or thermally inactivated Escherichia coli, Environ. Eng. Res., 2018, 23, 282–287 CrossRef.
- P. Stiefel, S. Schmidt-Emrich, K. Maniura-Weber and Q. Ren, Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide, BMC Microbiol., 2015, 15, 1–9 CrossRef PubMed.
- F. Ou, C. McGoverin, S. Swift and F. Vanholsbeeck, Rapid and cost-effective evaluation of bacterial viability using fluorescence spectroscopy, Anal. Bioanal. Chem., 2019, 411, 3653–3663 CrossRef CAS PubMed.
- M. Berney, F. Hammes, F. Bosshard, H. U. Weilenmann and T. Egli, Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry, Appl. Environ. Microbiol., 2007, 73, 3283–3290 CrossRef CAS PubMed.
- J. Robertson, C. McGoverin, F. Vanholsbeeck and S. Swift, Optimisation of the protocol for the liVE/DEAD®BacLightTM bacterial viability kit for rapid determination of bacterial load, Front. Microbiol., 2019, 10, 1–13 CrossRef PubMed.
- S. M. Stocks, Mechanism and use of the commercially available viability stain, BacLight, Cytometry, Part A, 2004, 61, 189–195 CrossRef CAS PubMed.
- F. Ou, C. McGoverin, S. Swift and F. Vanholsbeeck, Absolute bacterial cell enumeration using flow cytometry, J. Appl. Microbiol., 2017, 123, 464–477 CrossRef CAS PubMed.
- F. Ou, C. McGoverin, S. Swift and F. VanholsbeeckRapid evaluation of bacterial viability using the optrode - A near real time portable fluorimeter. in Optics InfoBase Conference Papers AW3C.6, OSA - The Optical Society, 2016, DOI:10.1364/ACOFT.2016.AW3C.6.
- M. Okshevsky and R. L. Meyer, Evaluation of fluorescent stains for visualizing extracellular DNA in biofilms, J. Microbiol. Methods, 2014, 105, 102–104 CrossRef CAS PubMed.
- M. Rosenberg, N. F. Azevedo and A. Ivask, Propidium iodide staining underestimates viability of adherent bacterial cells, Sci. Rep., 2019, 9, 1–12 CAS.
- A. Samanta, B. K. Paul and N. Guchhait, Photophysics of DNA staining dye Propidium Iodide encapsulated in bio-mimetic micelle and genomic fish sperm DNA, J. Photochem. Photobiol., B, 2012, 109, 58–67 CrossRef CAS PubMed.
- C. McGoverin, J. Robertson, Y. Jonmohamadi, S. Swift and F. Vanholsbeeck, Species Dependence of SYTO 9 Staining of Bacteria, Front. Microbiol., 2020, 11, 1–11 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1va00017a |
| This journal is © The Royal Society of Chemistry 2022 |