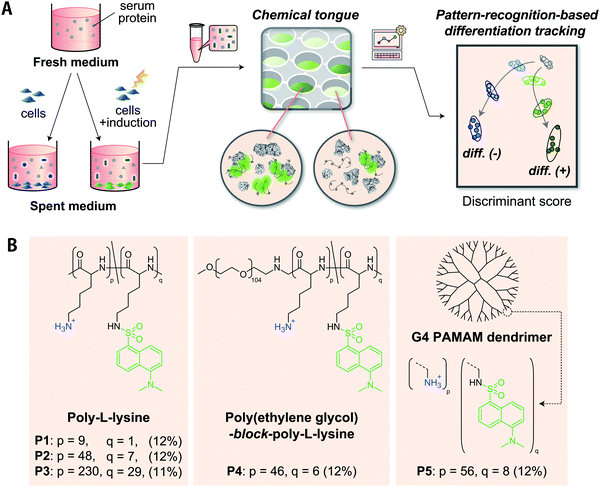
A polymer-based chemical tongue for the non-invasive monitoring of osteogenic stem-cell differentiation by pattern recognition of serum-supplemented spent media†
Shunsuke
Tomita
 *a,
Sayaka
Ishihara
a and
Ryoji
Kurita
*a,
Sayaka
Ishihara
a and
Ryoji
Kurita
 *ab
*ab
aHealth and Medical Research Institute, National Institute of Advanced Industrial Science and Technology, 1-1-1 Higashi, Tsukuba, Ibaraki 305-8566, Japan. E-mail: s.tomita@aist.go.jp; r.kurita@aist.go.jp
bFaculty of Pure and Applied Sciences, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8573, Japan
First published on 11th May 2022
Abstract
The development of non-invasive techniques to characterize cultured cells is invaluable not only to ensure the reproducibility of cell research, but also for quality assurance of industrial cell products for purposes such as regenerative medicine. Here, we present a polymer-based ‘chemical tongue’, i.e., a biosensing technique that mimics the human taste system, that is capable of non-invasively generating fluorescence response patterns that reflect the proteins secreted, and also partially consumed, by cultured cells, even from serum-supplemented media containing abundant interferants. Analysis of the spent media collected during cell culture using our chemical tongue, which consists of cationic polymers of different scaffolds appended with environmentally responsive dansyl fluorophores, led to the successful (i) identification of human-derived cell lines, (ii) monitoring of the differentiation process of stem cells, even at the stage when conventional staining was negative, and (iii) detection of cancer-cell contamination in stem cells. Since the characterization of cultured cells is usually performed via invasive methods that result in cell death, our chemical-tongue approach, which is of high practical utility, will offer a new means of addressing the growing demand for highly controlled cell production in the medical and environmental fields.
Introduction
Cell culture is now an indispensable technology in areas ranging from basic laboratory research to industrial applications. The global market for biopharmaceuticals is expected to reach approximately $400 billion by 2024, and nearly 70% of the recombinant proteins on the market are produced by mammalian cells.1 The scale of the stem-cell industry, with a particular focus on tissue engineering and regenerative medicine, has also grown remarkably over the past decade.2,3 In recent years, new stem cell applications have emerged, such as the production of cultivated meat to minimize climate impact4 and the development of models for understanding SARS-CoV-2 pathogenesis.5 Thus, the demand for cell-culture technologies for the robust and efficient supply of high-quality cell products has increased.In general, the states of cells in culture are characterized using biochemical assays,6 gene-expression profiling,7 and immunostaining.8 However, these techniques usually require that cultured cells are subjected to processes that damage the cells, such as enzymatic detachment from the substrate, fixation, lysis, and/or exposure to staining agents. Such treatments are problematic in that they cannot be used for continued monitoring of the cells because they must be performed at the endpoint, and further precludes their subsequent use for other purposes. The development of non-invasive alternative techniques would be invaluable not only to improve the quality of cell products, but also to facilitate routine cell maintenance and identity assurance in laboratories, cell banks, and manufacturing plants.
A key to the non-invasive characterization of cells is the detection of information presented externally by living cells. In this context, the so-called ‘secretome’, which is the entire set of proteins secreted into extracellular space, is an attractive target.9 Proteins secreted via various pathways indicate the intrinsic properties and condition of cultured cells, reflecting their physiological states and interplay with the environment. In-depth profiling of secretomes based on advanced proteomic techniques has found use in a wide variety of applications, including the discovery of cancer biomarkers10 and druggable targets11 as well as in alternatives to cell-based therapies.12,13
We have recently reported that chemical-tongue strategies that mimic the human taste system are useful for secretome recognition.14–16 The chemical-tongue strategy is a recently emerged analytical technique based primarily on nonspecific interactions, in contrast to conventional specific-recognition-based sensing using antibodies and enzymes.17 A typical chemical tongue consists of an array of environmentally responsive probes, which mimics taste receptor cells, and pattern-recognition techniques, which mimic information processing in the brain.18 The conversion of the multifaceted interactions between the array and the analytes into optical responses, combined with statistical analysis of the resulting multidimensional optical response patterns, has enabled the accurate identification or classification of various bioanalytes ranging from isolated proteins19–23 and cells20,24–27 to complex serum samples21,28–30 and fermented beverages.31–33 Using the recognition of complex secretome compositions by chemical tongues, we have successfully detected cell types,14,16 differentiation of adipose-derived mesenchymal stem cells (MSCs),14 fibroblast senescence,15 and drug responses in hepatoma cells16 in a non-invasive manner.
One of the most common issues in secretome analysis is the significant masking of target proteins by high-abundance serum proteins. The addition of serum (e.g., fetal bovine serum) containing nutrients, cell adhesion molecules, and growth factors is still often essential for stable and efficient cell culture.34 One means to avoid interfering of small amounts of secreted proteins is the use of a serum-free medium,11,13,35 which we have used in our previous studies.14–16 However, cells usually show optimal growth and viability when cultured in serum-supplemented medium. Serum starvation disturbs cell signaling and growth,36 thus affecting protein expression and secretory profiles, which can result in unintended experimental biases.35 Just as efforts have been made to address this issue in the proteomic analysis of secreted proteins,35,37 the development of chemical tongues to extract cell status information from serum-supplemented systems is a prerequisite for applications in more practical culture environments.
Here, we present chemical tongues that consist of polymeric probes that are capable of detecting compositional information on proteins secreted, and also partially consumed, by cultured cells despite being masked by a large amount of serum interferants (Fig. 1). Cationic polymers partially functionalized with environmentally responsive dansyl groups produced turn-on fluorescence signals when mixed with serum-supplemented spent media collected during cell culture. Information mining to overcome serum interferences was accomplished by producing fluorescence response patterns using various polymer scaffolds with amino groups (different repeat units, block-copolymerization, dendrimerization) and then selecting distinctive interactions from the responses by pattern-recognition techniques. The analysis of the spent media by a chemical tongue consisting of these polymer probes led to the successful (i) identification of human-derived cell lines, (ii) monitoring of the differentiation process of stem cells (even at the stage when conventional staining was negative), and (iii) detection of cancer-cell contamination in stem cells.
Experimental section
Materials and synthesis
A detailed list of the materials used in this study is given in the ESI.† Polymers modified with dansyl chloride (Dnc-polymers) were prepared according to literature procedures with slight modifications where necessary.38 Details of the synthesis and characterization of the Dnc-polymers are provided in the ESI.†Preparation of spent media
All cell lines were grown on a 10 cm diameter cell-culture dish (AGC Techno Glass Co.) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 vol% fetal bovine serum (FBS) and 1 vol% of a penicillin–streptomycin–neomycin (PSN) antibiotic mixture (DMEM + +) in a humidified 5% CO2 incubator at 37 °C. The cells cultured under the following conditions were washed twice with Dulbecco's phosphate-buffered saline (DPBS) (200 μL) and incubated with chemically defined CDCHO medium supplemented with 8 mM L-glutamine and 1.0 vol% FBS (300 μL) for 16–48 hours [for culture of the human lung adenocarcinoma epithelial cell line (A549; Table 1)] or 48 hours (others). Then, 250 μL of each cell-culture supernatant was collected and centrifuged at 3000 × g for 10 minutes. The supernatants were finally stored at −80 °C until use. The total protein concentrations in the spent media were quantified using the Bradford assay with Bradford reagent according to the manufacturer's instructions.| Cell line | Origin | Cell type |
|---|---|---|
| A549 | Lung | Cancerous cell |
| HepG2 | Liver | Cancerous cell |
| HeLa | Cervix | Cancerous cell |
| MG63 | Bone | Cancerous cell |
| MCF-7 | Breast | Cancerous cell |
| UE7T-13 | Bone marrow | Stem cell |
Culture of the A549 cells: A549 cells (0–6.0 × 104 cells per well) in DMEM + + were seeded on a 24-well tissue-culture plate (AGC Techno Glass Co.) and incubated for 48 hours in a humidified 5% CO2 incubator at 37 °C. Human-derived cell lines: Human-derived cell lines (6.0 × 104 cells per well) in DMEM + + were seeded on a 24-well tissue-culture plate and incubated as well. Mesenchymal stem cell line during osteogenic differentiation: Osteogenic differentiation of MSCs (UE7T-13)39 was carried out according to slightly modified literature procedures,14 using an osteogenic differentiation medium (0.1 μM dexamethasone, 10 mM β-glycerophosphate disodium salt hydrate, 0.2 mM (+)-sodium L-ascorbate, 0.005% DMSO in DMEM + +) prepared according to a previous report.40 UE7T-13 cells (6.0 × 104 cells per well) in DMEM + + were seeded on a 24-well tissue-culture plate and incubated as well. The cells were then incubated with the osteogenic differentiation medium or DMEM + + for 4, 8, and 12 days. The medium was changed every 72 hours. Mesenchymal stem cell lines contaminated with cancer-cell lines: UE7T-13 cells and human hepatoma carcinoma cells (HepG2) mixed in various proportions (total cell density: 6.0 × 104 cells per well) in DMEM + + were seeded on a 24-well tissue-culture plate and incubated as well.
Cell staining
Cell staining was carried out according to literature procedures.14 Prior to the staining of the UE7T-13 cells, the cultured cells were washed twice with DPBS (200 μL) and fixed with 4% paraformaldehyde (200 μL) for 15 minutes. The fixed cells were washed three times with distilled water (300 μL) and incubated in chilled methanol (300 μL) for 10 minutes. Finally, the cells were washed once with distilled water (200 μL) and soaked in 30 mM Alizarin Red S (the pH value was adjusted to ∼6.4 with KOH) (200 μL) for 15 minutes at 37 °C, washed with distilled water (200 μL), and then examined under an optical microscope (Primo Vert; Carl Zeiss) equipped with an AxioCam ERc5s camera (Carl Zeiss) and Axio Vision software (Carl Zeiss).Fluorescence responses of the polymers
Fluorescence measurements were performed using a SpectraMax GEMINI XPS (Molecular Devices). After incubation of the solutions prepared under the following conditions (30 °C, 10 min), the fluorescence spectrum (λex/λem = 360 nm/480–675 nm) or the fluorescence intensity (λex/λem = 360 nm/520 nm) was recorded at 30 °C.CDCHO medium: Solutions (200 μL) containing 2.0 μg mL−1 Dnc-polymers in 18–20 mM 3-morpholinopropanesulfonic acid (MOPS) buffer (pH = 7.4) or 18–20 mM 2-morpholinoethanesulfonic acid (MES) buffer (pH = 5.4) with 0–10.0 vol% CDCHO medium were prepared in each well of a 96-well NBSTM black microplate (Corning Inc.) using a PIPETMAX liquid handling system (Gilson Inc.). Human serum albumin (HSA): Solutions (200 μL) containing 2.0 μg mL−1 Dnc-polymers and HSA (0–32.0 μg mL−1) in 19 mM MOPS buffer (pH = 7.4) with 5 vol% CDCHO medium were prepared as well. FBS: Solutions (200 μL) containing 2.0 μg mL−1 Dnc-polymers and FBS (0–0.20 vol%) in 19 mM MOPS buffer (pH = 7.4) with 5.0 vol% CDCHO medium were prepared as well.
Chemical-tongue sensing
Proteins: Aliquots (187.5 μL) of solutions containing Dnc-polymers (2.1 μg mL−1) in 20 mM MOPS buffer (pH = 7.4) were deposited in the wells of a 96-well plate using a PIPETMAX system. After incubation (30 °C, 10 min), the fluorescence intensity was recorded using two different channels (Ch1: λex/λem = 320 nm/560 nm; Ch2: λex/λem = 360 nm/480 nm). Subsequently, aliquots (12.5 μL) of 320 μg mL−1 of the proteins in 4 mM MOPS buffer (pH = 7.4) with 80 vol% CDCHO medium were added to each well, and the fluorescence intensity was recorded after incubation (30 °C, 10 min). Spent medium: Aliquots (190.0 μL) of solutions containing Dnc-polymers (2.1 μg mL−1) in 20 mM MOPS buffer (pH = 7.4) or 20 mM MES buffer (pH = 5.4) were deposited in the wells of a 96-well plate using a PIPETMAX system. After incubation (30 °C, 10 min), the fluorescence intensity was recorded using two different channels (Ch1: λex/λem = 320 nm/560 nm; Ch2: λex/λem = 360 nm/480 nm). Subsequently, aliquots (10.0 μL) of the spent medium were added to each well, and the fluorescence intensity was recorded after incubation (30 °C, 10 min).These processes were performed six times for distinct samples to generate a training-data matrix. This training-data matrix was processed using linear discriminant analysis (LDA), hierarchical clustering analysis (HCA), and principal component analysis (PCA) in SYSTAT 13 (Systat Inc.). For holdout testing, four additional fluorescence patterns for each analyte were obtained and used as a test-data matrix. The test data were classified in groups generated by the remaining training matrix according to their shortest Mahalanobis distances. HCA dendrograms were created based on the Euclidean distances using the Ward method and a dataset standardized prior to analysis using the following equation: z = (x − μ)/σ, where z is the standardized score, x the raw score, μ the mean of the population, and σ the standard deviation of the population.
Results and discussion
Design of chemical tongue
Since approximately half of human proteins, including extracellular proteins, are negatively charged at pH ≈ 7,41 it would be reasonable to select cationic polymers that can interact electrostatically at multiple points as scaffold materials for recognizing proteins secreted from cultured cells. Their ability to bind tightly by multipoint contacts can selectively provide information on large constituents, such as proteins, from culture media that contain numerous molecules that differ with respect to size. We have previously demonstrated that cationic poly-L-lysine (PLL) modified with dansyl groups is useful for transducing the properties of proteins38 and cell surfaces42 into fluorescence patterns. Further advances in this strategy were assumed to be crucial for the successful application of chemical tongues to serum-supplemented cell culture media, which are more challenging and complex sample.As a strategy for creating an effective chemical tongue while keeping synthesis efforts as low as possible, we herein propose an approach for the diversification of scaffold materials (Fig. 1B). PLL with different repeat units (P1–P3), a block copolymer of polyethylene glycol (PEG) and PLL (P4), as well as a poly(amidoamine) (PAMAM) dendrimer (P5) were used; 11–12% of the amino groups were modified with environmentally responsive dansyl groups. The use of polymeric scaffolds with different numbers of repeat units (i.e., P1 to P3) was expected to aid in the selective extraction of information from proteins of different sizes because of their different multipoint interaction properties. The introduction of PEG segments into PLL (P4) should limit the size of the complex with proteins, as has been exemplified using polyion complexes for micellization in drug delivery systems.43,44 Dendrimer P5, unlike the other flexible linear polymers, is spherical and rigid, thereby limiting its interaction modes. These differences in material properties should help to provide unique interaction patterns even for challenging analytes.
Given that the composition of secreted proteins reflects properties and states of cultured cells,9 we hypothesized the following: The synthesized Dnc-polymers interact with various secretory and serum proteins contained in the medium. The observed fluorescence signals correspond to the sum of their interactions. The five Dnc-polymers exhibit different affinities for individual proteins in the medium depending on their chemical structure. The thus obtained fluorescence patterns contain information on the composition of the proteins that was altered by the cell culture, and pattern-recognition techniques can sense these differences, allowing for an accurate identification of properties and states of target cells.
Evaluation of the suitability of synthetic polymers for medium samples
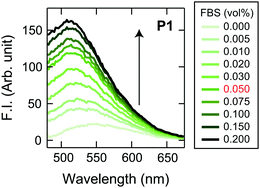
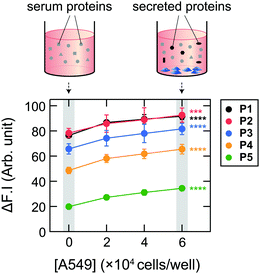
Initially, we examined the applicability of these Dnc-polymers in a chemical tongue. None of the Dnc-polymers responded to the cell culture medium itself (chemically defined CDCHO medium) even at 10.0 vol% (Fig. S1, ESI†), but their fluorescence intensity increased with increasing protein concentration in the presence of 5.0 vol% CDCHO medium (Fig. S2, ESI†). These responses should be due to a decrease in the polarity of the microenvironment around the dansyl groups on account of the binding of the Dnc-polymers to the proteins, as previously indicated.38,45 Furthermore, the responses of the Dnc-polymers varied depending on the type of protein, thereby allowing the generation of response patterns specific to the proteins (Fig. S3 and Dataset S1, ESI†).Since the protein properties can be successfully converted into turn-on fluorescence responses in serum-free medium, we attempted the application of the polymers to serum-supplemented medium. The addition of FBS to the Dnc-polymers caused an increase in fluorescence intensity, presumably due to nonspecific binding with the serum constituents (Fig. 2 and Fig. S4, ESI†). Based on these results, we determined the conditions for analyzing the serum-supplemented spent medium; spent CDCHO medium supplemented with 1.0 vol% FBS was added to aqueous solutions containing individual Dnc-polymers to give a final medium volume fraction of 5.0 vol%. Under these conditions, the Dnc-polymers exhibited some fluorescence enhancement, but when lung adenocarcinoma A549 cells (Table 1) were seeded and cultured in this medium for 48 h, the amount of fluorescence change increased significantly with increasing seeding density (Fig. 3). This increase is most likely due to the secretion of proteins from the cells into the medium. The changes in the fluorescence response depend on the culture time, suggesting gradual secretion into the medium (Fig. S5A, ESI†). We also confirmed that in the absence of cells, the responses changed little with increasing incubation time (Fig. S5B, ESI†).
Identification of cell lines based on an analysis of serum-supplemented spent media
After confirming that the information from the components secreted by the cultured cells was transduced to fluorescence signals even in the presence of serum interference, we attempted to recognize the individual cell lines using the chemical-tongue strategy. We selected six human-derived cell lines (Table 1) and compared them to the case when no cells were seeded. For the sensing procedure, each medium (5.0 vol%) was mixed with a specific Dnc-polymers (2.0 μg mL−1) in 19 mM MOPS buffer (pH = 7.4) in a 96-well plate. Fluorescence signals from each medium/polymer combination were recorded as the difference in fluorescence intensity before and after the addition of the analytes (I–I0) using two different channels [Ch1: λex/λem = 320 nm/560 nm; Ch2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 360 nm/480 nm], generating a dataset of 6 replicates × 5 polymers × 2 channels × 7 analytes. Because the change in polarity not only increases the fluorescence intensity of the dansyl groups but also results in a characteristic peak shift, we assumed that more diverse information could be produced by measuring the fluorescence signals at different wavelengths.
360 nm/480 nm], generating a dataset of 6 replicates × 5 polymers × 2 channels × 7 analytes. Because the change in polarity not only increases the fluorescence intensity of the dansyl groups but also results in a characteristic peak shift, we assumed that more diverse information could be produced by measuring the fluorescence signals at different wavelengths.
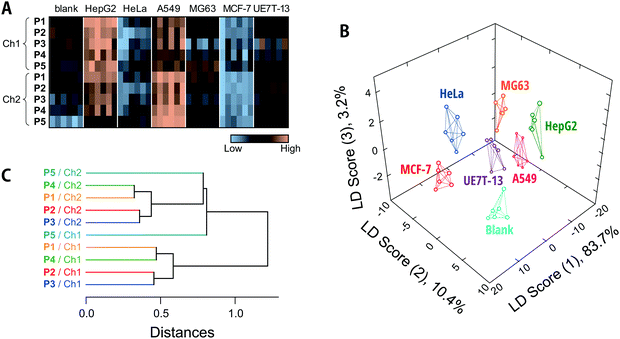
The fluorescence responses are summarized visually in the form of a heat map in Fig. 4A (for the raw data, see Dataset S2, ESI†). The response patterns of the spent media used to culture human cell lines were clearly different from those of fresh media and from each other. Overall, the responses to the media in contact with HepG2 and A549 were markedly higher than those for fresh media, whereas the Dnc-polymers exhibited similar- or even reduced-intensity responses to the other cells compared to their responses to fresh media. Since the total protein concentrations in the collected spent media were not reduced as a result of contact with the cells (Fig. S6, ESI†), the observed decreases in fluorescence response may be attributed to consumption of serum proteins by the cultured cells and/or secreted proteins that mask the interaction between the serum proteins and Dnc-polymers.
We subsequently subjected the response patterns to LDA to clarify the statistical differences among the generated response patterns. LDA is a representative pattern-recognition algorithm used to provide graphical output for describing the similarity of the data.17 In a three-dimensional LDA score plot (Fig. 4B), each point represents the fluorescence response pattern of a single analyte in the chemical tongue. The first three discriminant scores account for >97% of the total variance [score (1): 83.7%; score (2): 10.4%; score (3): 3.2%]. In other words, most of the information in the ten-dimensional data (5 polymers × 2 channels) is represented by the 3D plot. The plot provided seven well-separated clusters corresponding to the individual media. This result indicated statistically significant differences between the response patterns.
In order to quantitatively validate the applicability of the chemical tongue to discriminate the media, two different tests were performed, i.e., a leave-one-out cross-validation test and a holdout test,46 which afforded 98% and 96% classification accuracy, respectively (Dataset S2, ESI†). It should also be noted here that this discrimination did not derive solely from the amount of proteins secreted (Fig. S6, ESI†). Therefore, these results suggest that our Dnc-polymer-based chemical tongue has the ability to recognize intrinsic information from cell-specific protein secretions and potential consumption of serum proteins, even in the presence of large amounts of potentially interfering serum proteins. This ability is critical to the feasibility of chemical tongues for the non-invasive monitoring of cultured cells.
Then, unsupervised HCA17 was performed on the 10 elements (5 polymers × 2 channels) in an attempt to understand how the differences in the polymer scaffolds and detection channels affect pattern generation (Fig. 4C). In this dendrogram, the calculated distances between the sensing elements correspond to similarities in the fluorescence responses to the analyte media. Except in the case of P5/Ch1, clusters were observed for each detection channel (Ch1 and Ch2), indicating that the difference between the detection channels significantly contribute to the generation of unique responses. This trend is also replicated in the factor loadings calculated using PCA (Fig. S7, ESI†). The polymers P1/P4 and P2/P3 were clustered independently of the channel (Fig. 4C). The lower-molecular-weight P1 and P4 with a PEG segment are assumed to be less aggregative than P2 and P3, which may possibly be reflected in the responses. It should also be noted here that the relationship between these elements varies with the analyte and that differences in the scaffolds may contribute more (vide infra).
Monitoring the differentiation processes of stem cells
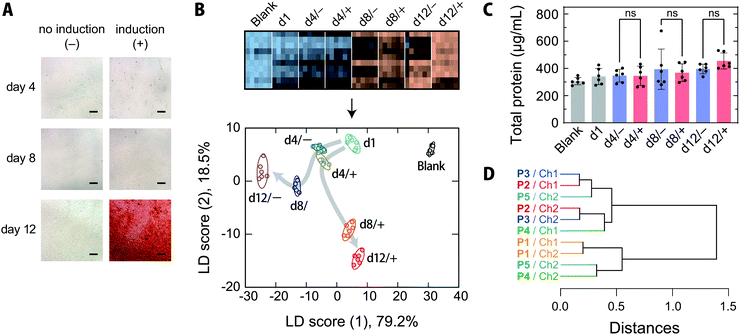
MSCs are capable of differentiating not only into mesodermal lineages, such as osteoblasts, chondrocytes, adipocytes, and myocytes, but also into ectodermal lineages including neurons in vitro.47,48 MSCs have been rapidly adopted in clinical trials, especially in the areas of regenerative medicine and cancer treatment, and are expected to address the unmet needs of modern medicine in the face of an aging society.47,49 Because the quality of MSCs is susceptible to differences in culture conditions, the processing of MSCs in medical applications, like other therapeutics, must be conducted in accordance with good manufacturing practices, and requires extensive regulatory efforts.47Thus, targeting the osteogenic differentiation process of human-bone-marrow-derived MSCs, which is closely related to the treatment of bone diseases,50 we examined whether our chemical tongue is capable of tracking the changes in cell state over time. When human-bone-marrow-derived MSCs (UE7T-13) were induced to differentiate into osteoblasts using a conventional method,14,40 calcium deposition, which is indicative of differentiation into osteoblasts, was first observed on day 12 (Fig. 5A). In contrast, UE7T-13 cells maintained in normal medium were not stained at all (Fig. 5A). Because the use of these stains involves destructive cell fixation, it would be desirable to be able to estimate the stage of induction non-invasively, ideally prior to endpoint staining. With this in mind, we carried out the recognition of spent media at different time points of differentiation induction (day 1, 4, 8, and 12) using our chemical tongue.
Chemical tongue analysis under the conditions used to distinguish cell types (Fig. 4) resulted in several clusters overlapping, with a leave-one-out cross-validation test and a holdout test giving accuracies of 83% and 84%, respectively, indicating poor discrimination performance (Fig. S8 and Dataset S3, ESI†). To further improve the accuracy, we focused on the pH of the aqueous solutions. We have previously reported that the response patterns of proteins can be effectively diversified under acidic conditions.38,51 These improvements are likely related to changes in the surface charge of many proteins upon decreasing the pH value. In the current case, some of the carboxyl groups in the proteins contained in the culture media are protonated at a weakly acidic pH, thereby weakening the electrostatic interactions between the cationic Dnc-polymers and the proteins, thus increasing the relative influence of other interactions such as hydrophobic interactions. Therefore, decreasing the pH value can be expected to allow capturing more multifaceted features of proteins. Accordingly, we then investigated the chemical-tongue analysis under weakly acidic conditions (pH = 5.4), where the polymers exhibit no responses to the CDCHO medium itself (Fig. S9, ESI†).
The resulting heatmap at pH = 5.4 shows a marked color change with increasing number of days elapsed (Fig. 5B and Fig. S8A, ESI†). Statistical analysis of the fluorescence response patterns using LDA revealed a clear separation of clusters with improved classification accuracies [96% and 94% for a leave-one-out cross-validation test and a holdout test, respectively; for the raw data, see Dataset S4, ESI†].
Furthermore, the LDA plot showed different cluster migration behavior for the normal-medium and differentiation-medium groups (Fig. 5B); the clusters moved primarily in the negative score (1) direction for the normal medium. In contrast, when osteogenic differentiation was induced, the migration was similar to that observed using the normal medium at day 4, but then changed direction and moved in the positive and negative score (1) and score (2) directions, respectively. There were no significant differences in the total protein concentrations in the spent media with or without induction at any of the measured time points (Fig. 5C), indicating that the reorientation of the cluster trajectory in the LDA plot was predominantly due to changes in the secretome compositions associated with induction.
Importantly, our chemical tongue captured information suggesting that the induction of differentiation was already underway at day 8, before it could be detected by the commonly used calcification-staining technique. The ability to distinguish changes in cell states, which are usually identified using multiple marker molecules, with a single technique is a distinct advantage of our chemical tongue, in addition to its non-invasive nature.
The HCA dendrogram (Fig. 5D) and a loading plot (Fig. S10, ESI†) of the chemical-tongue elements revealed that the contributions of the polymer scaffolds and detection channels are substantially different from those in the case of cell-type discrimination (Fig. 4C and Fig. S7, ESI†); the clusters of the scaffolds and channels were intermingled in the dendrogram, suggesting that the adoption of different polymer scaffolds plays a more important role in differentiation monitoring.
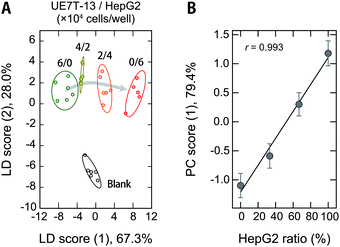
Detection of cell contamination
Cross-contamination by different cells remains a serious problem that reduces the reproducibility of cell studies,52,53 which is also an issue in the clinical utility of MSC-based products.54 For example, cross-contamination of sarcoma cell lines into MSCs has mistakenly been interpreted as MSC transformation, causing major confusion in the corresponding fields.55 As long as humans are involved in the experiments, these problems are inevitable, and it would thus be desirable to be able to detect contamination quickly and accurately even during routine culturing.Therefore, we used our polymer-based chemical tongue to detect the cross-contamination of cancer cells in MSCs. A LDA plot of serum-supplemented media exposed to model cell samples with different ratios of UE7T-13 and HepG2 cells provided separate clusters for each composition (Fig. 6A; for the raw data and a heat map, see Dataset S5 and Fig. S11, respectively, ESI†). A leave-one-out cross-validation test and a holdout test consistently afforded relatively high classification accuracies (97% and 90%, respectively) (Dataset S5, ESI†). In addition, a PCA analysis excluding the fresh medium data showed cluster migration proportional to the contamination ratio (r = 0.993; Fig. 6B), indicating the possibility of quantifying the degree of contamination via regression analysis based on machine-learning techniques. Further improvements in the system may allow not only the detection of contamination but also the simultaneous identification of the cell types.
Comparison with other techniques and practical applicability
Our polymer-based chemical tongue successfully discriminates cell types and states in a non-invasive fashion using serum-supplemented spent media. The advantages and challenges of this method are discussed below.Various attempts have been made to identify and monitor the states of cultured cells in a non-invasive manner. Representative examples include the use of nanostructured substrates for cell culture, such as spectroscopic detection of molecules released from cells collected on the underside of the substrate via nanosized-channels,56–58 as well as sensing of released molecules using nanoarrays based on electrochemistry59,60 and surface-enhanced Raman scattering (SERS).61 While these techniques represent powerful ways to effectively obtain information on cells non-invasively, they limit the environment in which cells can be cultured. As substrate-independent methods, multivariate analysis and machine learning combined with imaging techniques (e.g., hyperspectral imaging62 and Raman microspectroscopy)63–66 have been reported, as well as the detection of specific Raman peaks.67,68 These techniques enable label-free monitoring of stem-cell differentiation at the single-cell level. Our chemical-tongue-based method is expected to complement these state-of-the-art techniques because neither specialized cell-culture substrates nor advanced microscopic techniques are required.
In recent years, microscopic image-based cell-evaluation techniques combined with artificial intelligence are likely to be put into practical use, such as the selection of embryos fertilized in vitro.69,70 One advantage of these methods is that cell-morphology information, which is readily and rapidly obtained by optical microscopy, can be used. In contrast, chemical-information-based techniques, including our chemical tongues, are expected to be more accurate as they are based on direct information on biochemical reactions in cells, despite the disadvantage of taking time to obtain sufficient amounts of molecules.
In this study, chemical tongues were applied to evaluate spent CDCHO media supplemented with 1.0 vol% FBS. As Dnc-polymers respond selectively to large constituents such as proteins, this method can also be used in basal media other than CDCHO, e.g., DMEM and RPMI-1640. While 10 vol% FBS is used as a standard supplement to cell culture media in order to promote cell survival and proliferation, it has been reported that a low volume fraction of FBS is more effective for cell differentiation,71–73 including osteogenic differentiation,74 especially when essential factors are added. The addition of a small amount of transforming growth factor-β results in sufficient cell growth even at 1.0 vol% FBS.75 Our system should thus be useful in such cases. However, whether it can be used for general 10 vol% FBS is significant in terms of versatility. 1.0 vol% FBS contains about 300 μg mL−1 of proteins, and exposure to cells increases the concentration by up to 40% (Fig. 5C and Fig. S6, ESI†), thereby increasing the changes in the fluorescence signal only by ca. 20–80% (Fig. 3). Considering that the accuracy of the attempts shown in Fig. 4–6 are less than 100%, it can be estimated that this volume fraction of FBS is close to the upper limit of the current system. Nevertheless, we consider that the applicability of the chemical-tongue-based approach even to cases where the actual FBS concentration (1.0 vol%) was supplemented is a significant advance, at least from a proof-of-concept perspective.
As our chemical tongue is based on the pattern analysis of the composition of proteins secreted/consumed in the culture medium, this method has the potential to be expanded regardless of the stem-cell type [MSCs, embryonic stem cells (ESCs), or induced pluripotent stem cells (iPSCs)] or culture form (2D, 3D, or pellet). However, as with existing methods for the evaluation of cultured cells, differences in donor source, multistage differentiation processes, cell senescence, and culture environment are assumed to affect the identification accuracy. (i) The optimization of materials and analytical procedures to improve the sensitivity and accuracy of this method and (ii) the further study of the use of such improved chemical tongues to culture systems with higher volume fractions of FBS and to a variety of target cells and culture environments may possibly pave the way for practical applications to critical industrial challenges.
Conclusions
In summary, we have developed a chemical tongue, i.e., a biosensing technique that mimics the mammalian taste system. Our chemical tongue uses cationic polymers of different scaffolds appended with environmentally responsive dansyl fluorophores to generate fluorescence response patterns that reflect the secretory protein compositions unique to cultured cells, even in serum-supplemented media that contain abundant interferents. Our polymer-based chemical tongues are not only able to distinguish cell-line types, but also allow monitoring the differentiation process of mesenchymal stem cells, even at the stage when conventional staining is negative, and detection of cancer cell contamination of stem cells without damaging the cells. Importantly, this method does not involve a cumbersome sample-preparation process and can be performed quickly and easily with common laboratory equipment. Considering that the characterization of cultured cells is usually carried out via invasive methods that result in cell death, our chemical-tongue approach can be expected to provide a new means to aid the industrial production of cell products, whose demand is growing continuously in the medical and environmental fields.Author contributions
Conceptualization, funding acquisition, resources: S. T. and R. K.; formal analysis, methodology, visualization, writing – original draft: S. T.; data curation, investigation: S. T. and S. I.; writing – review & editing: all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially supported by JSPS KAKENHI grants JP17H04884 and JP20H02774 (S. T.), the AMED under grant number JP21wm0425004 (S. T.), and a special strategic grant from AIST (Japan) (R. K.)Notes and references
- R. O’Flaherty, A. Bergin, E. Flampouri, L. M. Mota, I. Obaidi, A. Quigley, Y. Xie and M. Butler, Biotechnol. Adv., 2020, 43, 107552 CrossRef.
- J. G. Hunsberger, T. Shupe and A. Atala, Stem Cells Transl. Med., 2018, 7, 564–568 CrossRef.
- Y. S. Kim, M. M. Smoak, A. J. Melchiorri and A. G. Mikos, Tissue Eng., Part A, 2019, 25, 1–8 CrossRef.
- C. Bomkamp, S. C. Skaalure, G. F. Fernando, T. Ben-Arye, E. W. Swartz and E. A. Specht, Adv. Sci., 2022, 9, 2102908 CrossRef CAS PubMed.
- C. R. Simoneau and M. Ott, Cell Stem Cell, 2020, 27, 859–868 CrossRef CAS.
- K. Präbst, H. Engelhardt, S. Ringgeler and H. Hübner, Methods Mol. Biol., 2017, 1601, 1–17 CrossRef.
- P. Baldi and G. W. Wesley Hatfield, DNA Microarrays and Gene Expression: From Experiments to Data Analysis and Modeling, Cambridge University Press, 2002 Search PubMed.
- A. Francisco-Cruz, E. R. Parra, M. T. Tetzlaff and I. I. Wistuba, Methods Mol. Biol., 2020, 2055, 467–495 CrossRef CAS.
- K. J. Brown, C. A. Formolo, H. Seol, R. L. Marathi, S. Duguez, E. An, D. Pillai, J. Nazarian, B. R. Rood and Y. Hathout, Expert Rev. Proteomics, 2012, 9, 337–345 CrossRef CAS PubMed.
- B. Rodrigues da Cunha, C. Domingos, A. C. Buzzo Stefanini, T. Henrique, G. Mussi Polachini, P. Castelo-Branco and E. H. Tajara, J. Cancer, 2019, 10, 4574–4587 CrossRef PubMed.
- M. Ding, H. Tegel, Å. Sivertsson, S. Hober, A. Snijder, M. Ormö, P.-E. Strömstedt, R. Davies and L. Holmberg Schiavone, SLAS Discovery, 2020, 25, 535–551 CrossRef.
- D. Z. Sun, B. Abelson, P. Babbar and M. S. Damaser, Nat. Rev. Urol., 2019, 16, 363–375 CrossRef PubMed.
- L. Daneshmandi, S. Shah, T. Jafari, M. Bhattacharjee, D. Momah, N. Saveh-Shemshaki, K. W.-H. Lo and C. T. Laurencin, Trends Biotechnol., 2020, 38, 1373–1384 CrossRef CAS.
- S. Tomita, M. Sakao, R. Kurita, O. Niwa and K. Yoshimoto, Chem. Sci., 2015, 6, 5831–5836 RSC.
- S. Tomita, H. Nomoto, T. Yoshitomi, K. Iijima, M. Hashizume and K. Yoshimoto, Anal. Chem., 2018, 90, 6348–6352 CrossRef CAS PubMed.
- H. Sugai, S. Tomita, S. Ishihara, K. Yoshioka and R. Kurita, Anal. Chem., 2020, 92, 14939–14946 CrossRef CAS.
- Z. Li, J. R. Askim and K. S. Suslick, Chem. Rev., 2019, 119, 231–292 CrossRef CAS PubMed.
- S. Tomita, Polym. J. DOI:10.1038/s41428-022-00636-w.
- M. De, S. Rana, H. Akpinar, O. R. Miranda, R. R. Arvizo, U. H.-F. Bunz and V. M. Rotello, Nat. Chem., 2009, 1, 461–465 CrossRef CAS.
- H. Pei, J. Li, M. Lv, J. Wang, J. Gao, J. Lu, Y. Li, Q. Huang, J. Hu and C. Fan, J. Am. Chem. Soc., 2012, 134, 13843–13849 CrossRef CAS.
- S. Xu, Y. Wu, X. Sun, Z. Wang and X. Luo, J. Mater. Chem. B, 2017, 5, 4207–4213 RSC.
- Z. Pode, R. Peri-Naor, J. M. Georgeson, T. Ilani, V. Kiss, T. Unger, B. Markus, H. M. Barr, L. Motiei and D. Margulies, Nat. Nanotechnol., 2017, 12, 1161–1168 CrossRef CAS PubMed.
- H. Wang, M. Chen, Y. Sun, L. Xu, F. Li and J. Han, Anal. Chem., 2022, 94, 2757–2763 CrossRef CAS PubMed.
- A. Bajaj, O. R. Miranda, I.-B. Kim, R. L. Phillips, D. J. Jerry, U. H.-F. Bunz and V. M. Rotello, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10912–10916 CrossRef CAS PubMed.
- S. Rana, S. G. Elci, R. Mout, A. K. Singla, M. Yazdani, M. Bender, A. Bajaj, K. Saha, U. H.-F. Bunz, F. R. Jirik and V. M. Rotello, J. Am. Chem. Soc., 2016, 138, 4522–4529 CrossRef CAS.
- Y. Tao, M. Li and D. T. Auguste, Biomaterials, 2017, 116, 21–33 CrossRef CAS PubMed.
- M. Jiang, A. N. Chattopadhyay, Y. Geng and V. M. Rotello, Chem. Commun., 2022, 58, 2890–2893 RSC.
- W. J. Peveler, R. F. Landis, M. Yazdani, J. W. Day, R. Modi, C. J. Carmalt, W. M. Rosenberg and V. M. Rotello, Adv. Mater., 2018, 30, 1800634 CrossRef PubMed.
- N. D.-B. Le, A. K. Singla, Y. Geng, J. Han, K. Seehafer, G. Prakash, D. F. Moyano, C. M. Downey, M. J. Monument, D. Itani, U. H.-F. Bunz, F. R. Jirik and V. M. Rotello, Chem. Commun., 2019, 55, 11458–11461 RSC.
- Y. Liu, J. Zhang, X. Zhao, W. Li, J. Wang, Y. Gao, Y. Cui, S. Xu and X. Luo, Chem. Commun., 2020, 56, 4074–4077 RSC.
- J. Han, M. Bender, K. Seehafer and U. H.-F. Bunz, Angew. Chem., Int. Ed., 2016, 55, 7689–7692 CrossRef CAS.
- J. Han, C. Ma, B. Wang, M. Bender, M. Bojanowski, M. Hergert, K. Seehafer, A. Herrmann and U. H.-F. Bunz, Chem, 2017, 2, 817–824 CAS.
- G. Macias, J. R. Sperling, W. J. Peveler, G. A. Burley, S. L. Neale and A. W. Clark, Nanoscale, 2019, 11, 15216–15223 RSC.
- Z. Yang and H.-R. Xiong, in Biomedical Tissue Culture, InTech, 2012 Search PubMed.
- J. Schira-Heinen, L. Grube, D. M. Waldera-Lupa, F. Baberg, M. Langini, O. Etemad-Parishanzadeh, G. Poschmann and K. Stühler, Biochim. Biophys. Acta, Proteins Proteomics, 2019, 1867, 140237 CrossRef CAS PubMed.
- J. J. Adler, D. E. Johnson, B. L. Heller, L. R. Bringman, W. P. Ranahan, M. D. Conwell, Y. Sun, A. Hudmon and C. D. Wells, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17368–17373 CrossRef CAS PubMed.
- K. Eichelbaum, M. Winter, M. Berriel Diaz, S. Herzig and J. Krijgsveld, Nat. Biotechnol., 2012, 30, 984–990 CrossRef CAS PubMed.
- S. Tomita, S. Ishihara and R. Kurita, ACS Appl. Mater. Interfaces, 2017, 9, 22970–22976 CrossRef CAS PubMed.
- T. Mori, T. Kiyono, H. Imabayashi, Y. Takeda, K. Tsuchiya, S. Miyoshi, H. Makino, K. Matsumoto, H. Saito, S. Ogawa, M. Sakamoto, J.-I. Hata and A. Umezawa, Mol. Cell. Biol., 2005, 25, 5183–5195 CrossRef CAS PubMed.
- S. Kern, H. Eichler, J. Stoeve, H. Klüter and K. Bieback, Stem Cells, 2006, 24, 1294–1301 CrossRef CAS.
- A. Kurotani, A. A. Tokmakov, K.-I. Sato, V. E. Stefanov, Y. Yamada and T. Sakurai, BMC Mol. Cell Biol., 2019, 20, 36 CrossRef PubMed.
- H. Sugai, S. Tomita, S. Ishihara and R. Kurita, ACS Sens., 2019, 4, 827–831 CrossRef CAS PubMed.
- Y. Lee, T. Ishii, H. Cabral, H. J. Kim, J.-H. Seo, N. Nishiyama, H. Oshima, K. Osada and K. Kataoka, Angew. Chem., Int. Ed., 2009, 48, 5309–5312 CrossRef CAS PubMed.
- A. Kim, Y. Miura, T. Ishii, O. F. Mutaf, N. Nishiyama, H. Cabral and K. Kataoka, Biomacromolecules, 2016, 17, 446–453 CrossRef CAS.
- Y.-D. Zhuang, P.-Y. Chiang, C.-W. Wang and K.-T. Tan, Angew. Chem., Int. Ed., 2013, 125, 8282–8286 CrossRef.
- M. Okada, H. Sugai, S. Tomita and R. Kurita, Sensors, 2020, 20, 5110 CrossRef CAS.
- A. Andrzejewska, B. Lukomska and M. Janowski, Stem Cells, 2019, 37, 855–864 CrossRef PubMed.
- S. Lou, Y. Duan, H. Nie, X. Cui, J. Du and Y. Yao, Biochimie, 2021, 185, 9–21 CrossRef CAS.
- Y. Guo, Y. Yu, S. Hu, Y. Chen and Z. Shen, Cell Death Dis., 2020, 11, 349 CrossRef.
- P. Kangari, T. Talaei-Khozani, I. Razeghian-Jahromi and M. Razmkhah, Stem Cell Res. Ther., 2020, 11, 492 CrossRef.
- S. Tomita, H. Sugai, M. Mimura, S. Ishihara, K. Shiraki and R. Kurita, ACS Appl. Mater. Interfaces, 2019, 11, 47428–47436 CrossRef CAS.
- A. Capes-Davis, G. Theodosopoulos, I. Atkin, H. G. Drexler, A. Kohara, R. A.-F. MacLeod, J. R. Masters, Y. Nakamura, Y. A. Reid, R. R. Reddel and R. I. Freshney, Int. J. Cancer, 2010, 127, 1–8 CrossRef CAS.
- L. P. Freedman, M. C. Gibson, S. P. Ethier, H. R. Soule, R. M. Neve and Y. A. Reid, Nat. Methods, 2015, 12, 493–497 CrossRef CAS PubMed.
- J. Q. Yin, J. Zhu and J. A. Ankrum, Nat. Biomed. Eng., 2019, 3, 90–104 CrossRef CAS.
- A. Torsvik, G. V. Røsland, A. Svendsen, A. Molven, H. Immervoll, E. McCormack, P. E. Lønning, M. Primon, E. Sobala, J.-C. Tonn, R. Goldbrunner, C. Schichor, J. Mysliwietz, T. T. Lah, H. Motaln, S. Knappskog and R. Bjerkvig, Cancer Res., 2010, 70, 6393–6396 CrossRef CAS PubMed.
- Y. Cao, M. Hjort, H. Chen, F. Birey, S. A. Leal-Ortiz, C. M. Han, J. G. Santiago, S. P. Paşca, J. C. Wu and N. A. Melosh, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E1866–E1874 CAS.
- R. Wen, A.-H. Zhang, D. Liu, J. Feng, J. Yang, D. Xia, J. Wang, C. Li, T. Zhang, N. Hu, T. Hang, G. He and X. Xie, ACS Appl. Mater. Interfaces, 2019, 11, 43936–43948 CrossRef CAS.
- P. Mukherjee, E. J. Berns, C. A. Patino, E. Hakim Moully, L. Chang, S. S.-P. Nathamgari, J. A. Kessler, M. Mrksich and H. D. Espinosa, Small, 2020, 16, 2000584 CrossRef CAS PubMed.
- J.-H. Lee, H. K. Choi, L. Yang, S.-T. D. Chueng, J.-W. Choi and K.-B. Lee, Adv. Mater., 2018, 30, 1802762 CrossRef PubMed.
- J.-H. Lee, J. Luo, H. K. Choi, S.-T. D. Chueng, K.-B. Lee and J.-W. Choi, Nanoscale, 2020, 12, 9306–9326 RSC.
- J.-H. Choi, T.-H. Kim, W. A. El-Said, J.-H. Lee, L. Yang, B. Conley, J.-W. Choi and K.-B. Lee, Nano Lett., 2020, 20, 7670–7679 CrossRef CAS.
- N. Mehta, S. Shaik, A. Prasad, A. Chaichi, S. P. Sahu, Q. Liu, S. M.-A. Hasan, E. Sheikh, F. Donnarumma, K. K. Murray, X. Fu, R. Devireddy and M. R. Gartia, Adv. Funct. Mater., 2021, 31, 2103955 CrossRef CAS PubMed.
- E. Brauchle, A. Knopf, H. Bauer, N. Shen, S. Linder, M. G. Monaghan, K. Ellwanger, S. L. Layland, S. Y. Brucker, A. Nsair and K. Schenke-Layland, Stem Cell Rep., 2016, 6, 188–199 CrossRef PubMed.
- S. Rangan, H. G. Schulze, M. Z. Vardaki, M. W. Blades, J. M. Piret and R. F.-B. Turner, Analyst, 2020, 145, 2070–2105 RSC.
- C.-C. Hsu, J. Xu, B. Brinkhof, H. Wang, Z. Cui, W. E. Huang and H. Ye, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18412–18423 CrossRef CAS PubMed.
- J. Geng, W. Zhang, C. Chen, H. Zhang, A. Zhou and Y. Huang, Anal. Chem., 2021, 93, 10453–10461 CrossRef CAS PubMed.
- I. R. Suhito, Y. Han, J. Min, H. Son and T.-H. Kim, Biomaterials, 2018, 154, 223–233 CrossRef CAS PubMed.
- I. R. Suhito, Y. Han, Y.-S. Ryu, H. Son and T.-H. Kim, Biosens. Bioelectron., 2021, 178, 113018 CrossRef CAS PubMed.
- N. Zaninovic and Z. Rosenwaks, Fertil. Steril., 2020, 114, 914–920 CrossRef CAS PubMed.
- M. VerMilyea, J. M.-M. Hall, S. M. Diakiw, A. Johnston, T. Nguyen, D. Perugini, A. Miller, A. Picou, A. P. Murphy and M. Perugini, Hum. Reprod., 2020, 35, 770–784 CrossRef CAS PubMed.
- B. Langley, M. Thomas, A. Bishop, M. Sharma, S. Gilmour and R. Kambadur, J. Biol. Chem., 2002, 277, 49831–49840 CrossRef CAS.
- K. A. D’Amour, A. D. Agulnick, S. Eliazer, O. G. Kelly, E. Kroon and E. E. Baetge, Nat. Biotechnol., 2005, 23, 1534–1541 CrossRef PubMed.
- Y. Hu, J. Ji, J. Xia, P. Zhao, X. Fan, Z. Wang, X. Zhou, M. Luo and P. Gu, Neurosci. Lett., 2013, 534, 90–95 CrossRef CAS PubMed.
- E. Arslan, M. O. Guler and A. B. Tekinay, Biomacromolecules, 2016, 17, 1280–1291 CrossRef CAS PubMed.
- N. J. Gunja, R. K. Uthamanthil and K. A. Athanasiou, Biomaterials, 2009, 30, 565–573 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and synthesis, responses of polymers to various analytes, identification of proteins, total protein concentrations in spent media, loading plots, optical pattern recognition for differentiation induction at pH = 7.4. See DOI: https://doi.org/10.1039/d2tb00606e |
| This journal is © The Royal Society of Chemistry 2022 |