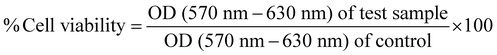Cationic lipopolymeric nanoplexes containing the CRISPR/Cas9 ribonucleoprotein for genome surgery†
Deepak Kumar
Sahel
a,
Mohd
Salman
bcd,
Mohd
Azhar
e,
Sangam Giri
Goswami
e,
Vivek
Singh
bd,
Manu
Dalela
f,
Sujata
Mohanty
 f,
Anupama
Mittal
a,
Sivaprakash
Ramalingam
e and
Deepak
Chitkara
f,
Anupama
Mittal
a,
Sivaprakash
Ramalingam
e and
Deepak
Chitkara
 *a
*a
aDepartment of Pharmacy, Birla Institute of Technology and Science, Pilani (BITS Pilani), Pilani Campus, Vidya Vihar, Pilani, Rajasthan 333031, India
bProf. Brien Holden Eye Research Center, Champalimaud Translational Centre for Eye Research, L.V. Prasad Eye Institute, Kallam Anji Reddy Campus, L V Prasad Marg, Hyderabad, 500 034, India
cManipal Academy of Higher Education, Manipal, Karnataka 576104, India
dCenter for Ocular Regeneration (CORE), LV Prasad Eye Institute, Hyderabad, Telangana, India
eCSIR-Institute of Genomics and Integrated Biology, New Delhi 110025, India
fStem Cell Facility, DBT-Centre of Excellence for Stem Cell Research, All India Institute of Medical Sciences (AIIMS), New Delhi 110029, India
First published on 20th July 2022
Abstract
sgRNA/Cas9 ribonucleoproteins (RNPs) provide a site-specific robust gene-editing approach avoiding the mutagenesis and unwanted off-target effects. However, the high molecular weight (∼165 kDa), hydrophilicity and net supranegative charge (∼−20 mV) hinder the intracellular delivery of these RNPs. In the present study, we have prepared cationic RNPs lipopolymeric nanoplexes that showed a size of 117.3 ± 7.64 nm with +6.17 ± 1.04 mV zeta potential and >90% entrapment efficiency of RNPs. Further, these RNPs lipopolymeric nanoplexes showed good complexation efficiency and were found to be stable for 12 h with fetal bovine serum. These RNPs lipopolymeric nanoplexes did not induce any significant cytotoxicity in HEK293T cells, and were efficiently uptaken via a clathrin-mediated pathway with optimal transfection efficiency and nuclear localization after 48 h. Further, HEK293T cells having the mGFP insert were used as a cell line model for gene editing, wherein the loss of the mGFP signal was observed as a function of gene editing after transfection with mGFP targeting RNPs lipopolymeric nanoplexes. Further, the T7 endonuclease and TIDE assay data showed a decent gene editing efficiency. Additionally, the lipopolymeric nanoplexes were able to transfect muscle cells in vivo, when injected intra-muscularly. Collectively, this study explored the potential of cationic lipopolymeric nanoplexes for delivering gene-editing endonucleases.
Introduction
Clustered regularly interspaced short palindromic repeat (CRISPR) is a versatile and precise genome editing tool, providing ample therapeutic opportunity in ailments with genetic and non-genetic causes.1,2 Up to now, out of the three available adaptive delivery forms of CRISPR (plasmid, mRNA, and direct Cas9 protein), protein delivery is the most effective genome editing platform due to the advantages like lesser off-target effects, lower insertional mutagenesis, short persistence of the Cas9 protein and lower immunogenic responses. The CRISPR/Cas9 system consists of two main components: the first one is the CRISPR-associated protein (Cas9) with endonuclease activity and the second one is a single guide RNA (sgRNA) with a complementary target gene sequence. Therefore, the Cas9 protein along with sgRNA is being delivered in the form of a ribonucleoprotein (RNP) to edit genes at the desired site by causing a double-strand break (DSB) that leads to activation of cellular DNA repair using non-homologous end-joining (NHEJ) and homology-directed repair (HDR). Despite precise and effective genome editing, the large molecular weight of the Cas9 protein (∼165 kD), hydrophilicity, and supra-negative charge (∼−20 mV), along with its sensitive and fragile nature, hinder the delivery of RNPs in vitro as well as in vivo.Previously, viral vectors were the only choice for delivering CRISPR/Cas9 tools which have a safety concern for therapeutic genome editing. Moreover, AAVs suffer from limited packaging capacity (4.7 kb) for commonly used SpCas9. Hence, attempts have been made to provide non-viral vectors, where RNPs were delivered using cationic polymeric or lipidic nanocarriers for efficient gene editing with certain limitations such as limited payload capacity, cytotoxicity, lack of tissue-specific targeting, instability, and limited in vivo application. Cationic lipid-based non-viral vectors also provided ample delivery opportunities; for instance, Wang et al. have recently reported a bio-reducible lipid nanocarrier complex for protein-based Cas9 genome editing.3 It was produced through electrostatic interaction of cationic lipids and super-negatively charged complexes via protein–protein fusion. Besides, Zuris et al. have demonstrated that negatively charged Cas9 endonuclease proteins could be fused with anionic supercharged proteins or anionic nucleic acids followed by complexation with the cationic lipids. They efficiently delivered Cre recombinase, TALEN- and Cas9-based transcriptional activators, and Cas9:sgRNA nuclease complexes into cultured human cells. Further, up to 80% of genome modification was observed with Cas9:sgRNA complexes as compared to DNA transfection.4 Initially, lipids were widely explored for CRISPR/Cas9 delivery due to their unique properties. Zhen et al. delivered CRISPR/Cas9 for the treatment of prostate cancer by using cationic liposomes containing poly(ethylene glycol)-grafted 1,2-distearoyl-sn-glycero-3-phosphatidylethanolamine.5 Further, core–shell nanoparticles consisting of PEG phospholipid-modified cationic lipid encapsulated Cas9/sgRNA plasmid targeting Polo-like kinase 1 (PLK-1) gene showed an in vitro transfection of 47.4% in A375 cells. Further, these nanoparticles showed significant downregulation of the PLK-1 protein and suppression of the tumor growth in melanoma tumor-bearing mice.6 In another study, Cas9-sgRNA RNPs directed against the dipeptidyl peptidase-4 gene (DPP-4) for modulating glucagon-like peptide 1 function were delivered using nano-liposomes that disrupted DPP-4 gene expression and declined the DPP-4 enzyme activity in type 2 diabetes mellitus (T2DM) db/db mice resulting in normalized blood glucose levels.7 In another study, cationic lipids were used to deliver sgRNA/Cas9 RNPs in MCF-7 cells to knockout the MDR1 gene, responsible for the efflux of DOX. The results showed an increase in drug uptake by four-fold relative to the untreated cells by decreasing the multidrug resistance mutation 1 (MDR1) gene-mediated resistance.8 Gene editing proteins need to be delivered into the nucleus for their activity; therefore, the delivery vector should possess the capability of functional delivery. In another study, Chen et al. prepared Cas9 RNPs complexed polymeric biodegradable nanocapsules having a hydrodynamic size of 25 nm for robust gene editing in vivo in murine retinal pigment epithelium (RPE) tissue and skeletal muscle after local administration.9 Polymeric nanocarriers also offer the freedom of chemical modification in the structure according to the requirement. For example, Lu et al. reported that micelles prepared using a polymer having an imidazole ring facilitate escape of cargo from the endosome.10 In another study, polyethylene glycol monomethyl ether (mPEG) conjugated chitosan was explored for the delivery of the CRISPR/Cas system. PEGylated chitosan of low and medium molecular weight was complexed with the pSpCas9-2A-GFP plasmid, and it was observed that low molecular weight PEGylated chitosan showed optimal transfection at a N/P ratio of 20, while PEGylated medium molecular weight chitosan showed optimal transfection at a N/P ratio of 5.11 Liu et al. reported poly(ethylene glycol)-b-poly-(lactic acid-co-glycolic acid) (PEG-PLGA)-based cationic lipid-assisted polymeric nanoparticles (CLANs) for delivering the CRISPR/Cas9 plasmid (pCas9) that efficiently disrupted the CML-related BCR-ABL fusion gene and increased the survival of a CML mouse model.12 Likewise, different hybrid polymers, as well as lipids, have been already screened for in vivo delivery of CRISPR/Cas components.2,13,14
We have previously reported that cholesterol and morpholine grafted amphiphilic cationic polymeric nanocarriers efficiently deliver miRNA-34a into the cancer cell by escaping the lysosomal acidic environment.15,16 These cationic polymers, being positively charged, could be electrostatically complexed with the negatively charged sgRNA/Cas9 RNPs to form stable nanoplexes. Herein, we report a robust gene-editing strategy based on the delivery of these anionic RNPs using cationic amphiphilic lipopolymeric nanoplexes. The double emulsion solvent evaporation method was used to prepare the cationic lipopolymeric nanoplexes followed by the formation of RNPs lipopolymeric nanoplexes through electrostatic complexation. Characterization of RNPs lipopolymeric nanoplexes was done using particle size, zeta potential, and gel retardation assay. Further, the release behavior of RNPs was determined using a heparin competition assay followed by the evaluation of the enzymatic activity and stability in the presence of fetal bovine serum. Fluorescence microscopy assays were performed to evaluate the transfection efficiency as well as the uptake mechanism of sgRNA/Cas9 RNPs lipopolymeric nanoplexes in HEK293T cells. Further, confocal microscopy was performed to examine intracellular trafficking and concomitant transport of RNPs to the nucleus using the CASFISH (CRISPR/Cas mediated in situ hybridization) experiment. Moreover, a fluorescence quenching-based mGFP gene editing assay was performed in mGFP-HEK293T cells to evaluate the gene-editing efficiency of SpCas9 RNPs lipopolymeric nanoplexes. T7 endonuclease assay revealed the quantitative gene-editing efficiency of the RNPs lipopolymeric nanoplexes and an intramuscular in vivo transfection assay in mice was used to verify the in vivo durability and performance of lipopolymeric nanoplexes.
Experimental section
Materials
OptiMEM™ reduced serum media, fetal bovine serum (FBS), Dulbecco's Modified Eagle's Medium (DMEM), Snakeskin dialysis tubing™ (3.5 kD), Micro BCA™ Protein Assay Kit, MEGAscript™ T7 Transcription Kit, Hoechst, CRISPRMax and DAPI (4′,6-diamidino-2-phenylindole) were obtained from ThermoFischer Scientific (Massachusetts, USA). T7 endonuclease I was purchased from Biolab (Delhi, India), while the genomic DNA purification kit was purchased from Promega (Delhi, India). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was purchased from Merck (Mumbai India). All the primers were purchased from Imperial Life Science (ILS, Delhi, India). N,N-Dimethyldipropylenetriamine (DP), benzyl bromide, tin(II) 2-ethylhexanoate, cholesterol, methoxy poly(ethylene glycol) (mPEG, 5000 Da), hydroxybenzotriazole (HOBt), Bis(hydroxymethyl) propionic acid, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC·HCl), 4-(2-aminoethyl)morpholine, HEPES buffer and heparin sodium salt from porcine intestinal mucosa were purchased from Sigma Aldrich (St. Louis, MO). The pET-dSpCas9-eGFP plasmid was obtained from Dr. Debojyoti (Senior Scientist, CSIR-IGIB, New Delhi, India) as a kind gift. The remaining solvents and chemicals were of analytical grade and were procured from local vendors.Synthesis of the cationic polymer, mPEG-b-(CB-{g-Cationic chain; g-Chol; g-Morph}) (8)
For the synthesis of the cationic polymer, a previously reported multistep reaction scheme was adopted with slight modifications, as shown in Fig. S1 (ESI†).15 Briefly, a cyclic monomer (1), 2-methyl-2-benzyloxycarbonylpropylene carbonate (MBC), was synthesized by mixing 2,2-bis (hydroxymethyl) propionic acid (BHMP) with potassium hydroxide (KOH) followed by the addition of benzyl bromide in dimethylformamide at 100 °C for 15 h. The product obtained, benzyl 2,2-bis (methylol) propionate, was purified and recrystallized using toluene and further reacted with triphosgene in dichloromethane and pyridine mixture to get the cyclic monomer, MBC (1). Furthermore, the cyclic monomer obtained was dried and used in monowave directed ring-opening polymerization (ROP) with mPEG (molecular weight: 5000 Da) (2) at 135 °C for 35 minutes in the presence of tin(II) 2-ethylhexanoate (10 mol% of mPEG) as a catalyst. The polymer, mPEG-b-p(CB) (3), obtained was purified using isopropanol (IPA) and diethyl ether (DEE). To obtain the polymer with free carboxyl (–COOH) groups i.e., mPEG-b-p(CB-{g-COOH}) (4), the benzylic moiety of the polymer mPEG-b-p(CB) (3) was reduced by dissolving it in a tetrahydrofuran and methanol (THF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH, 1
MeOH, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) mixture containing palladium on carbon (Pd/C) in the presence of hydrogen gas at 45 psi pressure for 6 h. After the reaction, the reaction mixture was centrifuged at 6500 rpm for 10 min and the supernatant was collected and dried under vacuum to obtain the sticky and transparent mPEG-b-p(CB-{g-COOH}) polymer (4). Different pendant groups, viz., dimethyldipropylenetriamine (5), 4-(2-aminoethyl) morpholine (6) and cholesterol (7), were grafted to the free carboxyl groups of mPEG-MCC (4) using EDC/HOBt coupling chemistry. Briefly, the mPEG-MCC polymer (4) was dissolved in DMF for 1 h in the presence of N,N-diisopropylethylamine (DIPEA) followed by the addition of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC·HCl) and hydroxybenzotriazole (HOBt) at 0 °C. Further, the reaction was carried out in a nitrogen atmosphere followed by the addition of N,N-dimethyldipropylenetriamine (5), 4-(2-aminoethyl) morpholine (6) and cholesterol (7) and the reaction was carried out for 48 h under a nitrogen atmosphere followed by dialysis (M.W cut-off 3.5 kDa) against purified water for 6 h wherein the medium was replaced after every 2 h. The contents of the dialysis bag were freeze-dried to obtain the mPEG-b-(CB-{g-cationic chain; g-Chol; g-Morph}) (8) polymer.15
1 v/v) mixture containing palladium on carbon (Pd/C) in the presence of hydrogen gas at 45 psi pressure for 6 h. After the reaction, the reaction mixture was centrifuged at 6500 rpm for 10 min and the supernatant was collected and dried under vacuum to obtain the sticky and transparent mPEG-b-p(CB-{g-COOH}) polymer (4). Different pendant groups, viz., dimethyldipropylenetriamine (5), 4-(2-aminoethyl) morpholine (6) and cholesterol (7), were grafted to the free carboxyl groups of mPEG-MCC (4) using EDC/HOBt coupling chemistry. Briefly, the mPEG-MCC polymer (4) was dissolved in DMF for 1 h in the presence of N,N-diisopropylethylamine (DIPEA) followed by the addition of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC·HCl) and hydroxybenzotriazole (HOBt) at 0 °C. Further, the reaction was carried out in a nitrogen atmosphere followed by the addition of N,N-dimethyldipropylenetriamine (5), 4-(2-aminoethyl) morpholine (6) and cholesterol (7) and the reaction was carried out for 48 h under a nitrogen atmosphere followed by dialysis (M.W cut-off 3.5 kDa) against purified water for 6 h wherein the medium was replaced after every 2 h. The contents of the dialysis bag were freeze-dried to obtain the mPEG-b-(CB-{g-cationic chain; g-Chol; g-Morph}) (8) polymer.15
All the intermediates, monomer/polymers (1, 3, and 4), as well as the final polymer (8), were characterized by 1H NMR using a Bruker (400 MHz) NMR spectrophotometer. The nitrogen content of polymer 8 was analyzed by using Elemental Analyzer (Vario EL cube by Elementar Analyser system), wherein the sample was weighed accurately and burned in the presence of oxygen. The combustion products of different elements, i.e. water, nitric oxide and carbon dioxide, were collected by different traps. The % of elements (C, H and N) in the polymer 8 was calculated from the masses of combustion products.
Preparation of ribonucleoprotein complexes (RNPs)
sgRNA required for the RNPs preparation was synthesized by the in vitro transcription (IVT) method using a dsDNA template. Briefly, targeted genes were screened for the protospacer adjacent motif (PAM) site and specific sgRNAs were designed using CHOPCHOP/CRISPOR software. Forward (FP) and reverse primers (RP) were designed, screened and compared for off-targets followed by synthesis of dsDNA using PCR. Thereupon, the in vitro transcription kit (MEGAscript™ T7 Transcription Kit, Thermo Scientific) was used to synthesize sgRNA as per the manufacturer's protocol.9Further, the catalytically active CRISPR-associated protein, Streptococcus Pyogenes Cas9 (SpCas9), was expressed in Escherichia coli Rosetta2 (DE3) (Novagen) using a pET-based expression vector as described previously by Jinek et al.17 Briefly, pET-28b-Cas9-His (Addgene #47237) or pET-dSpCas9-eGFP vector transformed E. coli Rosetta2 (DE3) cells were cultured up to an OD600 of 0.6 in lysogeny broth (LB) medium containing 50 mg L−1 kanamycin at 37 °C and protein expression was induced by the addition of 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG). The Cas9 expressing E. coli Rosetta2 (DE3) cells were harvested by centrifugation and the pellet was resuspended in lysis buffer (20 mM tris–Cl, pH 8.0, 500 mM NaCl, 5% glycerol, 1 mM DTT, 1X protease inhibitor cocktail (Roche), 100 μg mL−1 lysozyme) and lysed by probe sonication and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The pellet obtained was discarded, and the supernatant was affinity purified using Ni-NTA beads (Roche) followed by size-exclusion chromatography using a HiLoad Superdex 200 16/600 column (GE Healthcare). The purified spCas9 protein was analyzed using the BCA kit (Thermo Scientific) for its concentration4 and was stored at −80 °C in HEPES buffer (50 mM, pH 7.5) containing 100 μM Tris (2-carboxyethyl) phosphine, 10% glycerol and 300 mM NaCl.
000 rpm for 15 min. The pellet obtained was discarded, and the supernatant was affinity purified using Ni-NTA beads (Roche) followed by size-exclusion chromatography using a HiLoad Superdex 200 16/600 column (GE Healthcare). The purified spCas9 protein was analyzed using the BCA kit (Thermo Scientific) for its concentration4 and was stored at −80 °C in HEPES buffer (50 mM, pH 7.5) containing 100 μM Tris (2-carboxyethyl) phosphine, 10% glycerol and 300 mM NaCl.
To prepare the sgRNA/Cas9 RNP complex, the obtained sgRNA and SpCas9 protein were taken in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mol ratio. The mixture was incubated for 10 min at room temperature and characterized by running on a 0.8% agarose gel.9,18
1 mol ratio. The mixture was incubated for 10 min at room temperature and characterized by running on a 0.8% agarose gel.9,18
Preparation of ribonucleoprotein lipopolymeric nanoplexes
An emulsion-based method was used for the preparation of blank lipopolymeric nanoplexes followed by their complexation with RNPs. Briefly, cationic lipopolymer 8 (3 mg) was dissolved in 600 μL of dichloromethane (DCM) followed by the addition of 100 μL of nuclease-free HEPES buffer (10 mM; pH 6.7). The mixture was sonicated at 20% amplitude for 30 seconds to obtain the primary emulsion (W/O). The primary emulsion was added dropwise to 3 mL of HEPES buffer (10 mM; pH 6.7) followed by probe sonication at 20% amplitude for 3.5 min on an ice bath to get the secondary emulsion (W/O/W). The organic phase was removed under vacuum (Büchi® Rotavapor®) and centrifuged at 5000 rpm for 5 min to obtain the blank nanoplexes in the supernatant. CRISPR/Cas9 RNP lipopolymeric nanoplexes were prepared by mixing the blank nanoplexes with sgRNA/Cas9 RNPs in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio (w/w) and incubated at room temperature for 30 min to allow electrostatic complexation. The nanoplexes were characterized using particle size and zeta potential analyzer (Malvern Zeta Sizer, Nano ZS) and high resolution-transmission electron microscopy (HR-TEM; TECNAI 200 Kv TEM, FEI Electron Optics, Eindhoven, Netherlands). The complexation efficiency (%) of sgRNA/Cas9 RNPs with the blank nanoplexes was determined at 2 and 5% theoretical loading (%) of the RNPs using bicinchoninic acid (BCA) assay as reported earlier.9 Briefly, RNPs lipopolymeric nanoplexes at 2 and 5% w/w theoretical loadings were complexed at room temperature for 30 min followed by determination of zeta potential using a Zetasizer (Malvern, Nano ZS). The nanoplexes were then centrifuged at 18
10 ratio (w/w) and incubated at room temperature for 30 min to allow electrostatic complexation. The nanoplexes were characterized using particle size and zeta potential analyzer (Malvern Zeta Sizer, Nano ZS) and high resolution-transmission electron microscopy (HR-TEM; TECNAI 200 Kv TEM, FEI Electron Optics, Eindhoven, Netherlands). The complexation efficiency (%) of sgRNA/Cas9 RNPs with the blank nanoplexes was determined at 2 and 5% theoretical loading (%) of the RNPs using bicinchoninic acid (BCA) assay as reported earlier.9 Briefly, RNPs lipopolymeric nanoplexes at 2 and 5% w/w theoretical loadings were complexed at room temperature for 30 min followed by determination of zeta potential using a Zetasizer (Malvern, Nano ZS). The nanoplexes were then centrifuged at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 45 min to pelletize, and the sgRNA/Cas9 RNPs concentration in the supernatant was determined using the BCA kit as per the manufacturer's protocol9 (Pierce™ BCA Protein Assay Kit, Thermo Scientific™).
000 rpm for 45 min to pelletize, and the sgRNA/Cas9 RNPs concentration in the supernatant was determined using the BCA kit as per the manufacturer's protocol9 (Pierce™ BCA Protein Assay Kit, Thermo Scientific™).
Gel retardation assay
Gel retardation assay was performed to determine the blank lipopolymeric nanoplexes to sgRNA/Cas9 RNPs ratio (μg) required to form the RNPs lipopolymeric nanoplexes. Briefly, a fixed amount of sgRNA/Cas9 RNPs (μg) was complexed with different amounts (in μg) of blank lipopolymeric nanoplexes in RNase-free HEPES buffer (10 mM; pH 6.7) and incubated for 30 minutes at room temperature. A loading dye (5 μL) was added to the samples, which were subsequently subjected to agarose gel electrophoresis. Gel electrophoresis was performed on a 0.8% agarose gel for 30 min at 110 V and visualized under the Gel Doc system (Gel Doc™XR + Gel Documentation system).18Heparin competition assay
Heparin competition assay was performed to understand the release behavior of sgRNA/Cas9 RNPs from the lipopolymeric nanoplexes. Heparin is a competitor anion used to release complexed DNA/RNAs from cationic polymers and lipids.19 Herein, RNPs lipopolymeric nanoplexes were incubated with different concentrations of heparin (0.005 to 0.5 IU) at 37 °C for 30 min, followed by the addition of a loading dye (5 μL) and agarose gel electrophoresis as mentioned above. Herein, naked RNPs and RNPs lipopolymeric nanoplexes without heparin were taken as controls.RNPs lipopolymeric nanoplexes stability in fetal bovine serum (FBS)
The stability of RNPs lipopolymeric nanoplexes was determined in the presence of FBS.20 Briefly, freshly prepared RNPs lipopolymeric nanoplexes were incubated with 20% FBS at 37 °C for predetermined time periods (i.e., 0, 2, 4, 6, 8, 12, 24, and 48 h). After incubation, EDTA (10 μL) was added to inactivate the FBS. Further, heparin (0.1 IU) was added to the samples, followed by incubation at 37 °C for another 1 h to release the RNPs from the lipopolymeric nanoplexes. Thereafter, all the samples were loaded on a 0.8% agarose gel, and electrophoresis was performed at 110 V for 30 min. RNPs treated with FBS and RNPs lipopolymeric nanoplexes without FBS were used as the positive and negative control, respectively. The gel was visualized under the Gel Doc™XR + Gel Documentation system.Endonuclease enzymatic activity of RNPs after release from lipopolymeric nanoplexes
Attrition of endonuclease activity is one of the major concerns with CRISPR/Cas9 RNP delivery via the nanoformulation strategy. To examine this property, the developed RNPs lipopolymeric nanoplexes were treated with heparin to release RNPs and evaluated for their DNA cleavage activity.21 Briefly, the desired amount of RNPs lipopolymeric nanoplexes containing 100 ng of Cas9 protein was treated with 0.1 IU of heparin followed by incubation at 37 °C for 30 min to release RNPs. Thereupon, 200 ng of DNA substrate (pCAGs-RFP-P2A-eGFP) having a Cas9 cleavage site was incubated with the released RNPs at 37 °C for 1 h. Herein, the DNA substrate alone and the DNA substrate treated with freshly prepared RNPs were used as the negative and positive control, respectively. Samples were loaded on a 0.8% agarose gel and electrophoresis was performed at 110 V for 30 min. The gel was visualized under the Gel Doc™XR + Gel Documentation system.Hemocompatibility assay
The developed RNPs lipopolymeric nanoplexes were evaluated for their compatibility with the mice's blood. Briefly, 2 mL of blood was collected via retro-orbital plexus from swiss albino mice and centrifuged at 2000 rpm for 5 min. The supernatant was discarded, and erythrocytes were washed with PBS followed by resuspension in normal saline. Furthermore, 1 mL of erythrocytes was taken in the microcentrifuge tube and treated with blank lipopolymeric nanoplexes, RNPs (200 nM) and RNPs lipopolymeric nanoplexes (containing 200 nM RNPs) followed by incubation for 1 h at room temperature. The untreated and Triton-X treated erythrocytes served as negative and positive control, respectively. After treatment, the samples were centrifuged at 2000 rpm for 5 min, followed by visual inspection and microscopic evaluation for hemolysis and measurement of absorbance of the supernatant at 415 nm using a plate reader (BioTeK Epoch). Hemolysis (%) was calculated w.r.t. Triton X that showed 100% hemolysis.Cell culture-based assay
HEK293T and mGFP-HEK293T cells were provided by Dr Debojyoti Chakraborty, CSIR-Institute of Genomics and Integrated Biology (CSIR-IGIB), New Delhi, as a kind gift. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (100× penicillin/streptomycin) and kept at 37 °C in a humidified atmosphere containing 5% CO2.Transfection efficiency
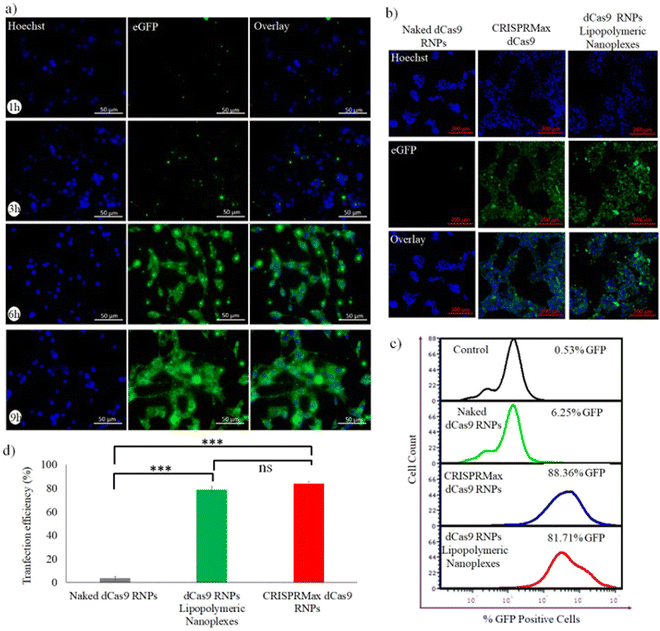
To evaluate the uptake of the RNPs in HEK293T cells, sgRNA/eGFP-dCas9 RNPs were used in the study. Briefly, HEK293T cells were seeded in 24-well cell culture plates (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) in DMEM (with 10% FBS) followed by incubation at 37 °C in 5% CO2 overnight to allow the cells to adhere. The next day, the cells were washed with PBS and the medium was replaced with optiMEM medium containing naked sgRNA/eGFP-dCas9 RNPs (200 nM eGFP-dCas9) and sgRNA/eGFP-dCas9 RNPs lipopolymeric nanoplexes (containing 200 nM eGFP-dCas9). In this study, CRISPRMax was used as a standard transfecting agent. After predetermined time periods (i.e., 1, 3, 6 and 9 h), the cells were washed with PBS and counterstained with the Hoechst dye (100 μg mL−1, for nuclear staining) followed by observation under a fluorescence microscope (Axio Vert A1, Zeiss).9 For quantitative measurement of transfection efficiency, the cells were analysed using flow cytometry (Beckman Coulter, USA) and the data were processed using CytExpert software (version 2.3).
000 cells per well) in DMEM (with 10% FBS) followed by incubation at 37 °C in 5% CO2 overnight to allow the cells to adhere. The next day, the cells were washed with PBS and the medium was replaced with optiMEM medium containing naked sgRNA/eGFP-dCas9 RNPs (200 nM eGFP-dCas9) and sgRNA/eGFP-dCas9 RNPs lipopolymeric nanoplexes (containing 200 nM eGFP-dCas9). In this study, CRISPRMax was used as a standard transfecting agent. After predetermined time periods (i.e., 1, 3, 6 and 9 h), the cells were washed with PBS and counterstained with the Hoechst dye (100 μg mL−1, for nuclear staining) followed by observation under a fluorescence microscope (Axio Vert A1, Zeiss).9 For quantitative measurement of transfection efficiency, the cells were analysed using flow cytometry (Beckman Coulter, USA) and the data were processed using CytExpert software (version 2.3).
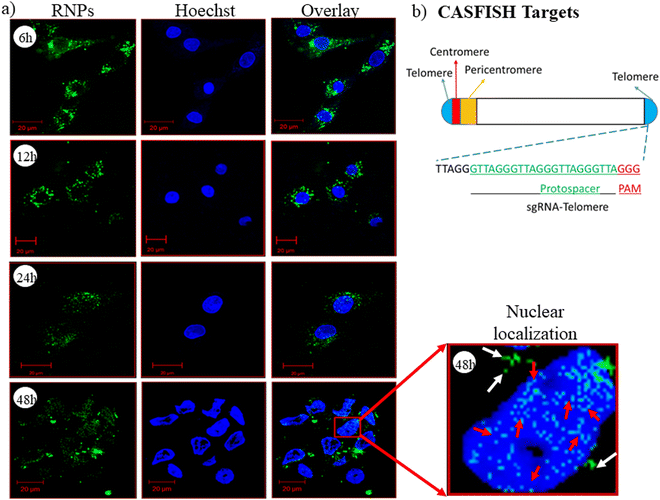
CASFISH based nuclear localization
The basic requirement for the nanocarrier delivering CRISPR/Cas9 RNPs is related to its efficiency of delivering RNPs to the nucleus in their native form. Therefore, to evaluate such efficiency of the developed lipopolymeric nanoplexes, a CASFISH experiment was performed.22 Briefly, RNPs (composed of sgRNA targeting the telomere region and eGFP-dCas9) were complexed with blank lipopolymeric nanoplexes and transfection was done in HEK293T cells followed by incubation for different predetermined time durations (6, 12, 24, and 48 h). Further, the cells were washed thrice with PBS and counterstained with Hoechst for nuclear staining and analyzed using CLSM (Carl Zeiss, Germany). The sequence of sgRNA designed for the telomere region is given in Table S2 (ESI†).Endocytosis uptake pathway
Briefly, HEK293T cells were seeded in 6-well cell culture plates (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) and incubated at 37 °C/5% CO2 overnight. After incubation, the cells were washed with PBS and the medium containing different endocytic inhibitors, including nystatin (27 μM), chlorpromazine (10 μM), methyl β-cyclodextrin (3 mM) and amiloride (1 mM), was added to the cells followed by incubation for 30 min at 37 °C/5% CO2. Further, the cells were washed with PBS and fresh optiMEM medium containing RNPs lipopolymeric nanoplexes (200 nM eGFP-Cas9) was added and the cells were incubated for 6 h followed by analysis using fluorescence microscopy. The cells were washed with PBS, stained with the Hoechst dye and observed under a fluorescence microscope (Axio Vert.A1, ZEISS).
000 cells per well) and incubated at 37 °C/5% CO2 overnight. After incubation, the cells were washed with PBS and the medium containing different endocytic inhibitors, including nystatin (27 μM), chlorpromazine (10 μM), methyl β-cyclodextrin (3 mM) and amiloride (1 mM), was added to the cells followed by incubation for 30 min at 37 °C/5% CO2. Further, the cells were washed with PBS and fresh optiMEM medium containing RNPs lipopolymeric nanoplexes (200 nM eGFP-Cas9) was added and the cells were incubated for 6 h followed by analysis using fluorescence microscopy. The cells were washed with PBS, stained with the Hoechst dye and observed under a fluorescence microscope (Axio Vert.A1, ZEISS).
In vitro cytotoxicity assay
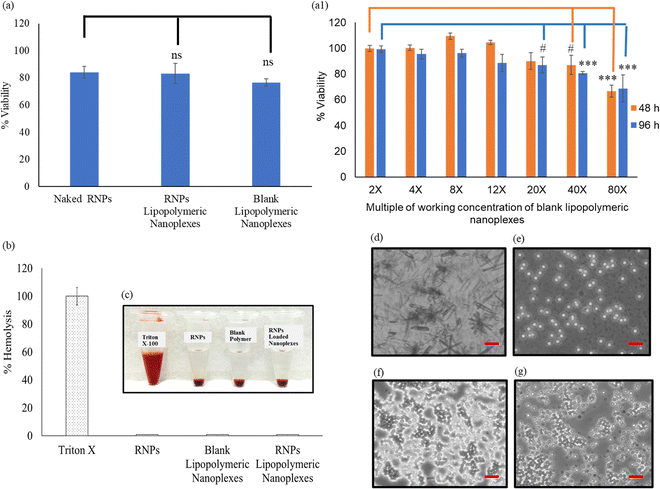
The cytotoxicity of sgRNA/Cas9 RNPs lipopolymeric nanoplexes was studied in HEK293T cells. Briefly, the cells were seeded in 96 well cell culture plates (5000 cells per well) and incubated at 37 °C/5% CO2 overnight. After incubation, the cells were treated with the naked sgRNA/dCas9 RNPs and sgRNA/dCas9 RNPs lipopolymeric nanoplexes equivalent to 200 nM per well of Cas9 followed by incubation for 48 h. The cells treated with blank nanoplexes and PBS were used as controls. After 48 h, cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Further, the RNP lipopolymeric nanoplexes were evaluated for their in vitro toxicity in HEK293T cells in a dose and time-dependent manner. For this, the cells were treated with RNPs lipopolymeric nanoplexes with different doses (ranging from 2× to 50× of the working dose) and the cell viability was determined after 48 h and 96 h. Briefly, the cells were washed with PBS and 200 μL of fresh DMEM (10% FBS) containing 0.5 mg mL−1 MTT was added to each well followed by incubation for 4 h at 37 °C/5% CO2. After incubation, the cells were washed with PBS followed by the dissolution of formazan crystals, formed as a result of MTT metabolism by mitochondria, in 200 μL of DMSO. Thereafter, the samples were observed at 570 nm and 630 nm under a microplate reader (BioTeK Epoch) and cell viability was calculated using the following equation:9,23,24mGFP gene disruption assay
The mGFP gene disruption by Cas9 RNPs was assayed using microscopy-based fluorescence analysis as reported earlier.3 The HEK293T cells with the mGFP gene (mGFP-HEK293T) integrated into the genome were seeded into 6-well cell culture plates (1 × 105 cells per well) and allowed to adhere for 24 h. The cells were then treated with RNPs lipopolymeric nanoplexes targeting the mGFP gene for 6 h; the medium was replaced with fresh medium and the cells were further cultured for 72 h. After 72 h, the cells were washed thrice with PBS and immediately observed with a fluorescence microscope (Axio Vert.A1, ZEISS) under the FITC channel with an excitation and emission wavelength of 488 nm and 525 nm, respectively. Herein, CRISPRMax was taken as the control. The sgRNA sequence used in this assay is shown in Table S2 (ESI†).T7 endonuclease assay
HEK293T cells were transfected with sgBPR-Cas9 RNPs lipopolymeric nanoplexes containing 200 nM of Cas9 protein. After 6 h, the medium was replaced with fresh medium followed by incubation for 48 h at 37 °C and 5% CO2. The cells were washed, harvested using trypsin/EDTA, centrifuged at 1200 rpm for 3 min and genomic DNA was isolated using the Wizard® Genomic DNA purification kit (Promega, India). The purified genomic DNA was amplified for the target site using PCR and 2 μg of purified PCR product was treated with 1 μL (10 U) of T7 Endo I and incubated at 37 °C for 15 min. The reaction was stopped by adding 2 μL of 0.25 M EDTA. The sample was loaded immediately on a 1.5% agarose gel for indel (%) analysis. Herein, genomic DNA without T7 endo 1 was taken as a negative control25 and CRISPRMax was taken as a standard transfecting agent. The primers used in this assay for sgRNA synthesis and PCR amplification are shown in Table S2 (ESI†). ImageJ software was used to process the gel image and the following formula was used to determine the indel efficiency:26| % Indel = 100 × (1 − (1 − fraction cleaved)1/2) |
Tracking of indels by decomposition (TIDE) assay
The real-time quantitative assessment of gene-editing was performed as reported earlier.27 Briefly, the HEK293T cells were transfected with RNPs lipopolymeric nanoplexes targeting the 5BPR gene followed by incubation for 48 h. Next, the cells were harvested, and the target gene site was PCR amplified, followed by PCR purification and Sanger sequencing. The sequenced PCR product was analyzed for indel efficiency using the TIDE Software (https://tide.nki.nl/).In vivo transfection
To evaluate the in vivo stability of the developed lipopolymeric nanoplexes, in vivo transfection assay was performed in mice after the approval of the protocol from Institutional Animal Ethics Commitee (IAEC; Protocol No-IAEC/RES/31/12) of BITS-Pilani, Pilani campus, Rajasthan (India). In brief, the mice were injected intra-muscularly with 50 μL of RNPs lipopolymeric nanoplexes containing 1 mg kg−1 of the eGFP-dCas9 protein and were kept under observation for 6 h. Thereafter, the mice were sacrificed and the muscle tissue from the site of injection was incised, frozen, and cryosectioned. The tissue was stained with DAPI and observed under a confocal microscope (CLSM, Carl Zeiss, Germany) to see the transfection.Statistical analysis
The statistical analyses were carried out using GraphPad Prism (USA). Student's t test and analysis of variance (ANOVA) followed by Tukey's test were used to determine the statistical differences between two or more groups, and the p value <0.05 was considered as statistically significant.Results
Synthesis of the cationic amphiphilic copolymer
The cationic amphiphilic copolymer, mPEG-b-p(CB-{g-Cationic chain; g-Chol; g-Morph}) (8), was synthesized using the multistep reaction scheme shown in Fig. S1 (ESI†), wherein the ring-opening polymerization of the cyclic monomer [MBC, (1)] was done with mPEG (2) as the chain initiator to obtain the mPEG-b-p(CB) (3) amphiphilic copolymer. The structure, composition, and molecular weight of mPEG-b-p(CB) were determined using 1H NMR (400 MHz, CDCl3) (shown in Fig. S2, ESI†) which revealed a molecular weight of 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240 Da with 70 units of MBC (Table S1, ESI†). The benzylic groups in the polycarbonate block of mPEG-b-p(CB) (3) were reduced using Pd/C catalyzed hydrogenation to obtain the mPEG-b-p(CB-{g-COOH}) (4) amphiphilic copolymer with free –COOH groups as confirmed using 1H NMR (400 MHz, DMSO-d6) with the disappearance of benzylic protons at δ 7.3 (e) (C6H5, m, 5H) (Fig. S2, ESI†). The mPEG-b-(CB-{g-COOH}) (4) amphiphilic copolymer was found to have a molecular weight of 16
240 Da with 70 units of MBC (Table S1, ESI†). The benzylic groups in the polycarbonate block of mPEG-b-p(CB) (3) were reduced using Pd/C catalyzed hydrogenation to obtain the mPEG-b-p(CB-{g-COOH}) (4) amphiphilic copolymer with free –COOH groups as confirmed using 1H NMR (400 MHz, DMSO-d6) with the disappearance of benzylic protons at δ 7.3 (e) (C6H5, m, 5H) (Fig. S2, ESI†). The mPEG-b-(CB-{g-COOH}) (4) amphiphilic copolymer was found to have a molecular weight of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 Da with 65 free –COOH groups (Table S1, ESI†) in the polycarbonate block. In the next step, N,N-dimethyl dipropylenetriamine (5), 4-(2-aminoethyl) morpholine (6) and cholesterol (7) were grafted on the free carboxyl groups of mPEG-b-(CB-{g-COOH}) (4) using EDC/HOBt coupling chemistry to yield the cationic amphiphilic copolymer mPEG-b-(CB-{g-Cationic chain; g-Chol; g-Morph}) (8). 1H NMR (400 MHz, DMSO-d6) (Fig. S2, ESI†) shows the protons corresponding to N,N-dimethyl dipropylenetriamine at δ 2.1 (e) (NH, s, 1H), δ 8.4 (d) (Amidic NH, s, 1H), 4-(2-aminoethyl) morpholine at δ 3.7 (i) (CH2, m, 4H), δ 8.4 (d) (Amidic NH, s, 1H) and cholesterol at δ 0.8–1.0 (f) (CH3, s, 6H), δ 4.25 (g) (CH, q, 1H), δ 5.14 (h) (CH, d, 1H). The molecular weight of the mPEG-b-(CB-{g-Cationic chain; g-Chol; g-Morph}) (8) amphiphilic copolymer was found to be 24,553 Da with 18 cationic chain units, 22 cholesterol units and 25 morpholine units (Table S1, ESI†). The elemental analysis showed 7.07%, 6.72% and 44.19% of nitrogen, hydrogen and carbon content, respectively, in the mPEG-b-(CB-{g-cation chain; g-Chol; g-Morph}) (8) amphiphilic copolymer (Table S1, ESI†).
650 Da with 65 free –COOH groups (Table S1, ESI†) in the polycarbonate block. In the next step, N,N-dimethyl dipropylenetriamine (5), 4-(2-aminoethyl) morpholine (6) and cholesterol (7) were grafted on the free carboxyl groups of mPEG-b-(CB-{g-COOH}) (4) using EDC/HOBt coupling chemistry to yield the cationic amphiphilic copolymer mPEG-b-(CB-{g-Cationic chain; g-Chol; g-Morph}) (8). 1H NMR (400 MHz, DMSO-d6) (Fig. S2, ESI†) shows the protons corresponding to N,N-dimethyl dipropylenetriamine at δ 2.1 (e) (NH, s, 1H), δ 8.4 (d) (Amidic NH, s, 1H), 4-(2-aminoethyl) morpholine at δ 3.7 (i) (CH2, m, 4H), δ 8.4 (d) (Amidic NH, s, 1H) and cholesterol at δ 0.8–1.0 (f) (CH3, s, 6H), δ 4.25 (g) (CH, q, 1H), δ 5.14 (h) (CH, d, 1H). The molecular weight of the mPEG-b-(CB-{g-Cationic chain; g-Chol; g-Morph}) (8) amphiphilic copolymer was found to be 24,553 Da with 18 cationic chain units, 22 cholesterol units and 25 morpholine units (Table S1, ESI†). The elemental analysis showed 7.07%, 6.72% and 44.19% of nitrogen, hydrogen and carbon content, respectively, in the mPEG-b-(CB-{g-cation chain; g-Chol; g-Morph}) (8) amphiphilic copolymer (Table S1, ESI†).
Preparation of ribonucleoprotein complexes (RNPs)
sgRNAs used in this study were synthesized via IVT reaction from dsDNA, obtained by the annealing of forward and reverse primers, as shown in Table S2 (ESI†). The synthesized sgRNAs were characterized using agarose gel electrophoresis confirming the size of 100 bp (Fig. 1(a)). The purified eGFP-SpCas9 protein was characterized using sodium dodecyl-sulfate polyacrylamide (SDS-PAGE) gel electrophoresis, where a dense band of approx. 200 kDa was observed (Fig. 1(b)). Thereafter, sgRNA and SpCas9 were complexed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Mol ratio to obtain RNPs. The retardation in the mobility of sgRNA after being complexed with the SpCas9 protein (as shown in Fig. 1(c)) is an indication of RNPs formation.
1 Mol ratio to obtain RNPs. The retardation in the mobility of sgRNA after being complexed with the SpCas9 protein (as shown in Fig. 1(c)) is an indication of RNPs formation.
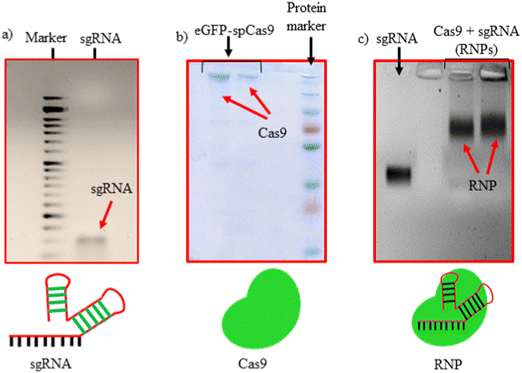 | ||
| Fig. 1 Characterization of RNPs formation. (a) sgRNA from IVT, (b) purified eGFP-Cas9 and (c) sgRNA/Cas9 RNPs complex. | ||
Preparation of lipopolymeric RNP nanoplexes
An emulsion-based method was used to prepare blank lipopolymeric nanoplexes from the synthesized cationic lipopolymer (mPEG-b-(CB-{g-cation chain; g-Chol; g-Morph})) in RNase free HEPES buffer (10 mM, pH 6.7). The blank lipopolymeric nanoplexes obtained were incubated with sgRNA/Cas9 RNPs for 30 minutes at room temperature to obtain RNPs lipopolymeric nanoplexes. Blank lipopolymeric nanoplexes and RNPs lipopolymeric nanoplexes showed a particle size of 73.75 ± 6.2 (PDI-0.240) and 117.3 ± 7.6 nm (PDI-0.399) nm, respectively, and a zeta potential of 16.2 ± 2.42 mV and 6.17 ± 1.04 mV, respectively (Fig. 2(a) and (b)). Furthermore, the encapsulation efficiency (%) of RNPs with the blank nanoplexes was determined at 5% theoretical loading (%) of the RNPs using bicinchoninic acid (BCA) assay. An encapsulation efficiency of >90% was observed at 5% theoretical loading indicating efficient complexation of the sgRNA/cas9 RNPs with the lipopolymeric nanoplexes. The morphology of the developed RNPs lipopolymeric nanoplexes was observed using transmission electron microscopy which indicated a spherical shape (Fig. 2(c)).Gel retardation assay
Gel retardation assay was performed to determine the blank lipopolymeric nanoplexes to RNPs ratio (w/w) required to form the RNPs lipopolymeric nanoplexes. A fixed amount of RNPs (in μg) was complexed with different amounts (in μg) of blank nanoplexes in RNase-free water and incubated for 30 minutes at room temperature. Gel electrophoresis results indicated a decrease in the mobility of RNPs when the amount of blank lipopolymeric nanoplexes was increased (Fig. 2(d)) with a complete complexation of the RNPs with 5× (w/w) of the blank lipopolymeric nanoplexes.Heparin competition assay and endonuclease activity of the released RNPs
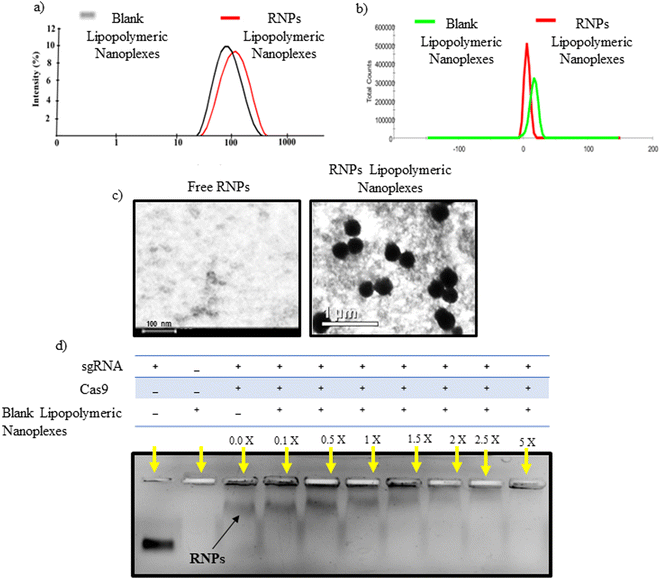
Heparin is a competitor anion used to release the complexed DNA/RNAs from electrostatic complexes with cationic polymers and lipids.28 We utilized a similar strategy to decomplex the RNPs lipopolymeric nanoplexes. It was observed that at 0.1 IU of heparin, RNPs got released from the lipopolymeric nanoplexes (Fig. 3(a)). Further, the released RNPs were able to cleave the DNA substrate indicating the retention of their endonuclease activity after complexation and decomplexation reactions with lipopolymeric nanoplexes (Fig. 3(b)).Stability of RNPs lipopolymeric nanoplexes in fetal bovine serum (FBS)
RNPs lipopolymeric nanoplexes were incubated with 20% FBS for predetermined time durations followed by treatment with heparin (0.1 IU) and visualization on the agarose gel. The data indicated that the RNPs were stable in FBS for 12 h when complexed with the lipopolymeric nanoplexes as compared to the naked sgRNA/Cas9 RNPs, which were degraded in FBS within 2 h (Fig. 3(c)).Cyto-compatibility study
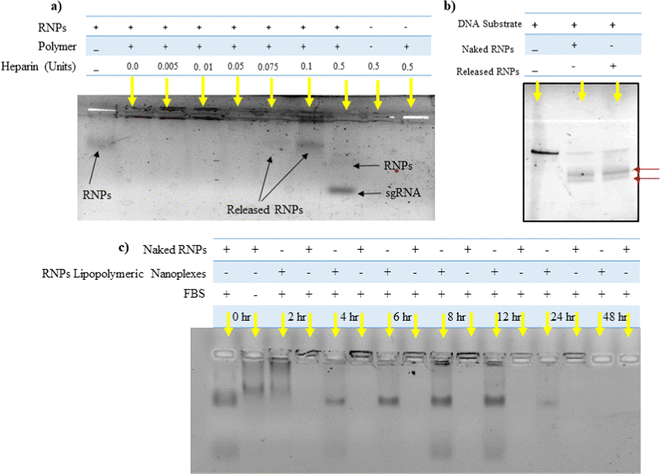
The toxicity profile of RNPs lipopolymeric nanoplexes was determined in HEK293T cells using MTT assay, wherein the cells were treated with naked RNPs (equivalent to 200 nM Cas9), blank lipopolymeric nanoplexes and RNPs lipopolymeric nanoplexes (equivalent to 200 nM Cas9) for 48 h. As per the observations, the lipopolymeric nanoplexes showed minimal toxicity at the working concentration in HEK293T cells (Fig. 4(a)). Additionally, the lipopolymeric nanoplexes showed non-significant toxicity up to 20× dose of the working concentration after 48 h (Fig. 4(a1)).Hemocompatibility study
Lipopolymeric nanoplexes were screened for their compatibility with mice erythrocytes; therefore, naked RNPs (equivalent to 200 nM Cas9), blank nanoplexes, and RNPs lipopolymeric nanoplexes (equivalent to 200 nM Cas9) were incubated with 1 mL of freshly collected mice erythrocytes for 1 h. Triton X was used as a positive control that causes 100% hemolysis. Less than 1% hemolysis was observed for naked RNPs, blank lipopolymeric nanoplexes, and RNPs lipopolymeric nanoplexes, indicating no significant hemolysis which was further corroborated with visual and microscopic evaluation (Fig. 4(b&c) and (d)–(g)).Transfection efficiency
For evaluating the capability of lipopolymeric nanoplexes to transfect HEK293T cells, the cells were incubated with RNP lipopolymeric nanoplexes containing eGFP-dCas9 (equivalent to 200 nM) for predetermined time periods (1, 3, 6 and 9 h) followed by observation under fluorescence microscope. The observations revealed that the RNPs, showing green fluorescence due to eGFP-dCas9, were delivered efficiently in a time-dependent manner by the lipopolymeric nanoplexes in HEK293T cells (Fig. 5(a)) and the maximum transfection was seen after 6 h (Fig. 5(b)). Herein, the blue colour indicates nuclear staining by the Hoechst dye and the green colour is from eGFP-dCas9. Further, as per the flow cytometry data the RNPs lipopolymeric nanoplexes showed 81.71% of transfection w.r.t. CRISPRMax, which showed 88.36% of transfection (Fig. 5(c and d)).CASFISH based nuclear localization
CRISPR/Cas9 RNPs should reach the nucleus with their intact endonuclease property, and the nanocarrier/vector delivering RNPs should not affect their native form. To evaluate this property of the developed nanoplexes, we have performed a CASFISH experiment, wherein RNPs containing eGFP-dCas9 and sgRNA (Telo-sgRNA) targeting the telomere region (TTAGGG repeats) were used. The CLSM images showed time-dependent nuclear localization of RNPs (Fig. 6(a)). After 6 h of treatment, the RNPs were seen in the cytoplasm (green color), while after 48 h, the RNP molecules were also observed in the nucleus. The results clearly indicated that the lipopolymeric nanoplexes delivered the RNPs into the cellular environment in an intact form, which were localized to the nucleus and bound to the telomere region efficiently (red arrow, Fig. 6(a)).Endocytic uptake pathway
To determine the uptake mechanism, HEK293T cells were treated with the endocytic uptake inhibitors, i.e., nystatin (caveolae inhibitor), chlorpromazine (clathrin pathway inhibitor), methyl β-cyclodextrin (lipid-mediated endocytic inhibitor) and amiloride (micropinocytic inhibitor) followed by treatment with RNPs lipopolymeric nanoplexes (equivalent to 200 nM eGFP-dCas9). The data showed that the RNPs lipopolymeric nanoplexes followed lipid-mediated as well as clathrin-based internalization since the uptake was inhibited majorly by chlorpromazine (clathrin pathway inhibitor) and methyl β-cyclodextrin (lipid-mediated endocytic inhibitor) (Fig. 7).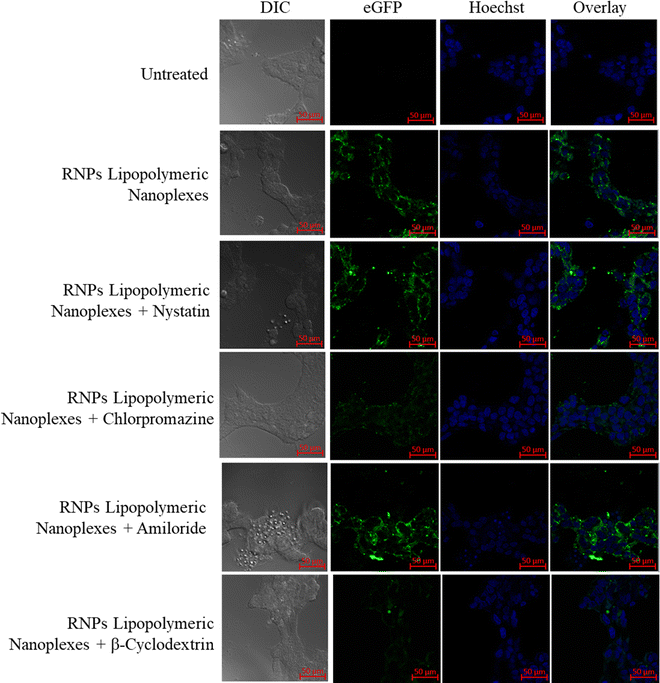 | ||
| Fig. 7 Uptake mechanism of eGFP-dCas9 RNPs lipopolymeric nanoplexes in the presence of endocytic uptake inhibitors. | ||
mGFP gene disruption assay
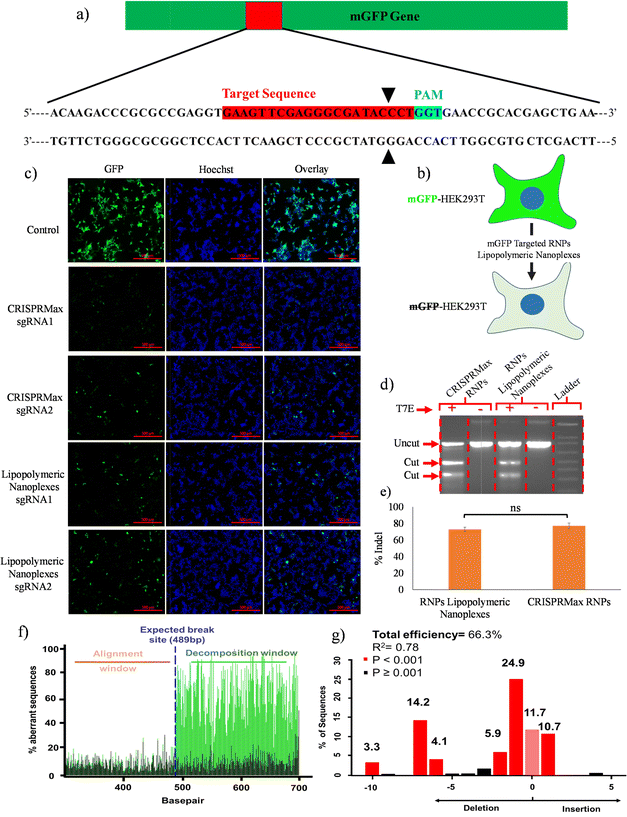
A qualitative gene disruption assay was performed using fluorescence microscopy in mGFP-HEK293T cells (Fig. 8b). The cells were treated with mGFP-sgRNA/Cas9 RNPs lipopolymeric nanoplexes for 72 h followed by an examination of mGFP protein expression. Fluorescence microscopic data showed a decrease in mGFP intensity as compared to the control group indicating the disruption of the mGFP gene in mGFP-HEK293T cells (Fig. 8(c)). Overall, these data suggested the efficient delivery of Cas9 RNPs by lipopolymeric nanoplexes.T7 endonuclease assay
The in vitro gene editing efficiency of Cas9 RNPs delivered via lipopolymeric nanoplexes was determined as previously described.25 In this experiment, RNPs lipopolymeric nanoplexes targeting the BPR2 gene were delivered in HEK293T cells for 48 h. T7 endonuclease digestion indicated ∼70% of indel efficiency in the HEK293T cells treated with RNPs lipopolymeric nanoplexes and CRISPRMax RNPs (Fig. 8(d) and (e)). Overall, these data suggested the efficient delivery of Cas9 RNPs by lipopolymeric nanoplexes.Tracking of indels by decomposition (TIDE) assay
The gene editing was further assessed using sequence trace decomposition or TIDE assay. Fig. 8(f) shows the aberrant nucleotide sequence signal of the test sample (green) w.r.t. that of the control sample (black) and Fig. 8(g) shows the indel spectrum and their frequencies determined by TIDE. Overall, the TIDE Software data showed 66.3% of indel cells treated with RNPs lipopolymeric nanoplexes.In vivo transfection
To evaluate the in vivo stability of the developed eGFP-dCas9 RNPs lipopolymeric nanoplexes, in vivo transfection was performed in mice after intra-muscular injection. As shown in Fig. 9, the naked eGFP-dCas9 RNPs disappear or undergo degradation in the muscle tissue. On the other hand, the eGFP-dCas9 RNPs lipopolymeric nanoplexes are transfected inside the muscle cells (green color) after 6 h of injection. These findings revealed the capability of the developed lipopolymeric nanoplexes to work in vivo efficiently.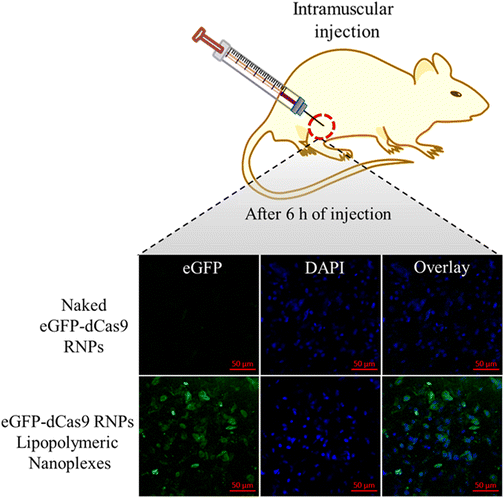 | ||
| Fig. 9 In vivo transfection of eGFP-dCas9 RNPs lipopolymeric nanoplexes after intra-muscular injection in mice. | ||
Discussion
Owing to its ample therapeutic potential for manipulating genetic material in prokaryotes and eukaryotes, the CRISPR/Cas9 system (especially RNPs) is now a center of attraction. Delivery technologies for the CRISPR/Cas9 gene-editing system often require viral vectors that pose safety concerns for therapeutic genome editing. Ample interest has been generated to utilize non-viral vectors with established safety profiles to deliver the CRISPR/Cas9 tools. Several approaches have been taken for the intracellular delivery of the CRISPR/Cas9 system, such as plasmids (having the Cas9 and sgRNA construct), Cas9-encoded mRNA, or direct Cas9 RNP delivery. Cas9 RNPs offer advantages due to their low insertion mutagenesis, short persistence of the Cas9 protein, low immunogenicity, and lesser off-target effect, thus making them attractive for gene editing application in vitro and in vivo.29 However, Cas9 RNPs has several limitations in terms of delivery owing to their large molecular weight (∼165 kD) and supra-negative charge. Attempts have been made to design nanocarriers consisting of cationic polymers and lipids to deliver Cas9 RNPs intracellularly.1 Recently, cationic polymers have become a feasible approach to deliver Cas9 RNPs via electrostatic interaction since this approach was previously explored for delivering nucleotides. PEI polymers have also been used alone or in combination with liposomes for Cas9 protein delivery in vivo to help induce endosomal escape.30 Sun et al. reported a polymeric core–shell nanoparticle with a PEI coating on a DNA nanoclew loaded with a Cas9–sgRNA complex.31 Furthermore, Chen et al. synthesized a glutathione cleavable polymeric nanocapsule using in situ polymerization. These nanocapsules were found to have a particle size of 25 nm with in vitro and in vivo gene editing efficiency without apparent toxicity.9 Previously we have shown that the blank lipopolymeric nanoplexes prepared using cationic amphiphilic copolymers effectively complexed the negatively charged miRNA-34a and delivered it efficiently in MCF-7 and 4T1 cells.15 The outcomes of the work consolidated the role of cationic amphiphilic copolymers in the delivery of negatively charged nucleic acids. Despite a Cas9 protein, guide RNA is an another major component of RNPs, which possesses a net negative charge and could provide opportunities to deliver these RNPs using cationic polymers by electrostatic complexation.In the current study, an emulsion solvent evaporation method was used to prepare the blank lipopolymeric nanoplexes with a positive surface charge that could complex negatively charged sgRNA/Cas9 RNPs. The copolymer mPEG-b-(CB-{g-Cationic chain; g-Chol; g-Morph}) (8) was successfully synthesized as indicated by 1H NMR spectroscopy that showed the appearance of amidic, cholesterol and morpholine proton peaks. The lipopolymer has a polycarbonate backbone, which possesses biodegradability and biocompatibility, and several pendant groups that impart multifunctionality to the polymer. The cationic chain provides positive charge, which facilitates its complexation with negatively charged RNPs. Additionally, the morpholine group is known for its endosomal escape properties as reported earlier.15 The blank lipopolymeric nanoplexes prepared using an emulsion-based method showed a zeta potential and particle size of +16.2 ± 2.42 mV and 73.75 ± 6.2 nm, respectively, which on complexation with the negatively charged RNPs showed an increase in nanoplex size and a decrease in zeta potential.15,21 Similar observations have been reported previously as well, wherein an increase in particle size and a decrease in zeta potential were considered as the primary indication of charge-based nano-complex formation.9 Further, gel retardation assay showed that 5X of polymer is sufficient to complex the RNPs (1×). The complexation of RNPs should be reversible and the same was confirmed by the heparin competition assay. Heparin, being a competitor anion, is well reported to release the RNA/DNA/plasmid complexes from cationic polymers or lipids.32 In the present study, heparin competition assay suggested that at a lower concentration of heparin, i.e. 0.005 to 0.075 IU, RNPs were observed to be in complex with the lipopolymeric nanoplexes, but as the concentration of heparin reached 0.5 IU, RNPs were released from nanoplexes (Fig. 3(a)). Such findings collectively consolidate the stability of RNPs lipopolymeric nanoplexes by strong electrostatic interactions. Being sensitive, the RNPs may lose their endonuclease activity on complexation,33 we evaluated the endonuclease activity of released RNPs21 which showed endonuclease activity similar to that of the freshly prepared RNPs, thereby indicating the potential of the developed lipopolymeric nanoplexes to deliver these RNPs in the intact form.
Cytotoxicity is one of the major concerns in using cationic carriers for gene delivery applications. We have previously reported the non-cytotoxic nature of the blank lipopolymeric nanoplexes in vitro in cancer cell lines and in vivo in swiss albino mice.34 To further test the RNPs lipopolymeric nanoplexes, MTT assay was performed in HEK293T cells wherein the developed RNPs lipopolymeric nanoplexes showed minimal toxicity up to a dose of 20× of the working concentration and the probable reason for the reduced cytotoxicity could be the reduction in the net charge on the nanoplexes after complexation with the RNPs. The blank nanoplexes are cationic and possess a net positive charge of 16.2 ± 2.42 mV, while upon complexation of RNPs with nanoplexes, the net charge gets reduced to 6.17 ± 1.04 mV. On the other hand, at a higher dose, the RNPs lipopolymeric nanoplexes were found to be toxic, and the possible reason could be the excessive cationic charge of the polymers. Previously, it has been reported that the cationic polymers or lipids show charge-dependent toxicity.35–37 Further, the lipopolymeric nanoplexes were non-toxic towards erythrocytes as indicated by the hemocompatibility assays (Fig. 4(b)–(g)). The nanocarrier systems possess a high transfection efficiency owing to their surface properties like zeta potential and targeted ligands, which modulate their internalization through endocytic transporters on the lipidic cellular membrane.38 Huang et al. reported that the nanoformulation having high zeta potential possesses more transfection efficiency as compared to formulations having zeta potential values that are near neutral.39 In a previous report, a cationic polymer with a zeta potential of ∼+39 mV successfully delivered miRNA-34a in MCF-7 and 4T1 cells. Similarly, in our current study, RNPs lipopolymeric nanoplexes were efficiently uptaken in HEK293T cells (Fig. 5), which could be attributed to the nano-size, cationic surface charge, and presence of the cholesterol group in the lipopolymer. We also examined the endocytic uptake pathway for RNPs lipopolymeric nanoplexes by fluorescence microscopy based analysis in the presence of endocytic inhibitors (Fig. 7). The receptor-mediated clathrin endocytosis is the most common cellular uptake pathway for plasma lipids and nutrients by covering almost 2% of the cellular surface. The uptake of nanoplexes was inhibited in the presence of a clathrin inhibitor (chlorpromazine), indicating the uptake through clathrin vesicles which could be attributed to the cholesterol group grafted in the hydrophobic chain of the polymer.40 Also, the presence of methyl β-cyclodextrin inhibits the clathrin-mediated endocytosis by extracting cholesterol from the plasma membrane and negatively effects the uptake of RNPs lipopolymeric nanoplexes, consolidating the mechanistic role of cholesterol in the uptake.40 Since the CRISPR/Cas9 system works by its targeted endonuclease activity on the cellular genome, it needs to be delivered effectively and functionally inside the nucleus of the cell, bypassing many of the intracellular barriers, viz., cell membrane, endosomal–lysosomal acidic vesicle and nuclear membrane. Therefore, cytosolic transfection of RNPs alone cannot prove the downstream efficiency of RNPs lipopolymeric nanoplexes. To understand the gene-editing functionality of RNPs after being delivered to the cytoplasm, we have performed CASFISH assay, wherein a sgRNA (telo-sgRNA) targeting the telomere TTAGGG repeat region (shown in Fig. 6(b)) has been complexed with eGFP-dCas9 to form RNPs and delivered using polymeric nanoplexes. Since there are several repeats of TTAGGG in the internal loci on the long and short arms of chromosomes, many RNP binding sites are available in the nucleus. Confocal microscopy images revealed the efficient binding of RNPs in the nucleus indicated by green punctuate fluorescence observed after 48 h of incubation with the RNPs lipopolymeric nanoplexes (Fig. 6(a), red arrow). The green fluorescence at/before 12 h observed in the cytoplasm could be attributed to the time required for the endo–lysosomal escape of the RNPs lipopolymeric nanoplexes and the decomplexation of RNPs from nanoplexes. The CASFISH experiment was explored previously to find out a specific gene location by Deng et al.22 These findings could prove the hypothesis that the polymeric nanoplexes functionally delivered the RNPs in vitro into the cellular environment. Further, these lipopolymeric nanoplexes could be used for gene editing applications by targeting various diseases. Previously, the loss of GFP expression after knockout of the GFP gene using CRISPR in a GFP expressing cell line was used as an in vitro model for visual evaluation and qualitative gene editing.3 Utilizing this, we further evaluated the in vitro gene editing of the RNPs lipopolymeric nanoplexes by transfecting the mGFP-HEK293T cells with the RNPs targeting the mGFP gene using lipopolymeric nanoplexes. Loss of mGFP expression after 72 h of incubation indicated that the gene had been successfully edited by the RNPs. Additionally, the T7 endonuclease assay showed a good gene editing efficiency (∼70% indel), which was further confirmed quantitatively using the TIDE assay,27 where 66.3% of indel was observed. This is also an indication of the functional delivery of RNPs into the cellular environment. After in vitro examination, we have also evaluated the in vivo stability of the developed lipopolymeric nanoplexes via in vivo transfection assay. As per our observation, the RNPs lipopolymeric nanoplexes behave similarly and were able to transfect the muscle cell after 6 h of intramuscular injection. The main advantage of the system is that even at a low w/w ratio, the developed lipopolymeric nanoplexes were effective in delivering Cas9 RNPs with minimal cytotoxicity. Additionally, the lipopolymer nanoplexes could be further modified in terms of active targeting and could be used to enhance tissue-specific RNPs delivery in vivo. Collectively, the nanoplexes gave a good overall in vitro and in vivo outcomes and could be evaluated further in terms of their in vivo gene editing potential in disease models.
Conclusion
The synthesized cationic lipopolymer has a free –COOH moiety, which makes it feasible for grafting with pendant functional groups, namely cationic chains, morpholine, and cholesterol. The presence of these pendant groups makes this polymer an efficient delivery vehicle due to the specific role of these groups at various stages of the intracellular fate of the cargo. Rapid complex formation with RNPs, good payload capacity, and efficient transfection are the key advantages of this system. Since the complexation occurs on the surface, it could be a disadvantage at the same time. Although CASFISH data showed nuclear localization, nonetheless further investigation will be required for gene editing efficiency in vivo in a disease model. The RNPs lipopolymeric nanoplexes were formed at an appropriate polymer/RNPs ratio, due to the cationic nature of the polymer, their toxicity may be a challenge during in vivo application. Our initial hemocompatibility data indicated the non-toxic nature of the complexes; however, a detailed in vivo toxicity assessment is warranted. Overall, the cationic lipopolymer could be a suitable non-viral nanocarrier for CRISPR/Cas9 RNPs for gene editing applications.Conflicts of interest
There are no conflicts of interest to declare about the data presented in this publication.Acknowledgements
The funding support from the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India, to DC, VS, and AM through project grant (BT/PR26897/NNT/28/1489/2017) is duly acknowledged. We sincerely acknowledge the Indian Council of Medical Research (ICMR) for financial support through Senior Research Fellowship (SRF) to DKS (File no. 45/66/2019-Nan/BMS) and JRF to MS (No. 3/1/3/JRF-2019/HRD-063(25389)). We gratefully acknowledge the All India Institute of Medical Sciences (AIIMS), New Delhi, for providing instrumental support for the Cryo Transmission Electron Microscopy (cryo-TEM) study. We would like to thank Dr Debojyoti Chakraborty and Dr Souvik Maiti, CSIR-IGIB (New Delhi, India), for extending protocols for CRISPR/Cas experiments and necessary facilities. We would also like to thank Mr. Suman Kumar from CIF, BITS-Pilani, Pilani campus, for confocal imaging. VS would also like to thank Hyderabad Eye Research Foundation (HERF), Hyderabad, India, for all its support.References
- D. K. Sahel, A. Mittal and D. Chitkara, J. Pharmacol. Exp. Ther., 2019, 370, 725–735 CrossRef CAS.
- A. Lohia, D. K. Sahel, M. Salman, V. Singh, I. Mariappan, A. Mittal and D. Chitkara, Asian J. Pharm. Sci., 2022 DOI:10.1016/j.ajps.2022.02.001.
- M. Wang, J. A. Zuris, F. Meng, H. Rees, S. Sun, P. Deng, Y. Han, X. Gao, D. Pouli and Q. Wu, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2868–2873 CrossRef CAS PubMed.
- J. A. Zuris, D. B. Thompson, Y. Shu, J. P. Guilinger, J. L. Bessen, J. H. Hu, M. L. Maeder, J. K. Joung, Z.-Y. Chen and D. R. Liu, Nat. Biotechnol., 2015, 33, 73–80 CrossRef CAS PubMed.
- S. Zhen, Y. Takahashi, S. Narita, Y.-C. Yang and X. Li, Oncotarget, 2017, 8, 9375 CrossRef PubMed.
- L. Zhang, P. Wang, Q. Feng, N. Wang, Z. Chen, Y. Huang, W. Zheng and X. Jiang, NPG Asia Mater., 2017, 9, e441 CrossRef CAS.
- E. Y. Cho, J.-Y. Ryu, H. A. R. Lee, S. H. Hong, H. S. Park, K. S. Hong, S.-G. Park, H. P. Kim and T.-J. Yoon, J. Nanobiotechnol., 2019, 17, 1–12 CrossRef PubMed.
- J. S. Ha, J. Byun and D.-R. Ahn, Sci. Rep., 2016, 6, 1–7 CrossRef PubMed.
- G. Chen, A. A. Abdeen, Y. Wang, P. K. Shahi, S. Robertson, R. Xie, M. Suzuki, B. R. Pattnaik, K. Saha and S. Gong, Nat. Nanotechnol., 2019, 14, 974–980 CrossRef CAS PubMed.
- D. Lu, Y. An, S. Feng, X. Li, A. Fan, Z. Wang and Y. Zhao, AAPS PharmSciTech, 2018, 19, 2610–2619 CrossRef CAS.
- H. Zhang, T. F. Bahamondez-Canas, Y. Zhang, J. Leal and H. D. Smyth, Mol. Pharmaceutics, 2018, 15, 4814–4826 CrossRef CAS.
- Y. Liu, G. Zhao, C.-F. Xu, Y.-L. Luo, Z.-D. Lu and J. Wang, Biomater. Sci., 2018, 6, 1592–1603 RSC.
- L. Farbiak, Q. Cheng, T. Wei, E. Álvarez-Benedicto, L. T. Johnson, S. Lee and D. J. Siegwart, Adv. Mater., 2021, 33, 2006619 CrossRef CAS PubMed.
- A. Noureddine, A. Maestas-Olguin, E. A. Saada, A. E. LaBauve, J. O. Agola, K. E. Baty, T. Howard, J. K. Sabo, C. R. S. Espinoza and J. A. Doudna, Acta Biomater., 2020, 114, 358–368 CrossRef CAS PubMed.
- S. Sharma, S. Mazumdar, K. S. Italiya, T. Date, R. I. Mahato, A. Mittal and D. Chitkara, Mol. Pharmaceutics, 2018, 15, 2391–2402 CrossRef CAS PubMed.
- S. Sharma, S. Pukale, D. K. Sahel, P. Singh, A. Mittal and D. Chitkara, Mater. Sci. Eng., C, 2021, 128, 112305 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- S. Acharya, A. Mishra, D. Paul, A. H. Ansari, M. Azhar, M. Kumar, R. Rauthan, N. Sharma, M. Aich and D. Sinha, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20959–20968 CrossRef CAS.
- B. Krieg, M. Hirsch, E. Scholz, L. Nuhn, I. Tabujew, H. Bauer, S. Decker, A. Khobta, M. Schmidt and W. Tremel, Pharm. Res., 2015, 32, 1957–1974 CrossRef CAS PubMed.
- T. Dowler, D. Bergeron, A.-L. Tedeschi, L. Paquet, N. Ferrari and M. J. Damha, Nucleic Acids Res., 2006, 34, 1669–1675 CrossRef CAS PubMed.
- K. Lee, M. Conboy, H. Park, F. Jiang, H. Kim, M. Dewitt, V. Mackley, K. Chang, A. Rao, C. Skinner, T. Shobha, M. Mehdipour, H. Liu, W. C. Huang, F. Lan, N. L. Bray, S. Li, J. E. Corn, K. Kataoka, J. A. Doudna, I. Conboy and N. Murthy, Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair, Nat. Biomed. Eng., 2017, 1, 889–901 CrossRef CAS PubMed.
- W. Deng, X. Shi, R. Tjian, T. Lionnet and R. H. Singer, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11870–11875 CrossRef CAS PubMed.
- Y. Li, J. Bolinger, Y. Yu, Z. Glass, N. Shi, L. Yang, M. Wang and Q. Xu, Biomater. Sci., 2019, 7, 596–606 RSC.
- S. Sharma, S. S. Pukale, D. K. Sahel, D. S. Agarwal, M. Dalela, S. Mohanty, R. Sakhuja, A. Mittal and D. Chitkara, AAPS PharmSciTech, 2020, 21, 1–21 CrossRef.
- Y. Rui, D. R. Wilson, J. Choi, M. Varanasi, K. Sanders, J. Karlsson, M. Lim and J. J. Green, Sci. Adv., 2019, 5, eaay3255 CrossRef CAS.
- F. Ran, P. D. Hsu, J. Wright, V. Agarwala, D. A. Scott and F. Zhang, Nat. Protoc., 2013, 8, 2281–2308 CrossRef CAS.
- E. K. Brinkman, T. Chen, M. Amendola and B. Van Steensel, Nucleic Acids Res., 2014, 42, e168 CrossRef PubMed.
- M. Bertschinger, G. Backliwal, A. Schertenleib, M. Jordan, D. L. Hacker and F. M. Wurm, J. Controlled Release, 2006, 116, 96–104 CrossRef CAS PubMed.
- D. Luther, Y. Lee, H. Nagaraj, F. Scaletti and V. Rotello, Expert Opin. Drug Delivery, 2018, 15, 905–913 CrossRef CAS PubMed.
- X. Wang, D. Niu, C. Hu and P. Li, Curr. Pharm. Des., 2015, 21, 6140–6156 CrossRef CAS PubMed.
- R. Mout, M. Ray, G. Yesilbag Tonga, Y.-W. Lee, T. Tay, K. Sasaki and V. M. Rotello, ACS Nano, 2017, 11, 2452–2458 CrossRef CAS PubMed.
- T. Okuda, T. Niidome and H. Aoyagi, J. Controlled Release, 2004, 98, 325–332 CrossRef CAS PubMed.
- J. Qiao, W. Sun, S. Lin, R. Jin, L. Ma and Y. Liu, Chem. Commun., 2019, 55, 4707–4710 RSC.
- S. Sharma, S. Pukale, D. K. Sahel, P. Singh, A. Mittal and D. Chitkara, Mater. Sci. Eng., C, 2021, 128, 112305, DOI:10.1016/j.msec.2021.112305.
- H. Lv, S. Zhang, B. Wang, S. Cui and J. Yan, J. Controlled Release, 2006, 114, 100–109 CrossRef CAS PubMed.
- S. Cui, Y. Wang, Y. Gong, X. Lin, Y. Zhao, D. Zhi, Q. Zhou and S. Zhang, Toxicol. Res., 2018, 7, 473–479 CrossRef CAS PubMed.
- X. Wei, B. Shao, Z. He, T. Ye, M. Luo, Y. Sang, X. Liang, W. Wang, S. Luo and S. Yang, Cell Res., 2015, 25, 237–253 CrossRef CAS PubMed.
- S. Mazumdar, D. Chitkara and A. Mittal, Acta Pharm. Sin. B, 2021, 11, 903–924 CrossRef CAS PubMed.
- X. Huang, W. Liao, G. Zhang, S. Kang and C. Y. Zhang, Int. J. Nanomed., 2017, 12, 2215 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: The synthesis scheme of the cationic lipopolymer along with its characterization, i.e.1H NMR data are incorporated. See DOI: https://doi.org/10.1039/d2tb00645f |
| This journal is © The Royal Society of Chemistry 2022 |