Easy preparation of a liposome-mediated protein delivery system by freeze–thawing a liposome–protein complex
Hiroyuki
Koide
 *a,
Hiroki
Ochiai
a,
Hikaru
Suzuki
a,
Shinya
Hirata
a,
Midori
Watanabe
a,
Sei
Yonezawa
a,
Takehisa
Dewa
*a,
Hiroki
Ochiai
a,
Hikaru
Suzuki
a,
Shinya
Hirata
a,
Midori
Watanabe
a,
Sei
Yonezawa
a,
Takehisa
Dewa
 b,
Naoto
Oku
c and
Tomohiro
Asai
a
b,
Naoto
Oku
c and
Tomohiro
Asai
a
aDepartment of Medical Biochemistry, University of Shizuoka School of Pharmaceutical Sciences, 52-1 Yada, Suruga-ku, Shizuoka, Shizuoka 422-8526, Japan
bDepartment of Life Science and Applied Chemistry, Graduate School of Engineering, Nagoya Institute of Technology, Gokiso-cho, Showa-ku, Nagoya, Aichi 466-8555, Japan
cFaculty of Pharma-Science, Teikyo University, 2-11-1 Kaga, Itabashi-ku, Tokyo 173-8605, Japan
First published on 25th April 2022
Abstract
Homeostasis can be achieved by adding a protein supplement; however, an appropriate vector is required to deliver the protein into the cell because of the low stability of proteins in the blood and low cell membrane permeability. Here we report an easy one-step method of encapsulating proteins into liposomes for delivery. We used negatively charged superoxide dismutase (SOD) and a polycation liposome as protein and liposome models, respectively. Liposome-encapsulated SOD was prepared by freeze–thawing the SOD–liposome complex (lipoplexes). The amount of immobilized SOD within the lipoplex significantly increased on freeze–thawing. Surprisingly, subjecting the single-layered lipoplexes to freeze–thawing produced multilayered liposomes with SOD localized between the lipid layers. The amount of SOD delivered intracellularly significantly increased by freeze–thawing compared with that delivered by lipoplexes without freeze–thawing. SOD, liposomes, and endosomes were separately localized in the cells. The freeze–thawed lipoplex-encapsulated SOD samples were intravenously injected in mice. The SOD biodistribution was dramatically changed compared with the injection of free SOD or lipoplex. SOD was detached from the lipoplex in the bloodstream after the injection of non-freeze–thawed lipoplex, whereas the encapsulation of SOD in the liposomes upon freeze–thawing enabled the stable circulation of SOD with the liposomes in the bloodstream. This work paves the way for the application of the freeze–thawing technology for the easy one-step encapsulation of proteins into liposomes for protein delivery.
Introduction
Proteins play essential roles in cellular processes, including signal transduction, catalysis of the metabolic response, and host defense. Accordingly, protein depletion can cause serious disorders, such as cancer, inflammation, and multiple organ dysfunction, as a result of homeostasis imbalance.1,2 Supplementation of the depleted protein can help recover homeostasis;3 however, proteins have low stability in blood, cell membrane permeability, and endosomal escape ability, making it challenging to deliver proteins into the cell.4,5 The targets of protein drugs currently in use, such as antibodies, hormones, cytokines, and enzymes, are generally extracellular molecules.3,6 Therefore, the development of a highly efficient strategy for protein delivery into cells is of pivotal importance.To date, diverse nanomaterials such as liposomes (also known as lipid nanoparticles [LNPs]7,8), polymeric micelles,9 gold nanoparticles,10 inorganic nanoparticles,11,12 and polymeric nanoparticles13 have been used for protein delivery.5,14 Particularly, cationic LNPs are suitable for both in vitro and in vivo protein delivery because they electrostatically bind to the negatively charged cell membrane and enhance cellular uptake.7,15 Additionally, LNPs demonstrate high endosomal escape ability by damaging the endosomal membrane and proton sponge effect, which allows the delivery of the protein into the cytoplasm. Therefore, using LNPs can be a promising strategy for protein delivery. Attaching proteins on the LNP surface is easy; however, protein–LNP binding generally involves noncovalent interactions including electrostatic interactions,16 which are disrupted in the bloodstream, resulting in protein detachment and degradation. To tackle this issue, protein encapsulation within LNPs is attracting increasing research attention as an efficient protein delivery system. Recently, proteins have been encapsulated within LNPs by mixing functional lipids dissolved in organic solvents and proteins dissolved in water.7,17,18 The main advantage of using LNPs as an encapsulation system is that it allows us to precisely control the particle size with high reproducibility.19 In addition, no chemical modification of the proteins is required. Unfortunately, the use of organic solvents often leads to loss of protein functionality. Moreover, dialysis or other purification methods and unconventional instruments are required for the preparation of the delivery system. Although other systems, such as peptide- or polymer-conjugated proteins, have been developed for protein delivery,15,20,21 these methods may affect the original protein function. Therefore, an easy one-step protein encapsulation method for efficient protein delivery is required.
Our group previously reported the one-step method for the easy encapsulation of small interfering RNA (siRNA) with a molecular weight (M.W.) of ca. 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 between the lipid layers of multilayered liposomes by freeze–thawing a mixture of siRNA and single-layered liposome complexes.22,23 The amount of immobilized siRNA in the liposomes, cellular uptake, and the siRNA knockdown effect significantly increased after freeze–thawing compared with those using free siRNA and a nonfreeze–thawed complex. This indicates that freeze–thawing is an attractive strategy for the easy encapsulation of siRNA into liposomes. However, to the best of our knowledge, the encapsulation of high-molecular-weight molecules, such as proteins, into liposomes using the freeze–thawing method to improve intracellular protein delivery has not been attempted yet.
000 between the lipid layers of multilayered liposomes by freeze–thawing a mixture of siRNA and single-layered liposome complexes.22,23 The amount of immobilized siRNA in the liposomes, cellular uptake, and the siRNA knockdown effect significantly increased after freeze–thawing compared with those using free siRNA and a nonfreeze–thawed complex. This indicates that freeze–thawing is an attractive strategy for the easy encapsulation of siRNA into liposomes. However, to the best of our knowledge, the encapsulation of high-molecular-weight molecules, such as proteins, into liposomes using the freeze–thawing method to improve intracellular protein delivery has not been attempted yet.
This work describes an easy one-step method for protein encapsulation into liposomes by freeze–thawing a protein–liposome complex (lipoplex). Superoxide dismutase (SOD, M.W. ∼600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was used as a target protein model, and dicetyl phosphate–diethylenetriamine (DCP–DETA)-based polycation liposome was used as a liposome model for delivery. In addition, the amount of SOD delivered into the cell markedly increased using the freeze–thawed lipoplex compared with the use of nonfreeze–thawed lipoplex. SOD showed stable circulation when the freeze–thawed lipoplex was injected due to the encapsulation. These results suggest that the developed freeze–thawing strategy is an attractive and suitable approach for the easy encapsulation of proteins within liposomes for protein delivery.
000) was used as a target protein model, and dicetyl phosphate–diethylenetriamine (DCP–DETA)-based polycation liposome was used as a liposome model for delivery. In addition, the amount of SOD delivered into the cell markedly increased using the freeze–thawed lipoplex compared with the use of nonfreeze–thawed lipoplex. SOD showed stable circulation when the freeze–thawed lipoplex was injected due to the encapsulation. These results suggest that the developed freeze–thawing strategy is an attractive and suitable approach for the easy encapsulation of proteins within liposomes for protein delivery.
Experimental
Materials
Cholesterol, dioleoylphosphatidylethanolamine (DOPE), dipalmitoylphosphatidylcholine (DPPC), N-(carbonyl-methoxy polyethylene glycol 2000)-1,2-distearoyl-sn-glycero-3-phosphoethanolamine (mPEG2000–DSPE), and N-(carbonyl-methoxy polyethylene glycol 6000)-1,2-distearoyl-sn-glycero-3-phosphoethanolamine (mPEG6000–DSPE) were kindly donated by Nippon Fine Chemical Co. (Hyogo, Japan). DCP–DETA was synthesized as described previously.22 Chloroform, tert-butyl alcohol, acetonitrile, and Dulbecco's modified Eagle's medium (DMEM, high glucose) were purchased from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan). SOD was purchased from Cosmo Bio (Tokyo, Japan). N-Hydroxysuccinimide (NHS) and gold nanoparticles (Au NPs) were purchased from Cytodiagnostics (Burlington, Canada). Sulfo-Cy5 NHS ester was purchased from Lumiprobe (Maryland, USA). Float-A-Lyzer G2 dialysis device (M.W. cut-off: 3.5–5 kDa) was purchased from Spectrum Laboratories Inc., Rancho Dominguez, (CA, USA). LysoTrackerTM Red DND-99, Permafluor aqueous mounting medium, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate [DiIC18(3)], and fetal bovine serum (FBS) were purchased from Thermo Fisher Scientific (MA, USA). Cell Counting Kit-8 (CCK-8) was purchased from Dojindo Lab (Kumamoto, Japan). All other reagents were of analytical grade.Measurement of SOD activity
SOD activity after the freeze–thawing was measured using a SOD assay kit according to the manufacturer's instructions. In brief, 20 μL of each sample was incubated with CCK-8 working (200 μL) and enzyme (20 μL) solutions for 20 min at 37 °C. Finally, the absorbance at 450 nm was measured to determine the SOD activity.Preparation of freeze–thawed lipoplex
DCP–DETA, DOPE, and cholesterol were dissolved in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1:1 in tert-butyl alcohol and freeze–dried. Liposomes were prepared by hydrating a lipid mixture with nanopore water at 60 °C. The liposomes were then freeze–thawed twice and extruded 21 times through a polycarbonate membrane filter with a pore size of 100 nm (Nucleopore, Maidstone, UK). Liposomes and SOD in a molar ratio of 3000
1:1 in tert-butyl alcohol and freeze–dried. Liposomes were prepared by hydrating a lipid mixture with nanopore water at 60 °C. The liposomes were then freeze–thawed twice and extruded 21 times through a polycarbonate membrane filter with a pore size of 100 nm (Nucleopore, Maidstone, UK). Liposomes and SOD in a molar ratio of 3000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were incubated for 20 min at room temperature (r.t.) under gentle shaking conditions (750 rpm) to form a liposome–SOD complex (lipoplex). The lipoplex was then freeze–thawed twice using liquid nitrogen and water bath (60 °C). For the preparation of fluorescent-labeled liposomes, DiIC18(3) was added to the initial solution. The particle size, polydispersity index (PDI), and ζ-potential were measured using a Zetasizer Nano ZS (Malvern, Worcs, UK) in 1 mM phosphate-buffered saline (PBS; pH = 7.4). A hydrated lipoplex was prepared by hydrating the freeze–dried liposomes with nanopore water containing SOD (40 μg mL−1) and sized by extrusion. The liposome, lipoplex, freeze–thawed lipoplex, and hydrated lipoplex samples were stored at 4 °C for 7 days to investigate their stability. The particle sizes were then measured using a Zetasizer Nano ZS (Malvern, Worcs, UK) in 1 mM PBS (pH = 7.4)
1 were incubated for 20 min at room temperature (r.t.) under gentle shaking conditions (750 rpm) to form a liposome–SOD complex (lipoplex). The lipoplex was then freeze–thawed twice using liquid nitrogen and water bath (60 °C). For the preparation of fluorescent-labeled liposomes, DiIC18(3) was added to the initial solution. The particle size, polydispersity index (PDI), and ζ-potential were measured using a Zetasizer Nano ZS (Malvern, Worcs, UK) in 1 mM phosphate-buffered saline (PBS; pH = 7.4). A hydrated lipoplex was prepared by hydrating the freeze–dried liposomes with nanopore water containing SOD (40 μg mL−1) and sized by extrusion. The liposome, lipoplex, freeze–thawed lipoplex, and hydrated lipoplex samples were stored at 4 °C for 7 days to investigate their stability. The particle sizes were then measured using a Zetasizer Nano ZS (Malvern, Worcs, UK) in 1 mM PBS (pH = 7.4)
Amount of immobilized SOD in the samples
The lipoplex and freeze–thawed lipoplex samples were ultracentrifuged (435![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, 15 min, 4 °C) to remove the unbound SOD, and the precipitate was dissolved in 1% Triton-X100 and incubated for 5 min at 55 °C. The SOD bound to liposome was measured at a wavelength of 220 nm using a high-performance liquid chromatography (HPLC) system (Prominence, Shimazdu, Japan) along with a TSK-gel ODS-100Z column (particle size 3 μm, 4.6 mm i.d. × 150 mm, Tosoh, Japan) at a flow rate of 1.0 mL min−1 and a temperature of 40 °C. The LC gradients for the mobile phases A and B (A: 0.1% trifluoroacetic acid (TFA) in H2O, B: 0.1% TFA in acetonitrile) were as follows: 0–3 min, 20%–36% B; 3–10 min, 36%–43% B; 10–13 min, 43%–95% B; 13–16 min, 95% B; 16–20 min, 95–20% B; 20–32 min, 20% B.
000×g, 15 min, 4 °C) to remove the unbound SOD, and the precipitate was dissolved in 1% Triton-X100 and incubated for 5 min at 55 °C. The SOD bound to liposome was measured at a wavelength of 220 nm using a high-performance liquid chromatography (HPLC) system (Prominence, Shimazdu, Japan) along with a TSK-gel ODS-100Z column (particle size 3 μm, 4.6 mm i.d. × 150 mm, Tosoh, Japan) at a flow rate of 1.0 mL min−1 and a temperature of 40 °C. The LC gradients for the mobile phases A and B (A: 0.1% trifluoroacetic acid (TFA) in H2O, B: 0.1% TFA in acetonitrile) were as follows: 0–3 min, 20%–36% B; 3–10 min, 36%–43% B; 10–13 min, 43%–95% B; 13–16 min, 95% B; 16–20 min, 95–20% B; 20–32 min, 20% B.
Change in particle size after freeze–drying
The lipoplex and freeze–thawed lipoplex samples were freeze–dried with or without 0.3 M sucrose and then resuspended with nanopure water. The particle size was measured using a Zetasizer Nano ZS (Malvern, Worcs, UK) in 1 mM PBS (pH = 7.4).Determining of the amount of SOD encapsulated within the freeze–thawed lipoplex
The lipoplex and freeze–thawed lipoplex samples were incubated with mPEG2000–DSPE for 30 min at 50 °C (30 mol% against phospholipid molar ratio) for polyethylene glycol (PEG) modification, and the PEGylated lipoplex and freeze–thawed lipoplex were ultracentrifuged (435![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, 15 min, 4 °C). The precipitate was dissolved in 1% Triton-X100 and incubated for 5 min at 55 °C to measure the SOD amount by HPLC.
000×g, 15 min, 4 °C). The precipitate was dissolved in 1% Triton-X100 and incubated for 5 min at 55 °C to measure the SOD amount by HPLC.
Localization of SOD in the lipoplex
SOD was conjugated with NHS-conjugated Au NPs (5 nm) according to the manufacture's protocol. In brief, 1 mg of SOD was dissolved in 200 μL of resuspension buffer. Further, 48 μL of this resulting SOD solution was mixed with reaction buffer (60 μL); this solution (90 μM) was incubated with NHS-conjugated Au NPs for 2 h at 37 °C. The reaction was quenched using a buffer (10 μL). Subsequently, the Au NP-conjugated SOD was purified by spin column (100 kDa. 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, 30 min, 4 °C). The amount of SOD molecules per Au NPs was determined as 16 by measuring the absorbance at 515 nm. Further, 5 μL of the lipoplex and freeze–thawed lipoplex samples prepared with Au NP-conjugated SOD were placed on a grid (Nisshin EM, Tokyo, Japan) and dried under a stream of warm air thrice. The obtained sample was negatively stained with 1 w/v% ammonium molybdate for 1 min and dried for 30 min and observed using an HT7700 transmission electron microscopy (TEM) system (Hitachi High-Technologies, Tokyo, Japan). The images were recorded using a charge-coupled camera at 1024 × 1024 pixels (Advanced Microscopy Techniques, Woburn, MA, USA).
000×g, 30 min, 4 °C). The amount of SOD molecules per Au NPs was determined as 16 by measuring the absorbance at 515 nm. Further, 5 μL of the lipoplex and freeze–thawed lipoplex samples prepared with Au NP-conjugated SOD were placed on a grid (Nisshin EM, Tokyo, Japan) and dried under a stream of warm air thrice. The obtained sample was negatively stained with 1 w/v% ammonium molybdate for 1 min and dried for 30 min and observed using an HT7700 transmission electron microscopy (TEM) system (Hitachi High-Technologies, Tokyo, Japan). The images were recorded using a charge-coupled camera at 1024 × 1024 pixels (Advanced Microscopy Techniques, Woburn, MA, USA).
Cell culturing
Colon26 NL-17 (C26-NL17) carcinoma cells were cultured in DMEM high glucose (Wako Pure Chemical Industries, Ltd) containing 10% FBS (AusGeneX, Oxenford, Australia), 100 units per mL penicillin G (MP Biomedicals, Irvine, CA), and 100 μg mL−1 streptomycin (MP Biomedicals) in a 5% CO2 incubator at 37 °C.Cellular uptake of liposomes and SOD
C26-NL17 cells were seeded onto a 24-well plate at a density of 1.5 × 104 cells per well. The free SOD, lipoplex, and freeze–thawed lipoplex samples were each added to the cells (final phospholipid and SOD concentrations were 3 μM and 1 nM, respectively). Cy5-conjugated SOD (Cy5–SOD) and DiI-labeled liposomes were used in this experiment. After 24 h, the cells were washed twice with PBS and lysed with 200 μL of 1 w/v% n-octyl-β-D-glucoside (Dojindo, Kumamoto, Japan) containing the following protease inhibitors: 1 mM phenylmethylsulfonyl fluoride, 2 μg mL−1 leupeptin, 2 μg mL−1 aprotinin, and 2 μg mL−1 pepstatin A (Sigma-Aldrich). The fluorescent intensity was then measured to determine the cellular uptake of liposomes and SOD.Cytotoxicity of the lipoplex
C26-NL17 cells were seeded onto a 24-well plate at a density of 1.5 × 104 cells per well, and the free SOD, lipoplex, and freeze–thawed lipoplex samples were each added to the cells (final phospholipid and SOD concentrations were 3 μM and 1 nM, respectively). After 24 h, viable cells were counted using the CCK-8 kit (Dojindo, Kumamoto, Japan) according to the manufacturer's instructions. Further, the absorbance was measured using an Infinite® M200 plate reader (Tecan Group, Männedorf, Switzerland) at a test wavelength of 450 nm and a reference wavelength of 630 nm.Localization of SOD, liposomes, and endosomes in the cells
C26-NL17 cells were seeded onto a glass-bottom 24-well plate (AGC Techno Glass Co. Ltd, Shizuoka, Japan) at a density of 1.0 × 104 cells per well. The lipoplex and freeze–thawed lipoplex samples were each added to the cells (final phospholipid and SOD concentrations were 3 μM and 1 nM, respectively). In this experiment, Cy5–SOD and DiI-labeled liposomes were used. After 24 h, the late endosomes were stained with LysoTracker (60 nM), and the localization of SOD, liposomes, and endosomes in the cells was observed using an A1R+ confocal laser scanning microscope (Nikon, Tokyo, Japan).Experimental animals
BALB/c male mice were purchased from Japan SLC Inc. (Shizuoka, Japan). All animals were raised according to the Animal Facility Guidelines of the University of Shizuoka. All animal experiment protocols were approved by the Animal and Ethics Review Committee of the University of Shizuoka.Biodistribution of SOD after intravenous injection of the lipoplex
BALB/c male mice (6-weeks-old) were intravenously injected with SOD, PEGylated lipoplex, or PEGylated freeze–thawed lipoplex. Cy5–SOD was used to determine the SOD biodistribution; further, Cy5–SOD biodistribution was measured using an in vivo imaging system (Xenogen IVIS Lumina System) coupled with Living Image software for data acquisition (Xenogen Corp., Alameda, CA, USA). The mice were sacrificed under deep anesthesia 24 h after the injection. Finally, the fluorescence activity of the heart, lungs, liver, spleen, and kidneys was measured.Statistical analyses
The differences in the groups were evaluated by performing an analysis of variance with the Tukey post hoc test.Results and discussion
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and sized to 100 nm using an extruder. DOPE was used to enhance the endosomal escapability.24,25 SOD was attached to the liposome surface via electrostatic interaction upon incubation for 20 min at r.t. The lipoplex was then freeze–thawed twice. The particle size, PDI, and ζ-potentials of liposome, the lipoplex, and the freeze–thawed lipoplex are shown in Table 1. As per Table 1, the values for liposome and the lipoplex were similar. In contrast, the particle size of the freeze–thawed lipoplex was significantly larger. This is most likely due to the increase in the interior aqueous phase during the freezing step,26 after which the liposomal membrane was reconstituted during the thawing step.
1 and sized to 100 nm using an extruder. DOPE was used to enhance the endosomal escapability.24,25 SOD was attached to the liposome surface via electrostatic interaction upon incubation for 20 min at r.t. The lipoplex was then freeze–thawed twice. The particle size, PDI, and ζ-potentials of liposome, the lipoplex, and the freeze–thawed lipoplex are shown in Table 1. As per Table 1, the values for liposome and the lipoplex were similar. In contrast, the particle size of the freeze–thawed lipoplex was significantly larger. This is most likely due to the increase in the interior aqueous phase during the freezing step,26 after which the liposomal membrane was reconstituted during the thawing step.
| Size (d. nm) | PDI | ζ-Potential (mV) | |
|---|---|---|---|
| Liposome | 148 ± 10 | 0.219 ± 0.013 | +29 ± 1 |
| Lipoplex | 147 ± 11 | 0.215 ± 0.015 | +29 ± 1 |
| Freeze–thawed lipoplex | 234 ± 18 | 0.241 ± 0.026 | +33 ± 1 |
| Hydrated lipoplex | 128 ± 8 | 0.070 ± 0.022 | +21 ± 3 |
SOD encapsulation within lipid nanoparticles
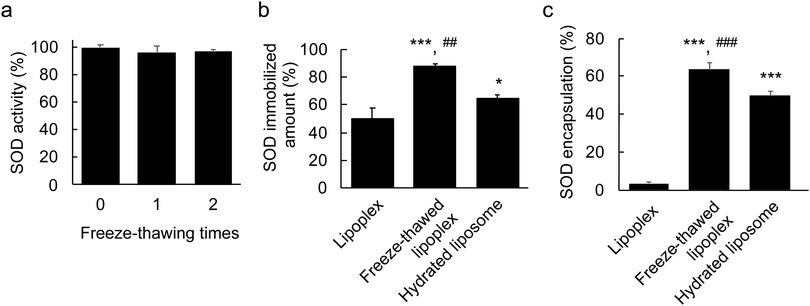
First, the stability of free SOD after the freeze–thawing process was investigated to ensure that its structure was not disrupted as this would result in loss of activity. The SOD sample was freeze–thawed twice and its activity was measured using an assay kit after each freeze–thaw cycle (Fig. 2a). No significant decrease was observed in the SOD activity, which demonstrated the stability of SOD under freeze–thawing treatment.Second, the SOD-immobilizing capacity of the lipoplex after freeze–thawing was evaluated. A small molecule can be stochastically encapsulated within liposomes by hydrating a freeze–dried lipid with a drug-containing solution.27 Accordingly, in this experiment, the freeze–dried lipid was hydrated with SOD-containing water to prepare a hydrated lipoplex as a control for the liposome-encapsulated SODs. The size, PDI, and ζ-potential of the hydrated lipoplex were ∼128 nm, ∼0.07, and ∼ + 21 mV, respectively (Table 1). The size of hydrated lipoplex was slightly smaller than that of liposome and lipoplex, suggesting that the SOD encapsulated within the hydrated lipoplex binds to the internal lipid layer, and the lipoplex then shrinks. The lipoplex, the freeze–thawed lipoplex, and the hydrated lipoplex samples were purified by ultracentrifugation, and the amount of immobilized SOD in each sample was determined to be ∼50%, ∼90%, and ∼60%, respectively, by HPLC (Fig. 2b). This indicates that the freeze–thawing process increased the SOD-immobilizing ability of the lipoplex.
To achieve high delivery efficiency, SOD must be encapsulated within the liposome. To investigate whether this was the case, the lipoplex was modified using PEG. PEG modification of nanoparticles is known to inhibit protein absorption in the bloodstream due to the formation of a steric barrier on the surface of the nanoparticle.28,29 Similarly, the SOD bound to the liposome surface was expected to detach upon PEGylation. Thus, 30 mol% of mPEG2000–DSPE was attached onto the lipoplex, the freeze–thawed lipoplex, and the hydrated lipoplex. The amount of immobilized SOD was measured by HPLC after purification (Fig. 2c) and was found to be ∼3.5% in the lipoplex after PEGylation, indicating that all SOD molecules had detached from the liposome surface. The amount of immobilized SOD from the hydrated lipoplex was ∼30%, which suggests that ∼50% of SOD was located on the hydrated lipoplex, whereas the remaining ∼50% was encapsulated within the lipoplex. Moreover, >60% of the SOD immobilized in the freeze–thawed lipoplex remained even after PEGylation. Eventually, the SOD encapsulation percentage in the freeze–thawed liposome was ∼55%. These results indicate that a large amount of SOD was encapsulated within the liposome by freeze–thawing the lipoplex. In general, liposomes exhibit a bilayer structure with functional lipids located in the outer and inner membranes. The nonfreeze–thawed lipoplex contains SOD only on the surface, whereas SOD binds to the outer and inner lipid layers because it is encapsulated within the liposome upon freeze–thawing. Therefore, the amount of immobilized SOD within the freeze–thawed lipoplex increased compared with that of the nonfreeze–thawed lipoplex. According to these results, freeze–thawing lipoplexes is an effective and easy approach to encapsulate proteins within liposomes.
To demonstrate the stability of the liposome, lipoplex, freeze–thawed lipoplex, and hydrated lipoplex samples, each sample was stored at 4 °C for 7 days, and the percentage of size change was determined. The percentages of particle size change of the liposome, lipoplex, freeze–thawed lipoplex, and hydrated lipoplex samples were 1 ± 2%, 1 ± 5%, 1 ± 4%, and 4 ± 2%, respectively. These results indicate that prepared lipoplexes can be stored for at least 1 week without any characteristic changes. We then demonstrated the particle size change after the freeze–drying. The lipoplex and freeze–thawed lipoplex samples were freeze–dried with or without sucrose (final concentration of sucrose; 0.3 M), and these samples were resuspended with nanopure water. The sizes of lipoplex and freeze–thawed lipoplex samples after freeze–drying without sucrose were increased by 770% and 400% after resuspension, indicating that each lipoplex was aggregated after the resuspension. However, the size of the lipoplex and freeze–thawed lipoplex samples did not increase after freeze–drying with 0.3 M sucrose (lipoplex; 12 ± 1%, freeze–thawed lipoplex; 2 ± 12%), indicating sucrose stabilized lipid membrane and inhibits lipoplex aggregation.30
Localization of SOD in the lipoplex
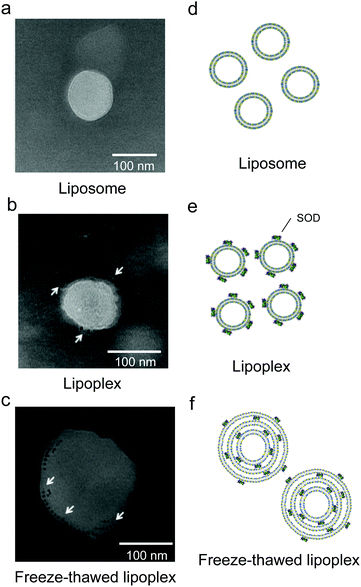
Having confirmed that SOD was encapsulated within the liposome upon freeze–thawing, the lipoplex morphology and localization of SOD before and after freeze–thawing were investigated by TEM observation of a lipoplex and a freeze–thawed lipoplex prepared using Au NP-conjugated SOD (Fig. 3). The liposome exhibited a single-layer structure before (Fig. 3a) and after (Fig. 3b) the preparation of the lipoplex. In the latter, however, Au NPs were observed on the liposome surface. Meanwhile, the freeze–thawed lipoplex containing Au NP-conjugated SOD formed a multilayered structure, with the Au NPs located between the lipid layers (Fig. 3c). These results indicate that freeze–thawing a protein/single-layered liposome complex leads to the formation of a multilayered liposome wherein the proteins are located between the lipid layers. Although the mechanism underlying the formation of the multilayered structure and the encapsulation of the protein between the lipid layers has not been investigated in detail, this process might occur via the disruption of the liposome membrane in the freezing step and its subsequent reconstitution during the thawing step. As SOD is surrounded by cationic lipids during the thawing step, the reconstitution of the liposome structure is presumed to occur around the SOD molecules, which would then be distributed between the lipid layers. Further studies are needed to clearly elucidate the mechanism.SOD and liposome cellular uptake
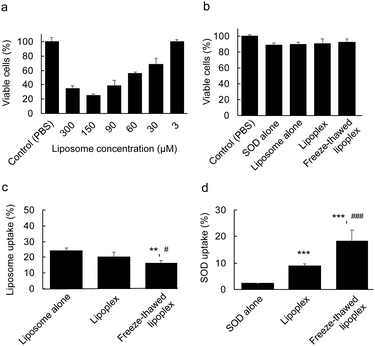
The effects of the encapsulation of SOD between the layers of the multilayered lipoplex on the cytotoxicity and cellular uptake of the liposome and SOD was investigated. We first demonstrated the dose-dependent cytotoxicity of liposome alone. C26NL17 cells were incubated with only liposomes at each concentration for 24 h at 37 °C. The viable cells were then determined by WST-8 assay (Fig. 4a). The liposomes did not show any cytotoxicity at a concentration of 3 μM; therefore, we used this concentration for subsequent experiments. To evaluate the cytotoxicity of PBS, SOD, the liposome, the lipoplex, and the freeze–thawed lipoplex, C26NL17 cells were incubated with each sample for 24 h at 37 °C. Further, viable cells were detected by WST-8 assay (Fig. 4b). All groups showed a survival rate of >90%, indicating the lack of cytotoxicity of these samples at the tested concentration. To study the cellular uptake of the liposomes, C26NL17 cells were incubated with the free liposome, the lipoplex, or the freeze–thawed lipoplex for 24 h (Fig. 4c) using the DiI-labeled liposome. The liposomal uptake of lipoplex (∼20%) was slightly lower than that of the free liposome (∼24%), suggesting that the presence of SOD on the liposomal surface inhibits the liposomal uptake because of the high negative charge of SOD. However, the liposomal uptake of the freeze–thawed lipoplex (∼16%) was lower than that of the free liposome and the lipoplex because the surface charge of the lipoplex decreased after the freeze–thawing process. Additionally, the particle size of the lipoplex increased from ∼150 to ∼230 nm upon freeze–thawing. An increase in the liposome size is known to decrease the cellular uptake.31The cellular uptake ratio of SOD was determined (Fig. 4d). C26NL17 cells were each incubated with free SOD, lipoplex, and freeze–thawed lipoplex samples for 24 h using Cy5–SOD. The results showed that the cellular uptake of SOD was about four times larger in the cell culture containing the lipoplex than in the culture with free SOD. Although SOD alone cannot penetrate the cellular membrane because of its high M.W. and electrostatic repulsion, the lipoplex can effectively deliver SOD inside the membrane. Moreover, a two-fold increase in the cellular uptake of SOD was observed when using the freeze–thawed lipoplex compared with the nonfreeze–thawed lipoplex. As the difference in the amount of immobilized SOD within the freeze–thawed lipoplex (87%) and the lipoplex (50%) was lower than two, a certain amount of liposomal surface-bound SOD would be detached from the lipoplex in the culture medium. The encapsulation of SOD in the freeze–thawed lipoplex enabled the effective delivery of SOD into the cell without detachment from the lipoplex in the medium, resulting in a large amount of SOD being delivered by the freeze–thawed lipoplex than by the nonfreeze–thawed lipoplex.
Localization of SOD and liposome in the cells
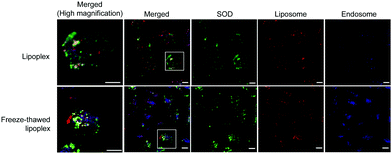
The localization of SOD, liposomes, and endosomes in the cells was investigated using Cy5–SOD and DiI-labeled liposome. C26NL17 cells were each incubated with the lipoplex and the freeze–thawed lipoplex for 24 h, and the late endosomes were stained with LysoTracker; then, the localization of SOD, liposomes, and endosomes was monitored by confocal laser microscopy (Fig. 5). Similar results between the cells treated with the lipoplex and the freeze–thawed lipoplex were obtained. SOD, liposomes, and endosomes were separately localized in the cells, indicating that both the lipoplex and the freeze–thawed lipoplex efficiently escaped from the endosome and that SOD was released from the freeze–thawed lipoplex to the cytoplasm after endosomal escape. Positively charged DCP–DETA interacts with the endosomal membrane, disrupting the membrane at an acidic pH and enhancing the endosomal escape. In addition, the presence of DOPE in the liposome enhances the fusing of the liposome with the endosomal membrane. We previously demonstrated that DCP–DETA or DOPE-deficient liposome and siRNA complexes did not induce gene silencing. This suggests that both lipids play important roles in the release of SOD from the endosome and the freeze–thawed lipoplex. However, as monitoring freeze–thawed lipoplexes in the cell after the endocytosis is challenging, the SOD release mechanism will be the investigated in future studied conducted by our group.Biodistribution of SOD after intravenous injection of the freeze–thawed lipoplex
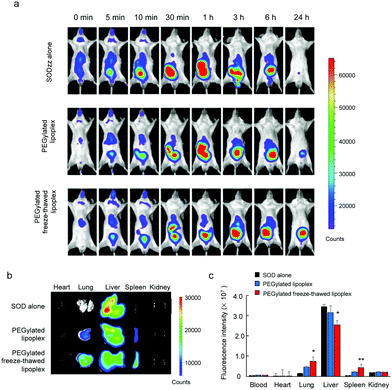
To study the SOD biodistribution after the intravenous injection of the lipoplex and the freeze–thawed lipoplex into living animals, the SOD, the lipoplex, and the freeze–thawed lipoplex samples were intravenously administered to BALB/c mice. Cy5–SOD was used to measure the SOD biodistribution. In addition, the lipoplex and freeze–thawed lipoplex samples were modified using mPEG6000–DSPE to improve their circulation time after the intravenous injection. The size, PDI, and ζ-potential of the PEGylated lipoplex and PEGylated freeze–thawed lipoplex were 130 ± 20 nm, 0.31 ± 0.13, −0.74 ± 4.0 mV, 175 ± 15 nm, 0.25 ± 0.02, and −2.34 ± 1.5 mV, respectively. Real time imaging of Cy5–SOD was demonstrated until 24 h after the intravenous injection of each sample (Fig. 6a). In addition, the fluorescent intensity of Cy5–SOD in each organ was calculated at 24 h after the injection (Fig. 6b and c). Cy5–SOD mainly accumulated in the liver, whereas only a small amount accumulated in other organs, such as lungs, spleen, and kidneys, after the intravenous injection of Cy5–SOD. Meanwhile, the SOD accumulation in the liver decreased after injecting the PEGylated nonfreeze–thawed lipoplex, whereas that in the lung and spleen significantly increased. However, the SOD biodistribution changed markedly after the PEGylated freeze–thawed lipoplex injection. Thus, the SOD accumulation significantly decreased in the liver and increased in the lung and spleen compared with the lipoplex injection. According to these results, the binding of SOD to the liposome in the lipoplex and its encapsulation in the freeze–thawed lipoplex modify the SOD biodistribution. SOD binds to the liposome via noncovalent (electrostatic) interactions; therefore, it can be displaced by other negatively charged and hydrophobic proteins circulating in the bloodstream, such as albumin and apolipoproteins.32,33 This would result in a similar biodistribution of SOD as that obtained after injecting free SOD. In contrast, the encapsulation of SOD in the freeze–thawed lipoplex prevents its detachment from the liposome, significantly changing its biodistribution. These results demonstrate that protein encapsulation is essential for an effective in vivo delivery.Conclusions
An easy one-step method for the encapsulation of proteins into liposomes using the freeze–thawing technology was developed in this study. Negatively charged SOD and a DCP–DETA-based positively charged liposome were used as the protein and liposome models, respectively. The SOD-immobilizing ability of a SOD–liposome complex (lipoplex) increased after freeze–thawing as a result of the encapsulation of SOD within the obtained multilayer liposome. Although the cellular uptake of liposome decreased with the freeze–thawed lipoplex compared with the nonfreeze–thawed lipoplex, the amount of SOD delivered increased significantly, and the SOD, liposomes, and endosomes were separately localized in the cell. The encapsulation of SOD prevented its detachment from the liposome in the bloodstream after the intravenous injection of the freeze–thawed lipoplex in mice, resulting in a SOD distribution that was different from that obtained after injecting free SOD or nonfreeze–thawed lipoplex. Although additional studies are required to optimize the amount of encapsulated SOD and to elucidate the mechanisms underlying the release of SOD from the freeze–thawed lipoplex in the cell and formation of the multilayer liposome structure, the present method provides is useful option for the encapsulation of proteins within liposomes and for protein delivery without requiring organic solvents or specific instruments.Author contributions
H. K., H. O., H. S., N. O. and. T. A. designed the research and H. K., H.O., H.S., and T. A. designed the experiments. H. K., H.O., H. S., M.W., H. S., S. Y., performed preformed in vitro and in vivo assay. T. D., synthesized DCP–DETA. H. K., N. O. and T. A. wrote the manuscript. All authors discussed the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by a Grant-in-Aid for Scientific Research B (19H04450) from the Japan Society for the Promotion of Science (JSPS).References
- I. S. Harris, J. E. Endress, J. L. Coloff, L. M. Selfors, S. K. McBrayer, J. M. Rosenbluth, N. Takahashi, S. Dhakal, V. Koduri, M. G. Oser, N. J. Schauer, L. M. Doherty, A. L. Hong, Y. P. Kang, S. T. Younger, J. G. Doench, W. C. Hahn, S. J. Buhrlage, G. M. DeNicola, W. G. Kaelin and J. S. Brugge, Cell Metab., 2019, 29, 1166 CrossRef CAS PubMed.
- M. E. Kotas and R. Medzhitov, Cell, 2015, 160, 816–827 CrossRef CAS PubMed.
- B. Leader, Q. J. Baca and D. E. Golan, Nat. Rev. Drug Discovery, 2008, 7, 21–39 CrossRef CAS PubMed.
- H. K. Shete, R. H. Prabhu and V. B. Patravale, J. Nanosci. Nanotechnol., 2014, 14, 460–474 CrossRef CAS PubMed.
- Y. W. Lee, D. C. Luther, J. A. Kretzmann, A. Burden, T. Jeon, S. M. Zhai and V. M. Rotello, Theranostics, 2019, 9, 3280–3292 CrossRef CAS PubMed.
- Z. Zhang, W. Shen, J. Ling, Y. Yan, J. Hu and Y. Cheng, Nat. Commun., 2018, 9, 1377 CrossRef PubMed.
- Y. Hirai, H. Hirose, M. Imanishi, T. Asai and S. Futaki, Sci. Rep., 2021, 11, 19896 CrossRef CAS PubMed.
- S. Martins, B. Sarmento, D. C. Ferreira and E. B. Souto, Int. J. Nanomed., 2007, 2, 595–607 CAS.
- G. H. Gao, M. J. Park, Y. Li, G. H. Im, J. H. Kim, H. N. Kim, J. W. Lee, P. Jeon, O. Y. Bang, J. H. Lee and D. S. Lee, Biomaterials, 2012, 33, 9157–9164 CrossRef CAS PubMed.
- P. Ghosh, X. C. Yang, R. Arvizo, Z. J. Zhu, S. S. Agasti, Z. H. Mo and V. M. Rotello, J. Am. Chem. Soc., 2010, 132, 2642–2645 CrossRef CAS PubMed.
- F. Scaletti, J. Hardie, Y. W. Lee, D. C. Luther, M. Ray and V. M. Rotello, Chem. Soc. Rev., 2018, 47, 3421–3432 RSC.
- D. C. Luther, R. Huang, T. Jeon, X. Z. Zhang, Y. W. Lee, H. Nagaraj and V. M. Rotello, Adv. Drug Delivery Rev., 2020, 156, 188–213 CrossRef CAS PubMed.
- H. Zhao, Z. Y. Lin, L. Yildirimer, A. Dhinakar, X. Zhao and J. Wu, J. Mater. Chem. B, 2016, 4, 4060–4071 RSC.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- J. A. Zuris, D. B. Thompson, Y. Shu, J. P. Guilinger, J. L. Bessen, J. H. Hu, M. L. Maeder, J. K. Joung, Z. Y. Chen and D. R. Liu, Nat. Biotechnol., 2015, 33, 73–80 CrossRef CAS PubMed.
- B. Chatin, M. Mevel, J. Devalliere, L. Dallet, T. Haudebourg, P. Peuziat, T. Colombani, M. Berchel, O. Lambert, A. Edelman and B. Pitard, Mol. Ther.--Nucleic Acids, 2015, 4, e244 CrossRef CAS PubMed.
- T. Wei, Q. Cheng, Y. L. Min, E. N. Olson and D. J. Siegwart, Nat. Commun., 2020, 11, 3232 CrossRef CAS PubMed.
- Y. Suzuki, H. Onuma, R. Sato, Y. Sato, A. Hashiba, M. Maeki, M. Tokeshi, M. E. H. Kayesh, M. Kohara, K. Tsukiyama-Kohara and H. Harashima, J. Controlled Release, 2021, 330, 61–71 CrossRef CAS PubMed.
- T. Terada, J. A. Kulkarni, A. Huynh, S. Chen, R. van der Meel, Y. Y. C. Tam and P. R. Cullis, Langmuir, 2021, 37, 1120–1128 CrossRef CAS PubMed.
- S. B. van Witteloostuijn, S. L. Pedersen and K. J. Jensen, ChemMedChem, 2016, 11, 2474–2495 CrossRef CAS PubMed.
- S. Futaki, J. V. V. Arafiles and H. Hirose, Chem. Lett., 2020, 49, 1088–1094 CrossRef CAS.
- H. Koide, A. Okamoto, H. Tsuchida, H. Ando, S. Ariizumi, C. Kiyokawa, M. Hashimoto, T. Asai, T. Dewa and N. Oku, J. Controlled Release, 2016, 228, 1–8 CrossRef CAS PubMed.
- A. Okamoto, H. Koide, N. Morita, Y. Hirai, Y. Kawato, H. Egami, Y. Hamashima, T. Asai, T. Dewa and N. Oku, J. Controlled Release, 2019, 295, 87–92 CrossRef CAS PubMed.
- S. Mochizuki, N. Kanegae, K. Nishina, Y. Kamikawa, K. Koiwai, H. Masunaga and K. Sakurai, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 412–418 CrossRef CAS PubMed.
- Z. Du, M. M. Munye, A. D. Tagalakis, M. D. Manunta and S. L. Hart, Sci. Rep., 2014, 4, 7107 CrossRef PubMed.
- P. R. Harrigan, T. D. Madden and P. R. Cullis, Chem. Phys. Lipids, 1990, 52, 139–149 CrossRef CAS PubMed.
- S. Ghanbarzadeh, H. Valizadeh and P. Zakeri-Milani, Bioimpacts, 2013, 3, 75–81 Search PubMed.
- A. S. Nosova, O. O. Koloskova, A. A. Nikonova, V. A. Simonova, V. V. Smirnov, D. Kudlay and M. R. Khaitov, MedChemComm, 2019, 10, 369–377 RSC.
- H. Hatakeyama, H. Akita and H. Harashima, Biol. Pharm. Bull., 2013, 36, 892–899 CrossRef CAS PubMed.
- W. F. Wolkers, H. Oldenhof, F. Tablin and J. H. Crowe, Biochim. Biophys. Acta, Biomembr., 2004, 1661, 125–134 CrossRef CAS PubMed.
- A. U. Andar, R. R. Hood, W. N. Vreeland, D. L. Devoe and P. W. Swaan, Pharm. Res., 2014, 31, 401–413 CrossRef CAS PubMed.
- H. Akita, T. Nakatani, K. Kuroki, K. Maenaka, K. Tange, Y. Nakai and H. Harashima, Int. J. Pharm., 2015, 490, 142–145 CrossRef CAS PubMed.
- K. Taguchi, Y. Okamoto, K. Matsumoto, M. Otagiri and V. T. G. Chuang, Pharmaceuticals, 2021, 14, 296 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |