Specific recognition of a target protein, cytochrome c, using molecularly imprinted hydrogels†
Chenchen
Liu
ab,
Takuya
Kubo
 *a and
Koji
Otsuka
*a and
Koji
Otsuka
 a
a
aDepartment of Material Chemistry, Graduate School of Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto, 615-8510, Japan. E-mail: kubo.takuya.6c@kyoto-u.ac.jp
bDepartment of Chemistry, Faculty of Science, Kyushu University, 744 Motooka, Nishi-ku, Fukuoka 819-0395, Japan
First published on 1st June 2022
Abstract
Protein imprinted hydrogel, which is one form of protein imprinted molecularly imprinted polymer (MIP), is an important material for enzyme-linked immunosorbent assay, drug delivery materials, sensors, separation materials, etc. To obtain a high protein recognition performance, it is essential to optimize the involved compositions. This work studies a copoly(poly(ethylene glycol) diacrylate/poly(ethylene glycol) acrylate), in short copoly(PEGDA/PEGA), based MIP hydrogel targeting cytochrome c recognition. The presented MIP hydrogel employs water-soluble PEGDA as the crosslinker, PEGA as the side chain, and sodium allylsulfonate as the functional monomer. The fabricated MIP hydrogels and non-imprinted polymer (NIP) hydrogels were treated as adsorbents for protein adsorption. Efforts were made targeting an optimized recognition performance. Factors including the template to functional monomer ratio, crosslinker length, crosslinker ratio of PEGDA/PEGA, ionic strength in the adsorption test, and presence of acidic modifier in the adsorption test were investigated. The results showed that a higher template to functional monomer ratio, a shorter crosslinker, and additional NaCl (20 mM) in the adsorption solvent provided a higher imprinting factor. A lower crosslinker ratio of no less than 6/4 offered a faster template removal; at the same time, the imprinting factor remained at a quite high level. Highly specific recognition of cytochrome c was realized with the presence of an optimized amount of HCl (10 mM) as an acidic modifier.
1. Introduction
Molecularly imprinting technology (MIT) is a facile, economic, and robust approach to the construction of molecular affinity structures for a specified molecule.1,2 Generally, the MIT involves a copolymerization of several types of monomers and functional monomers under the presence of template molecules. Owing to the complementary interactions between the monomers and template, the synthetic macromolecule bears the structural information of the template molecule, including the shape, size, and surface distribution pattern of functional groups. After removing the template, the resulting molecularly imprinted polymer (MIP) can show a specific affinity to the target molecule, which is evidenced by recognizable adsorption,3 structural reformation,4,5 or electrochemical characters.6–8 MIT is recognized as a powerful technology for being able to offer artificial affinity MIPs theoretically for any molecule – even one that doesn’t find a match in nature.9 The application of MIPs has evolved as sensors,10–12 drug delivery materials,13–15 sorbents,16–18 separation materials,19,20etc., for a variety of molecules.In the field of protein recognition, MIT has been realized as a promising tool because of its potential to overcome the issues from natural-source antibodies or receptors, which are scarce, unstable, or difficult to extract. However, the fabrication of protein imprinted MIPs is tricky due to the large size, structural complexity, and conformational instability of proteins.21 One difficulty is to construct rigid, large cavities for macromolecules. In the MIP material, there is supposed to be a densely crosslinked network for the conservation of the memorized structural information. Meanwhile, there should be enough space to facilitate the removal and rebinding of the protein macromolecule, such as pores in a porous material or pores in a polymer network.1 Another difficulty is the fabrication of MIPs in aqueous circumstances. Water-soluble proteins need to be imprinted and rebind in conditions close to their natural environment to guarantee conformational integrity.2 However, an organic porogen is commonly employed in most well-developed MIT methods. So far, it still requires a re-selection of MIP compositions, such as the functional monomer and crosslinker,22,23 to guarantee the effectiveness of polymerization and molecule–functional monomer interaction in an aqueous environment. Among the successful MIT methods for protein imprinting, including surface imprinting,19,24 epitope-mediated imprinting,25–27 and nanoparticle imprinting,28,29 the protein imprinted hydrogel has merits such as genuine aqueous environment, easy preparation, stimulus repressiveness, permeability, elasticity, etc. Protein imprinted hydrogels employ minimal steps in preparation by simply mixing the templates and monomers before polymerization. The obtained solid form substance can adsorb a large amount of water and has promising application prospects for its potential in smart materials30 and wearable soft materials.31–33
Thought-provoking works34,35 have found insightful compositional and structural strategies to tune the MIP-target association strength. The complementary interaction strength should be considered by a careful selection of affinity functional groups. Sulfonate groups have been recognized with a strong affinity with the amino residues that compose the protein.36 Our group investigated several types of sulfonate monomers, and the sodium allylsulfonate (SA) monomer-based MIP showed a superior affinity to cytochrome c compared with sodium p-styrenesulfonate and 2-acrylamido-2-methylpropane sulfonic acid.37 Continuing our previous studies, SA will be used for the preparation of a MIP hydrogel.
As mentioned above, a stiff organization of the MIP is also beneficial for the need to freeze the surface information. This can be realized by using dense crosslinkers, but a compromise has to be made to facilitate macromolecule diffusion. To achieve a balanced imprinting performance and molecular up-take efficiency, the fabrication of a protein imprinted hydrogel requires an optimization of the polymer network structure. The copoly(poly(ethylene glycol) diacrylate/poly(ethylene glycol) acrylate) copoly(PEGDA/PEGA) hydrogel is one type of crosslinked polymer network with PEGDA as a crosslinker and PEGA as side chains, which offers lots of opportunities in obtaining a high imprinting performance. Depending on the unit number (n) of PEG, a variety of commercially available PEGDA and PEGA monomers can be used, thereby providing a flexible way to tune the length of the network strands.38,39 The length of the network strands also relies on the crosslinker ratio. When n is equal to or more than 9, the PEGDA becomes water-soluble, thus allowing a wide range of crosslinking ratios of PEGDA/PEGA up to 10/0. As one type of PEG-based material, the copoly(PEGDA/PEGA) hydrogel is suitable for biomolecule imprinting and environmentally friendly applications owing to its features such as low toxicity and bio-compatibility. On the other hand, prosperous studies are using PEG-based materials, including PEG materials with ideally controllable structure,40 enhanced mechanical properties,41,42 and responsiveness to narrow temperature zone, pH,37,43–46 ionic strength,47etc. Thus, it is worthy to study the PEG-based MIPs to promote the diversity of MIP materials. Until now, the PEG-based hydrogels have found applications for carbohydrates, proteins, and small molecules, by showing specific adsorption,37 retention/affinity effect for separation,19 fluorescent response,48 and swelling-deswelling behavior49 to the targets.
Another strategy is the employment of counterions to modify the association strength. With the presence of a proper counterion, the proton transfer between the functional groups will be facilitated by the counterion memory effects.50,51 Studies have also indicated that MIPs are usually fabricated with an excessive amount of functional monomer, and the ionic competition effect from the counterion may reduce the unspecific adsorption. The ionic competition effect is evidenced by the reduction of gel swelling when the ionic strength is increased in the aqueous environment.47
In this work, efforts were made to discuss the parameters with a viewpoint on MIP's polymer structure and the counterion environment for recognition. We carried out a fundamental study with the copoly(PEGDA/PEGA) hydrogel38,52 for the fabrication of a protein imprinted MIP hydrogel. PEGDAs with different lengths (n = 9, 14, 23) were employed as crosslinkers, and PEGA with n = 9 was employed as the side chain. Cytochrome c was employed as a model template and the sodium allylsulfate was employed as a functional monomer to form ionic interactions with proteins. The obtained MIP hydrogels were used as adsorbents with cytochrome c and non-target protein solutions. Different factors were investigated for a high level of specific molecular adsorption, including the functional monomer-template ratio, crosslinker length, crosslinker ratio of PEGDA/PEGA, NaCl concentration, and HCl concentration in the adsorption test.
2. Experimental
2.1 Chemicals
The tris–HCl powder was purchased from Takara (Shiga, Japan). Trypsin from the bovine pancreas and cytochrome c from the equine heart were purchased from Sigma-Aldrich (St. Louis, USA). Sodium chloride, ammonium peroxodisulfate (APS), and N,N,N’,N’-tetramethylethylenediamine (TEMED) were purchased from Wako Pure Chemical Industries (Osaka, Japan). The PEGDA (namely 9G′, 14G′, and 23G′ when n equaled 9, 14 and 23, respectively) and PEGA (namely AM-90G, n = 9) were purchased from Shin-Nakamura Chemical (Wakayama, Japan). And the sodium allylsulfonate (SA) was from Tokyo Chemical Industry (Tokyo, Japan).2.2 Fabricating MIP hydrogels
The compositions of the MIP hydrogels are listed in Tables S1–S3 (ESI†). For reference, non-imprinted polymer (NIP) hydrogels were also fabricated without adding the protein template to the precursor solution. The prescribed compositions (except for TEMED) were added into one glass bottle and mixed by several repeats of pipetting. The mixture solution was degassed by performing a 20 min Argon bubbling and then a 10 min vacuum under 0.1 MPa. Immediately after degassing, the prescribed amount of TEMED was well mixed into the mixture. The mixture solution was quickly transferred into a slab gel mold and kept still for 24 h to complete the polymerization. The resulting gel was peeled off and cut into disks of 7 mm diameter. The gel was 1 mm in thickness and thus one piece of gel disk had an initial volume of around 38.5 μL.2.3 Template removal
To remove the templates from the MIP hydrogels, the gel pieces were immersed into 1 M NaCl with a volume more than 50 times the gel's volume. The washing solution with gels was kept at 40 °C temperature and refreshed every 24 h for a total of 6 times. Finally, the gels were conditioned in a buffer solution for another 24 h under 40 °C, followed by the adsorption test. This work employed different buffer solutions for the adsorption test. The buffer solutions for the last washing step were identical to the ones for the adsorption test. As a control reference, the NIP hydrogels were washed in the same manner.2.4 Adsorption test
The proteins, including cytochrome c, lysozyme, trypsin, and BSA, were dissolved into a buffer solution to make an adsorption solution. The buffer solution contained protein, pH 7.4 tris–HCl (1 mM otherwise indicated), and different concentrations of NaCl. The protein concentration was 0.018 mM unless otherwise indicated. To test the protein adsorption ability, the gels were put into the adsorption solution and kept shaking for 24 h. The supernatant of the adsorption solvent was examined under a UV spectrometer to detect the concentration of the remaining proteins. Each test was conducted with three repeats. In this study, although the removal of the template required a 7 day-long washing and was more efficient at 40 °C, the adsorption of protein into the gel would be completed within one day. For the detection of lysozyme, trypsin, and BSA the UV absorbance at 280 nm was examined; for cytochrome c the UV absorbance at 411 nm was examined. The adsorption amount and imprinting factor (IF) were calculated as follows:| Adsorption amount = (Cbefore − Cafter) × V | (1) |
 | (2) |
2.5 HPLC analysis
The HPLC analysis was carried out with an LC-40B X3 system (Shimadzu) and a reverse-phase column (Aeris wide pore XB-C8, 3.6 μm, 150 mm × 2.1 mm). The mobile phase A (MA) contained 0.1% TFA in H2O; the mobile phase B (MB) contained 0.1% TFA in acetonitrile. The samples were eluted followed by a gradient of 0–1 min 10% MB, 1–3 min 30% MB, 3–15 min 55% MB, 15–18 min 90% MB, and 18–25 min 10% MB. The flow rate was 0.2 mL min−1. The column temperature was 75 °C for enhancing the separation performance. The sample injection volume was 10 μL. And proteins were examined by UV adsorption at 280 nm.3. Results and discussion
3.1 Template removal
The importance of a proper template removal process cannot be emphasized more. Because the remaining templates may occupy the MIP sites to block the access of the target molecule, or unexpectedly be leaked into the adsorption circumstance to interfere with the molecular recognition performance. As previously reported, for an ionic gel, the mass density of the MIP network and the ionic strength of the washing solution influence the template removal efficiency. The mass density of the MIP structure is supposed to be low enough to allow a fast macromolecule elution, and also should be high enough to offer an operable mechanical strength. The ions in the washing solution will induce shrinkage of the ionic gel, and consequently, reduce the pore size of the polymer network. Thus, it is suggested to balance the composition of the washing solution until the ionic strength is strong enough for template elution, as well as moderate enough to keep the ionic gel from severe shrinkage. Based on our previous study, this work employed the mass density of 64.9 mg mL−1, and 1 M NaCl as a washing solution.37,47Through unsystematic experiments, we found that the temperature also affected the elution efficiency of the templates. Specifically, we achieved an efficient template removal by elevating the temperature to 40 °C. Our result showed that the gels eluted under 40 °C showed a higher IF than those eluted under room temperature (data not shown). As indicated in Fig. 1(a), after the MIP hydrogels were eluted with 1 M NaCl under 40 °C for two days, its colour fade was more significant compared to the one eluted under room temperature. Here we note that the colour fade was not a thermal-induced colour change, as a cytochrome c solution did not fade under 40 °C for up to 3 days. We would like to ascribe the fast colour fade of the MIP hydrogel to the faster dissociation of cytochrome c from the functional monomer under a high temperature because a dissociation reaction is an endothermic process. As a result, Fig. 1(b) and (c) shows typical gels after template removal and after cytochrome c rebinding, respectively. After removing the template, the MIP hydrogel swelled more significantly than the NIP hydrogel due to the loss of templates to confine the ionic repelling force. After cytochrome c rebinding, the MIP hydrogel and NIP hydrogel returned to close sizes.
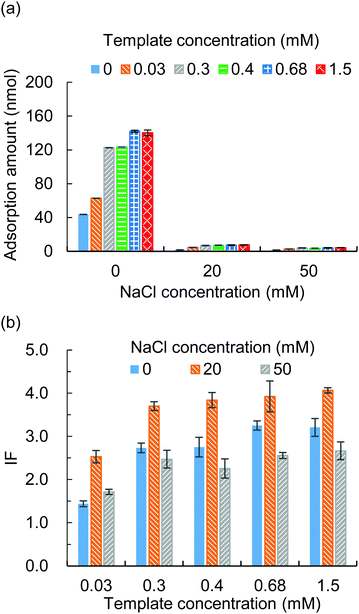
3.2 Template to functional monomer ratio and ionic strength in the adsorption solvent
The template to functional monomer ratio and the ionic strength in the adsorption solvent were studied for their effects on the imprinting factor (IF). The MIP hydrogels were prepared with different template to functional monomer ratios (Table S1, ESI†) and put into adsorption tests with different NaCl concentrations. The adsorption amounts of the MIP hydrogels and NIP hydrogels were examined and the IF was calculated. The results showed that a higher template to functional monomer ratio would improve the adsorption amount (Fig. 2(a)) and thus the IF (Fig. 2(b)). However, the increase in adsorption amount was gentle when the template concentration was higher than 0.3 mM. In addition, during the template removal process, we noticed that a higher template concentration required a longer washing time. Thus, we choose the template concentration of 0.3 mM for the following study.To modify the ionic strength, different concentrations of NaCl from 0 to 50 mM were added to the adsorption solvent. From Fig. 2(a) it was observed that the adsorption amount decreased along with the increase of the NaCl concentration, which proved that the presence of NaCl competed with the ionic interaction between the protein and functional monomer. As a result, the IF changed upon the change of NaCl concentration (Fig. 2(b)) and an optimum IF was found in the presence of 20 mM NaCl.
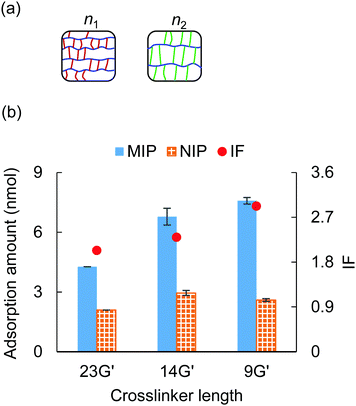
3.3 Crosslinker length
The effect of crosslinker length on the adsorption ability was investigated in this section. The crosslinkers employed here were 9G′ (n = 9), 14G′ (n = 14), and 23G′ (n = 23), and the crosslinker ratio of PEGDA/PEGA was fixed to 10/0 (Table S2, ESI†). Fig. 3(a) illustrates the ideal structure of PEGDA gels, given the same mass density, when using a shorter crosslinker, the side chain length decreases, and the main chain length increases.38 Thus, this structural difference might have an impact on the molecular recognition character of the MIP hydrogel.An adsorption test was carried out with the three types of PEGDA gels under 20 mM NaCl and 0.5 mM tris–HCl, and the results are shown in Fig. 3(b). It was observed that when the length of the crosslinker decreased, the MIP hydrogels’ adsorption ability to cytochrome c increased by a large margin, while the NIP hydrogels’ adsorption ability exhibited a slight difference. As a result, we found that the MIP hydrogels fabricated by shorter crosslinker 9G′ showed the highest specificity among the three types of gels. This result is consistent with the previous result that the shorter crosslinker provided a more rigid polymer structure and thus the MIP sites would be more robust.
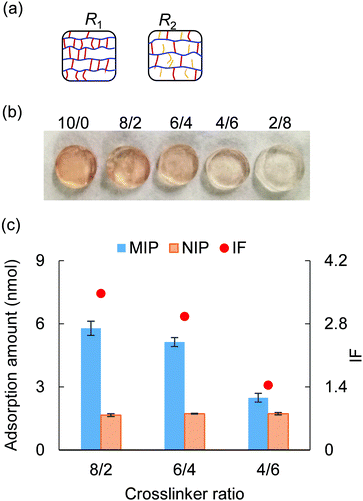
3.4 Crosslinker ratio
This section investigated the effect of crosslinker ratio PEGDA/PEGA on molecular recognition performance. The crosslinker ratio was changed ranging from 10/0 to 4/6, with 23G′ (PEGDA, n = 23) serving as the crosslinker and AM-90G (PEGA, n = 9) as the polymer chain (Table S3, ESI†). As illustrated in Fig. 4(a), given a fixed mass density, the lower the crosslinker ratio, the longer the polymer length between two crosslinking points, as well as the larger the pore size of the polymer network. Thus, using a less crosslinked gel might facilitate molecular elution and uptake. As typical evidence, after a 3 day 1 M NaCl washing, the less crosslinked gel exhibited a lighter colour (Fig. 4(b)). This indicated that the lower the crosslinker ratio was, the more efficient the elution of cytochrome c would be.After completing the template removal, each MIP and NIP hydrogel was immersed into 500 μL of a solution containing 0.03 mM cytochrome c, 20 mM NaCl, and 1 mM tris–HCl for the adsorption test. From the result of the adsorption test (Fig. 4(c)), we found that when the crosslinker ratio increased, the NIP hydrogels showed no significant variation in the adsorption ability, while the MIP hydrogels exhibited higher adsorption to cytochrome c. This suggested that the crosslinker ratio influenced the adsorption ability of MIP hydrogels because a larger population of rigid MIP cavities might form with a higher crosslinker ratio. By examining the IF, it was found that a quite specific recognition of cytochrome c (IF > 1.5) can still be obtained when the crosslinker was higher than 6/4. In brief, by using the crosslinker ratio of 6/4, a faster elution would be obtained while maintaining the specificity (IF > 1.5) of the molecular recognition.
3.5 Specificity
In an attempt to further investigate the effect of counterions on the MIP-protein recognition performance, HCl with a concentration ranging from 5 mM to 50 mM was employed in the adsorption solution as an acidic modifier. The MIP hydrogels with compositions shown in Table S2 (ESI†) (9G′) were employed as sorbents. Fig. 5 shows the result of the adsorption tests. For both the MIP and NIP hydrogels, the adsorption ability decreased with the increase of HCl, which is a similar trend when increasing NaCl concentration as the counterion (Fig. 2(a)). This trend suggested that the ion competition effect occurs to suppress the complementary interaction between the sulfonate group and protein.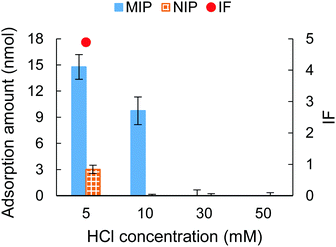 | ||
| Fig. 5 The adsorption amount and IF tested under different HCl concentrations. The IF was not shown when HCl was 10 mM (infinite value), 30 and 50 mM (invalid value). | ||
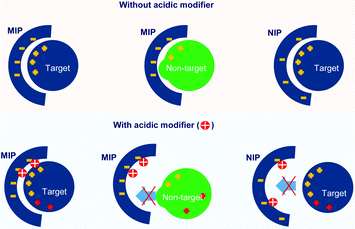
Fig. 5 also showed that the specificity for cytochrome c recognition was improved when using a low concentration of HCl (5 and 10 mM). Comparing the use of HCl with the use of NaCl, the NIP hydrogel showed a lower adsorption capacity when HCl was presented. This indicated that HCl showed a stronger capability to suppress unspecific adsorption. In particular, at 10 mM HCl, highly specific recognition of cytochrome c was obtained, evidenced by the unspecific adsorption of the NIP hydrogel being fully reduced. This evidenced the successful fabrication of imprinted cavities for cytochrome c. The effective suppression of cytochrome c adsorption to the hydrogels might be because the increase of HCl concentration caused a gradual protonation of the sulfonate group on the hydrogels. At the same time, the solution's pH decreased, and the surface charge of the amphiphilic protein was accordingly tuned to positive.53 As is shown in Fig. 6, this resulted in low interest, or even repulsion, between the acidic cytochrome c and the protonated hydrogels, which corresponded to the NIP hydrogels in 10–50 mM HCl and MIP hydrogels in 30–50 mM HCl.
When exposed to a pH range between 3.0 and 1.5, cytochrome c showed a reversible conformation change.54 Our result showed that despite the MIP hydrogels being synthesized under a pH 7.4 environment, they still showed adsorption ability under pH 2.0 (10 mM HCl). This indicated that the imprinted sites still matched with a certain portion of the unfolded conformation of cytochrome c.
3.6 Selectivity
The selectivity of the cytochrome c imprinted hydrogel was tested with non-target proteins of lysozyme, trypsin, and BSA. As is shown in Table S5 (ESI†), the cytochrome c, lysozyme, and trypsin have similar isoelectric points (pI) and carry positive charges at pH 7.4 or lower, where they may show affinity to the sulfonate functional monomer. Besides, unlike the larger proteins of trypsin and BSA, the molecular weight of lysozyme (14.4 kDa) is close to that of cytochrome c (12.3 kDa), which might pose a strong disguise to the molecular recognition. BSA has a pI of 4.7, thus carrying negative charges under pH 7.4 and positive charges under pH 2.0. The MIP hydrogels with compositions shown in Table S2 (ESI†) (9G′) were employed as sorbents. The adsorption solution had a volume of 1000 μL and contained 0.018 mM of each protein. 10 mM HCl was added to the adsorption solution. An HPLC test was carried out for the quantitative analysis of each protein in the mixture.The resulting chromatogram is shown in Fig. 7. It was found that the adsorption of cytochrome c was significantly higher than that of other non-target proteins, indicated by the decrease of the corresponding peak. The good selectivity of the MIP hydrogel to cytochrome c against other interfering proteins further demonstrated the high recognition performance of the imprinted cavities. To further investigate the mechanism behind the MIP-target recognition. A protein mixture sample containing 1 mM tris–HCl and 20 mM NaCl was also put into the test. The result in Fig. S1 (ESI†) showed that the unspecific adsorption to lysozyme increased considerably. It seemed like the imprinted cavities also accepted the molecules with a matching size in the neutral condition. This comparison test in Fig. S1 (ESI†) indicated that the presence of HCl played an important role in guaranteeing the selectivity of the imprinted sites. It was hypothesized that the HCl acted as a modifier to tune the surface distribution of functional groups on both the imprinted cavities and target protein so that the specificity of the complementary interaction was increased (Fig. 6).
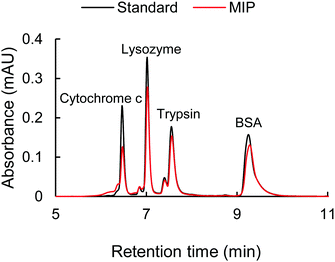 | ||
| Fig. 7 The chromatogram of the standard protein mixture with 0.018 mM of each protein (black line) and the protein mixture after the adsorption with MIP hydrogel (red line). | ||
4. Conclusions
This work involves a fundamental study of the copoly(PEGDA/PEGA) based MIP hydrogel. Cytochrome c was employed as a model template and the sodium allylsulfonate was employed as an ionic functional monomer. The effect of structural and compositional factors on the imprinting factor was investigated, including the template to functional monomer ratio, NaCl concentration in the adsorption solution (to adjust the ionic strength), crosslinker length, crosslinker ratio, and HCl concentration in the adsorption solution (as an acidic modifier). The results showed that a higher imprinting factor would be obtained when using a higher template to functional monomer ratio, 20 mM NaCl with 1 mM tris–HCl as the adsorption solution and a shorter crosslinker length. A lower crosslinker ratio facilitated the template removal; it was found that when using PEGDA/PEGA higher than 6/4, the imprinting factor remained at a relatively high value. Highly specific recognition of cytochrome c was realized using 10 mM HCl in the adsorption solution, where the unspecific adsorption was fully suppressed in the NIP. When testing the MIP in a protein mixture, it showed a higher adsorption to cytochrome c with the presence of lysozyme, trypsin and BSA. This work demonstrated the successful fabrication of imprinted sites in a PEG-based hydrogel and optimization of the molecular recognition performance of the MIP by tuning the structural and compositional parameters. The presented cytochrome c imprinted MIP material would be applied for specific adsorption and other uses, such as selective electrophoretic separation.Author contributions
Chenchen Liu: conceptualization, methodology, validation, investigation. Takuya Kubo: conceptualization, supervision, project administration. Koji Otsuka: visualization, project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was partly supported by the Adaptable and Seamless Technology transfer Program through Target-driven R&D (A-STEP) from Japan Science and Technology Agency (JST) [Grant Number JPMJTR214C] and the National Natural Science Foundation of China [Grant number 21804042].References
- L. Chen, S. Xu and J. Li, Chem. Soc. Rev., 2011, 40, 2922–2942 RSC.
- L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- A. Kloskowski, M. Pilarczyk, A. Przyjazny and J. Namiesnik, Crit. Rev. Anal. Chem., 2009, 39, 43–58 CrossRef CAS.
- W. Chen, Y. Ma, J. M. Pan, Z. H. Meng, G. Q. Pan and B. Sellergren, Polymers, 2015, 7, 1689–1715 CrossRef CAS.
- W. Bai, N. A. Gariano and D. A. Spivak, J. Am. Chem. Soc., 2013, 135, 6977–6984 CrossRef CAS PubMed.
- L. Jia, Y. Mao, S. Q. Zhang, H. Li, M. Qian, D. B. Liu and B. Qi, Microchem. J., 2021, 164, 105981 CrossRef CAS.
- A. Herrera-Chacon, X. Ceto and M. del Valle, Anal. Bioanal. Chem., 2021, 413, 6117–6140 CrossRef CAS PubMed.
- Y. Yoshimi, Y. Yagisawa, R. Yamaguchi and M. Seki, Sens. Actuators, B, 2018, 259, 455–462 CrossRef CAS.
- R. C. Advincula, Korean J. Chem. Eng., 2011, 28, 1313–1321 CrossRef CAS.
- O. S. Ahmad, T. S. Bedwell, C. Esen, A. Garcia-Cruz and S. A. Piletsky, Trends Biotechnol., 2019, 37, 294–309 CrossRef CAS PubMed.
- M. J. Whitcombe, I. Chianella, L. Larcombe, S. A. Piletsky, J. Noble, R. Porter and A. Horgan, Chem. Soc. Rev., 2011, 40, 1547–1571 RSC.
- L. Uzun and A. P. F. Turner, Biosens. Bioelectron., 2016, 76, 131–144 CrossRef CAS PubMed.
- A. E. Bodoki, B. C. Iacob and E. Bodoki, Polymers, 2019, 11, 2085 CrossRef CAS PubMed.
- B. Sellergren and C. J. Allender, Adv. Drug Delivery Rev., 2005, 57, 1733–1741 CrossRef CAS PubMed.
- Y. T. Qin, Y. S. Feng, Y. J. Ma, X. W. He, W. Y. Li and Y. K. Zhang, ACS Appl. Mater. Interfaces, 2020, 12, 24585–24598 CrossRef CAS PubMed.
- X. L. Sun, M. H. Wang, L. X. Yang, H. P. Wen, L. G. Wang, T. Li, C. L. Tang and J. J. Yang, J. Chromatogr. A, 2019, 1586, 1–8 CrossRef CAS PubMed.
- P. Panjan, R. P. Monasterio, A. Carrasco-Pancorbo, A. Fernandez-Gutierrez, A. M. Sesay and J. F. Fernandez-Sanchez, J. Chromatogr. A, 2018, 1576, 26–33 CrossRef CAS PubMed.
- A. Beltran, F. Borrull, P. A. G. Cormack and R. M. Marce, Trac, Trend. Anal. Chem., 2010, 29, 1363–1375 CrossRef CAS.
- R. Xing, S. Wang, Z. Bie, H. He and Z. Liu, Nat. Protoc., 2017, 12, 964–987 CrossRef CAS PubMed.
- B. C. Iacob, E. Bodoki and R. Oprean, Electrophoresis, 2014, 35, 2722–2732 CrossRef CAS PubMed.
- T. Khumsap, A. Corpuz and L. T. Nguyen, RSC Adv., 2021, 11, 11403–11414 RSC.
- L. Qin, X.-W. He, W. Zhang, W.-Y. Li and Y.-K. Zhang, Anal. Chem., 2009, 81, 7206–7216 CrossRef CAS PubMed.
- N. Bereli, M. Andaç, G. Baydemir, R. Say, I. Y. Galaev and A. Denizli, J. Chromatogr. A, 2008, 1190, 18–26 CrossRef CAS PubMed.
- C. Dong, H. Shi, Y. Han, Y. Yang, R. Wang and J. Men, Eur. Polym. J., 2021, 145, 110231 CrossRef CAS.
- L. Pasquardini and A. M. Bossi, Anal. Bioanal. Chem., 2021, 413, 6101–6115 CrossRef CAS PubMed.
- X. Wang, G. Chen, P. Zhang and Q. Jia, Anal. Methods, 2021, 13, 1660–1671 RSC.
- H. Nishino, C.-S. Huang and K. J. Shea, Angew. Chem., Int. Ed., 2006, 45, 2392–2396 CrossRef CAS PubMed.
- H. Zhang, Adv. Mater., 2020, 32, 1806328 CrossRef CAS PubMed.
- F. Canfarotta, A. Poma, A. Guerreiro and S. Piletsky, Nat. Protoc., 2016, 11, 443–455 CrossRef CAS PubMed.
- M. J. Whitcombe, Nat. Chem., 2011, 3, 657–658 CrossRef CAS PubMed.
- Q. W. Zhang, D. F. Jiang, C. S. Xu, Y. C. Ge, X. H. Liu, Q. Q. Wei, L. P. Huang, X. Q. Ren, C. D. Wang and Y. Wang, Sens. Actuators, B, 2020, 320, 128325 CrossRef CAS.
- O. Parlak, S. T. Keene, A. Marais, V. F. Curto and A. Salleo, Sci. Adv., 2018, 4, eaar2904 CrossRef CAS PubMed.
- Y. L. Liu, R. Liu, Y. Qin, Q. F. Qiu, Z. Chen, S. B. Cheng and W. H. Huang, Anal. Chem., 2018, 90, 13081–13087 CrossRef CAS PubMed.
- S. Shinde, A. Incel, M. Mansour, G. D. Olsson, I. A. Nicholls, C. Esen, J. Urraca and B. Sellergren, J. Am. Chem. Soc., 2020, 142, 11404–11416 CrossRef CAS PubMed.
- C. Pérez-Casas and A. K. Yatsimirsky, J. Org. Chem., 2008, 73, 2275–2284 CrossRef PubMed.
- T. Kubo, S. Arimura, T. Naito and K. Otsuka, Mol. Imprinting, 2015, 3, 18–25 CAS.
- T. Kubo, S. Arimura, Y. Tominaga, T. Naito, K. Hosoya and K. Otsuka, Macromolecules, 2015, 48, 4081–4087 CrossRef CAS.
- C. Liu, T. Kubo, T. Naito and K. Otsuka, ACS Appl. Polym. Mater., 2020, 2, 3886–3893 CrossRef CAS.
- X. Li, K. Khairulina, U.-I. Chung and T. Sakai, Macromolecules, 2014, 47, 3582–3586 CrossRef CAS.
- T. Sakai, T. Matsunaga, Y. Yamamoto, C. Ito, R. Yoshida, S. Suzuki, N. Sasaki, M. Shibayama and U. I. Chung, Macromolecules, 2008, 41, 5379–5384 CrossRef CAS.
- L. Jiang, C. Liu, K. Mayumi, K. Kato, H. Yokoyama and K. Ito, Chem. Mater., 2018, 30, 5013–5019 CrossRef CAS.
- A. Bin Imran, K. Esaki, H. Gotoh, T. Seki, K. Ito, Y. Sakai and Y. Takeoka, Nat. Commun., 2014, 5, 5124 CrossRef CAS PubMed.
- H. Q. Liu, Y. J. Li, R. Yang, X. J. Gao and G. G. Ying, PLoS One, 2016, 11, e0159296 CrossRef PubMed.
- Q. A. Ijaz, N. Abbas, M. S. Arshad, A. Hussain, Z. Shahiquz and Z. Javaid, J. Drug Delivery Sci. Technol., 2018, 43, 221–232 CrossRef CAS.
- W. Cui, R. Liu, H. Q. Jin, P. Lv, Y. Y. Sun, X. Men, S. N. Yang, X. Z. Qu, Z. Z. Yang and Y. N. Huang, J. Controlled Release, 2016, 225, 53–63 CrossRef CAS PubMed.
- J. J. Weng, Z. B. Huang, X. M. Pu, X. C. Chen, G. F. Yin, Y. P. Tian and Y. Song, Colloids Surf., B, 2020, 191, 110943 CrossRef CAS PubMed.
- Y. Tominaga, T. Kubo, K. Sueyoshi, K. Hosoya and K. Otsuka, J. Polym. Sci., Part A: Polym. Chem, 2013, 51, 3153–3158 CrossRef CAS.
- T. Kubo, N. Watanabe, C. Liu, S. Ikari, E. Kanao, T. Naito, T. Sano and K. Otsuka, Anal. Methods, 2021, 13, 3086–3091 RSC.
- T. Miyata, T. Hayashi, Y. Kuriu and T. Uragami, J. Mol. Recognit., 2012, 25, 336–343 CrossRef CAS PubMed.
- S. Wagner, C. Zapata, W. Wan, K. Gawlitza, M. Weber and K. Rurack, Langmuir, 2018, 34, 6963–6975 CrossRef CAS PubMed.
- M. Emgenbroich, C. Borrelli, S. Shinde, I. Lazraq, F. Vilela, A. J. Hall, J. Oxelbark, E. De Lorenzi, J. Courtois, A. Simanova, J. Verhage, K. Irgum, K. Karim and B. Sellergren, Chem. – Eur. J., 2008, 14, 9516–9529 CrossRef CAS PubMed.
- T. Kinoshita, Y. Ishigaki, K. Nakano, K. Yamaguchi, S. Akita, S. Nii and F. Kawaizumi, Sep. Purif. Technol., 2006, 49, 253–257 CrossRef CAS.
- R. Jain, R. Kumar, S. Kumar, R. Chhabra, M. C. Agarwal and R. Kumar, Arch. Biochem. Biophys., 2015, 585, 52–63 CrossRef CAS PubMed.
- M. Coletta, H. Costa, G. De Sanctis, F. Neri, G. Smulevich, D. L. Turner and H. Santos, J. Biol. Chem., 1997, 272, 24800–24804 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb00501h |
| This journal is © The Royal Society of Chemistry 2022 |