Detection of selective androgen receptor modulators (SARMs) in serum using a molecularly imprinted nanoparticle surface plasmon resonance sensor†
Alisha
Henderson
a,
Mark V.
Sullivan
a,
Rachel A.
Hand
 b and
Nicholas W.
Turner
b and
Nicholas W.
Turner
 *a
*a
aLeicester School of Pharmacy, De Montfort University, The Gateway, Leicester, LE1 9BH, UK. E-mail: nicholas.turner@dmu.ac.uk
bDepartment of Chemistry, University of Warwick, Library Road, Coventry, CV4 7AL, UK
First published on 6th June 2022
Abstract
Selective Androgen Receptor Modulators (SARMs) are a fairly new class of therapeutic compounds that act upon the androgen receptor. They proffer similar anabolic properties to steroids, but with a much-reduced androgenic profile. They have become a popular substance of abuse in competitive sport. Being relatively new, detection systems are limited to chromatographic methods. Here we present a surface plasmon resonance sensor for three commonly-used SARMS, Andarine, Ligandrol and RAD-140, using high-affinity molecularly imprinted nanoparticles (nanoMIPs) as the recognition element. Synthesised nanoMIPS exhibited dissociation constant (KD) values of 29.3 nM, 52.5 nM and 75.1 nM for Andarine, Ligandrol and RAD-140 nanoMIPs, respectively. Cross-reactivity of the particles was explored using the alternative SARMs, with the nanoMIPs demonstrating good specificity. Fetal Bovine Serum (FBS) was used to assess the ability of the SPR-based nanoMIP sensor to detect the target compounds in a comparable biological matrix, with observed KD values of 12.3 nM, 31.9 nM and 28.1 nM for Andarine, Ligandrol and RAD-140 nanoMIPs, respectively. Theoretical limits of detection (LoD) were estimated from a calibration plot in FBS and show that the nanoMIP-based sensors have the potential to theoretically measure these SARMs in the low to sub nM range. Crucially these levels are below the minimum required performance limit (MRPL) set for these compounds by WADA. This study highlights the power of modern molecular imprinting to rapidly address required molecular recognition for new compounds of interest.
Introduction
Selective androgen receptor modulators (SARMs) are a novel class of androgen receptor ligands that bind to androgenic receptors and display tissue-selective activation.1 SARMs are intended to have similar effects as androgenic drugs, but with more selectivity in their action, thus allowing SARMs to have more uses than those of anabolic steroids.2 SARMs have the potential to be used as alternatives therapies in treatment of diseases where steroidal androgens have been proposed as therapeutics. In example, the initial emphasis of the clinical development of SARMs was for treatment of muscular dystrophy.2 In example, Ponnusamy et al. highlights the potential use of SARMs in a Duchenne muscular dystrophy preclinical model, where the animals exhibited increases muscle mass and protein synthesis levels, comparable to that observed with oxandrolone, but without the off-target side effects.3These anabolic effects, combined with the lack of androgenic side effects have resulted in SARMs being of interest to the body building community, thus creating the potential for abuse among competitive athletes. While, there is a lack of FDA approval, various SARMs molecules are available for purchase online from unverifiable sources and due to their potential for abuse, in both amateur and elite sport, the World Anti-Doping Agency (WADA) included SARMs in the prohibited substance list in 2008.4 Despite this, a clear increase in SARMs use has been observed.5 The majority of sports drug testing approaches, that efficiently analyse for SARMs are based upon mass spectrometry methods.6 These methods rely on specific knowledge in regards to the drugs’ composition and metabolic fate within humans, which to date has been limiting in scientific literature. This has required drug candidates to be subjected to in-depth mass spectrometry studies, in order to provide enough analytical data.7
The use of androgen receptor-based bioassays is a developing strategy that is seen as a potential alternative to the mass spectroscopy approaches, but this is currently not routinely used by drug testing laboratories and shows that alternative methods for SARMs detection need to be developed.8 A recent review by Kintz5 highlights the complexity and sensitivity of any sensor needed given that a small oral administration of a SARM (e.g. ostarine) of 10 μg can lead to an observed urine concentration above the WADA minimum required performance level (MRPL) of 2 ng mL−1. SARMs are known to be still be present up to 60 days after dosage.6
Commonly SARMs are detected by chromatographic methods often linked with mass spectrometric detector systems, from a variety of media including hair, urine,9,10 nails11 and serum.12 In all these, low or sub nM detection (pg or ng per mL mg−1) is observed. As in all analytical detection there is always interest to consider biosensor platforms as an alternative platform due to their inherent benefits in cost, time and portability. This holds true for compounds such as anabolic agents where detection is not just in the clinical of sports setting, but increasingly is required in the workplace (or military services), where monitoring is used to ensure staff and personnel are complying with rules. However, a literature search at time of writing, yielded no examples of biosensor development studies for SARMs beyond application of cell bioassays13 to measure compound performance. This leaves an interesting opportunity to explore.
Surface Plasma Resonance (SPR) has proven itself to be a flexible system for development of biosensors, exhibiting a good level of stability and sensitivity.14 It has also become a technique by which the performance (affinity/selectivity) of newly developed affinity components is tested. SPR-based biosensors have been previously used successfully within the fields of clinical diagnosis, environmental contamination, and food safety, whereby antibodies and enzymes are used as recognition materials,14,15 due to their strong affinities, specificity towards target analytes and sensitive levels of detection.14,16 However, they offer limited reusability; can be costly and time-consuming to produce. Additionally, stability is not guaranteed.17,18
Molecularly imprinted polymers (MIPs) are synthetic recognition alternatives that have the potential to overcome the issues associated with biological-based recognition materials.19 They offer high affinity and selectivity, as well as being resistant to the extremes of environment making them ideal for working in biological systems.20,21 The general principle of molecular imprinting is relatively straight-forward, where a recognition site within a cross-linked matrix is formed around a target template, commonly through non-covalent interactions,20–22 which is then removed to leave an “imprint”. The recent development of MIP nanoparticles (nanoMIPs) has earned particular interest due their performance and improved applicability for biological systems, offering the possibility to improve traditional analytical techniques found in biochemistry, chemistry, biomedical and environmental fields.23,24
The physical properties and performance of these are now challenging their natural recognition element counterparts.25–27 MIPs have been shown to be compatible with SPR sensor systems,28,29 while nanoMIPs contain amino (–NH2) functionality has enabled easy immobilisation, using an EDC/NHS coupling method, onto the surface of gold SPR chips preconditioned with carboxyl (–COOH) groups.30 This opens up further potential possibilities of employing MIPs as the specific recognition elements in optical sensors.
Here we present, for the first time, the synthesis of nanoMIPs for three commonly-found SARMs, Andarine, Ligandrol, and RAD-140, presented in Fig. 1, using a solid-phase approach. We have used SPR to determine the affinities for the nanoMIPs, which were calculated using SPR kinetic studies, along with specificity via cross-reactivity studies, by using the non-templated SARMs as controls. We have then applied this to a sensor system, by spiking samples of fetal bovine serum, with the target SARMs enabled the exploration of the efficiency for the nanoMIPs and subsequent SPR sensor, within a biological matrix. To date, SARMs have not been targeted using molecular imprinting, making this work novel through its approach and potential application in both anti-doping and clinical environs.
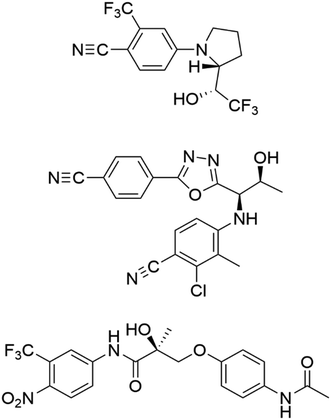 | ||
| Fig. 1 The structures of ligandrol (top), RAD-140 (centre) and andarine (bottom) – common SARMs used in both therapeutic and sports doping settings. | ||
Results and discussion
Using a solid-phase synthesis approach, adapted from the work of Poma et al.,23 molecularly imprinted nanoparticles (nanoMIPs) produced for the target SARMs, Andarine, Ligandrol and RAD-140. This is the first time that these compounds have been imprinted. The synthetic methods used generated MIP solutions of 91.1 ± 4.8 μg mL−1, 56.7 ± 2.8 μg mL−1 and 71.1 ± 3.7 μg mL−1 for the Andarine, Ligandrol and RAD-140 nanoMIPs, respectively. With approximately 100 mL of solution produced, the reaction provided enough material for this study. The yields matched those of prior work.31The size of the nanoMIPs were estimated using dynamic light scattering (DLS) and are presented in Fig. S1 (ESI†). The diameters of the particles are shown to be 52.7 ± 2.4 nm, 55.3 ± 3.1 nm and 48.8 ± 5.7 nm, polydispersity index values (PDI) of 0.185 ± 0.1 nm, 0.241 ± 0.2 nm, and 0.214 ± 0.1 nm, at 25 °C, for Andarine (Fig. S1A, ESI†), Ligandrol (Fig. S1B, ESI†) and RAD-140 (Fig. S1C, ESI†), respectively. The DLS curves shown in Fig. S1 (ESI†), display an excellent Gaussian distribution, further supporting this nanoMIP synthesis protocol produces regular homogenous particles, with Fig. S1B (ESI†) showing a particularly narrow size distribution range (compared with Fig. S1A and C, ESI†).
This methodology uses EDC/NHS coupling chemistry to enable deposition of the nanoparticles on the surface of the SPR chip. With a high percentage of amine-functionality within the nanoparticles (from NAPA, TBAm and NIPAm) this chemistry is favoured. The gold SPR chips used were pre-functionalised with a carboxymethyl dextran hydrogel layer as this provides a good deposition profile, due to the ease of activation of the hydrogel by the EDC/NHS.
Ethanolamine is used to deactivate any unwanted any unreacted carboxyl groups on the SPR chip surface, while also washing away any unbound nanoMIPs. Due to this selected deposition method a single layer of nanoMIPs is expected as the nanoMIPs being unable to bind to themselves. The initial deposition of nanoMIPs was added in excess in order to achieve full coverage on the chip, thus giving the potential maximum population of binding sites available per chip. By having a theoretical maximal receptor (binding population), standard models for ligand/receptor interactions can be applied. Given the limited size of the nanoMIPs we have applied a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 kinetic model.
1 kinetic model.
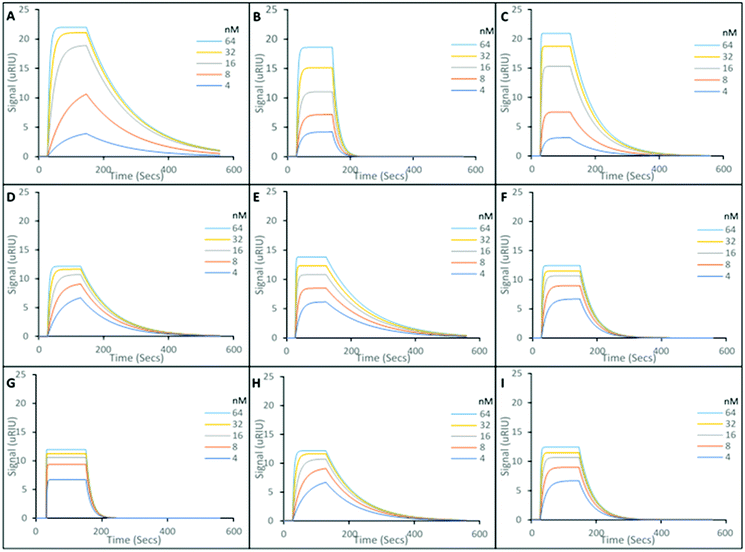
The SPR sensorgrams presented in Fig. 2, show the interactions of five different concentrations of the target molecules (Andarine, Ligandrol, and RAD-140), with their corresponding nanoMIPs (Fig. 2A, B and C, respectively), immobilised onto the surface of the sensor. From these curves and the application of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 model we are able to elucidate the overall equilibrium dissociation constant (KD) for the target interacting with their nanoMIPs, this is summarised in Table 1.
1 model we are able to elucidate the overall equilibrium dissociation constant (KD) for the target interacting with their nanoMIPs, this is summarised in Table 1.
| K D (M) | |||
|---|---|---|---|
| Andarine | Ligandrol | RAD-140 | |
| Andarine nanoMIP | 2.93 × 10−8 (± 0.43 × 10−8) | 2.99 × 10−6 (± 0.28 × 10−6) | 5.31 × 10−6 (± 0.37 × 10−6) |
| Ligandrol nanoMIP | 1.89 × 10−6 (± 0.26 × 10−6) | 5.25 × 10−8 (± 0.62 × 10−8) | 2.15 × 10−6 (± 0.13 × 10−6) |
| RAD-140 nanoMIP | 5.25 × 10−6 (± 0.62 × 10−6) | 1.38 × 10−6 (± 0.48 × 10−6) | 7.51 × 10−8 (± 0.64 × 10−8) |
The interaction of the SARMs molecules and their corresponding nanoMIPs were calculated with the KD values shown to be 29.3 nM, 52.5 nM and 75.1 nM for the Andarine, Ligandrol and RAD-140 nanoMIPs, respectively. The KD values found in this work are similar to those previously found for this type of nanoMIP, imprinted for small molecules. Poma et al. produced at nanoMIP for the target melamine, whereby a KD value of 63 nM, which in applicable terms is comparable to monoclonal antibodies23 and our own work using nanoMIPs shows similar for moxifloxacin, a fluoroquinolone antibiotic.26
To explore the specificity of the nanoMIPs, the cross-reactivity and non-specific binding was investigated by loading non-target SARMs onto the nanoMIP coated gold chip. Ligandrol and RAD-140 was used to test the Andarine nanoMIP (Fig. 2D and G, respectively); Andarine and RAD-140 for the Ligandrol nanoMIP (Fig. 2E and H, respectively); and finally, Andarine and Ligandrol was used for the RAD-140 nanoMIP (Fig. 2F and I, respectively). All experiments were performed in triplicate to produce an average. The KD's for the non-target SARM molecules interacting with the nanoMIPs were estimated via the Tracedrawer software, summarised in Table 1.
Where the nanoMIPs were loaded with a non-target molecule, KD values in μM range are observed, thus demonstrating target specificity with a 100-fold decrease in affinity. This is consistent with observed cross-reactivity in similarly produced nanoMIPs.
The SARMs under study have a range of medical applications, but have also due to their mode of action produce gains in body mass and protein synthesis, and as such they are gaining traction as body building supplements and have the potential for abuse amongst athletes, hence SARMs being included on the banned list by WADA. The methodology presented above (analysis in buffer) is suitable for testing of supplements as they can be prepared easily in a lab for testing however, the potential for screening in medical or sports environment is an attractive proposition. Towards this end we have explored the potential to detect SARMs in fetal bovine serum (FBS) as a mimic for testing in a blood sample. FBS was spiked at a series of concentrations of the target analytes between 4–64 nM and exposed to the nanoMIP-modified sensor surface.
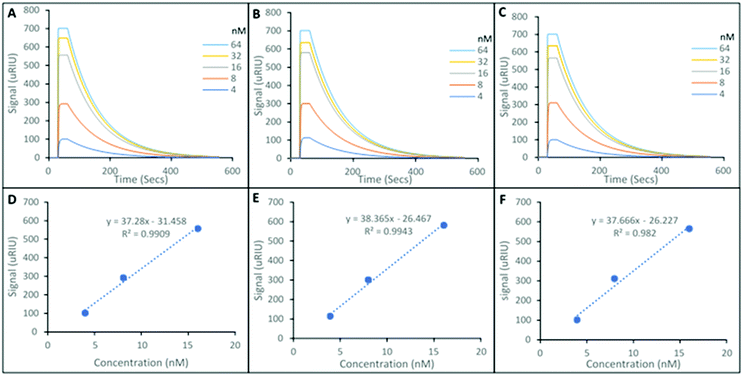
Fig. 3 shows representative SPR sensorgrams for the interactions of the SARMS in FBS, and their corresponding nanoMIP loaded SPR sensor chip. Andarine shown in Fig. 3A, Ligandrol shown in Fig. 3B and RAD-140 shown in Fig. 3C. These were repeated in triplicate and from this concentration calibrations were plotted, with Andarine, Ligandrol, and RAD-140 shown in Fig. 3D, E and F, respectively. These curves allowed for the estimation of a theoretical LOD. As a note, only three points were used to produce the calibration curves in Fig. 3D, E and F, this was due to these points being the linear proportion of the calibration. The full calibrations are presented in Fig. S2 (ESI†), showing the saturation that is also observed in the processed curves in Fig. 3(A)–(C).
When compared to the data in Fig. 2, the first obvious difference is the overall size of signal. There is a significant matrix effect observed. This is not surprising given the significant differences in density and optical properties between FBS and PBST and it is a common occurrence, and a known effect of SPR which refers to an optical phenomenon, and monitors changes in the refractive index.
This leads to a saturation of signal at higher concentrations, observed in the SPR sensorgrams in Fig. 3A–C), and was consistent across all replicates. Therefore, only the lower three concentrations (4, 8 and 16 nM) – the linear portion of the calibration were used to predict the calculated KD values and estimated theoretical lower LOD's. This is summarised in Table 2.
| (M) | |||
|---|---|---|---|
| Andarine | Ligandrol | RAD-140 | |
| Target reloaded from spiked FBS sample | 1.23 × 10−8 (± 0.18 × 10−8) | 3.19 × 10−8 (± 0.52 × 10−8) | 2.81 × 10−8 (± 0.34 × 10−8) |
| Theoretical LOD of target reloaded from spiked FBS sample | 8.44 × 10−10 | 6.90 × 10−10 | 6.96 × 10−10 |
As a team we have explored this type of sensor with other compounds in a range of matrices including river water and food samples.31
The KD values shown in Table 2, are consistent with those present in Table 1, with 29.3 nM, 52.5 nM and 75.1 nM (Table 1) compared with 12.3 nM, 31.9 nM and 28.1 nM (Table 2) for the Andarine, Ligandrol and RAD-140 nanoMIPs, respectively.
The slight differences observed are expected given the optical effect of the matrix and that the environmental differences in the injected sample will differ from PBST, in terms of pH, ionic strength etc., which will affect binding interactions.
This highlights that, as for many biosensor devices, recognition and affinity should be calibrated per application. For example, MIP-SPR detection of SARMs in urine should be possible, but the performance would have to be studied separately.
Overall, this data shows that the detection of SARMs is achievable from a biological matrix (FBS) with theoretical LOD limits are calculated as 0.84 nM, 0.69 nM and 0.70 nM for Andarine, Ligandrol and RAD-140, respectively, showing that the nanoMIPs produced have the ability to detect low concentrations of the target molecules.
This equates to detection in the ×10−10 g mL−1, is under the MRPL limit of 2 ng mL−1 required by WADA6 and is within the lower limit of detection of published mass spectroscopic techniques (0.2–10 ng mL−1).32
Experimental
Materials and equipment
All chemicals and solvents were analytical quality or high-performance liquid chromatography (HPLC) grade and were used as found without further purification.Acrylic acid (AA), 3-aminopropyltrimethyloxy-silane (APTMS), ammonium persulfate (APS), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), glutaraldehyde (GA), glycine, N-(3-aminopropyl)methacrylamide hydrochloride (NAPA), N,N′-methylenebisacrylamide (BIS), N-hydroxysuccinimide (NHS), N-isopropylacrylamide (NIPAm), sodium dodecyl sulphate (SDS), N-tert-butylacrylamide (TBAm), and tetramethylethyldiamide (TEMED), were all purchased from Sigma-Aldrich (Poole, Dorset, UK).
Acetone, acetonitrile (dry), dipotassium phosphate, disodium phosphate, ethanolamine, ethylenediaminetetraacetic acid (EDTA), methanol, potassium chloride, sodium hydroxide and Tween 20 were all purchased from Fisher Scientific UK (Loughborough, Leicester, UK).
Andarine, Ligandrol, and RAD-140 were purchased from Biosynth Carbosynth, Compton, Berkshire, UK.
Glass beads (75 μm diameter) were purchased from Microbeads AG, (Brugg, Switzerland) and used as found. Carboxymethyl Dextran Hydrogel Surface Sensor chips were purchased from Reichert Technologies Life Sciences, Buffalo, New York, USA.
Phosphate buffered saline (PBS) was made at 10 mM with pH 7.4. The running buffer (PBST) was a phosphate buffered saline made at 10 mM, pH 7.4, with 0.01% Tween 20 added. This small amount of Tween20 (0.01%) is present in order to reduce the amount of non-specific binding during rebinding studies.
Preparation of template derivatized glass beads
The preparation of the glass beads was as described in our prior work.26 In summary, for 30 g of beads, these were treated by boiling them for fifteen minutes in 4 M sodium hydroxide (24 mL) followed by washing in distilled water until the solution was pH 7. Following this, they were rinsed with acetone and dried at 80 °C.The beads were placed into a (3%, v/v) solution of APTMS in anhydrous toluene (12 mL) at 60 °C overnight, washed with acetone and methanol and oven-dried (150 °C for 30 minutes).
The selected template (9 mg of compound) was dissolved in 15 mL of 7% glutaraldehyde, PBS solution and purged with nitrogen. To this mixture 30 g of pre-prepared beads was added under nitrogen, at room temperature and left for 15 hours. The beads were then washed in water and dried under vacuum at ambient. These were used directly.
Solid-phase synthesis of SARMs imprinted nanoMIPs
This process is similar to our prior work,26 using a standardised mixture suggested by Canfarotta.24 Based on this work we maintained a polymerisation mixture of NIPAm (20 mg), NAPA (7 mg), BIS (1 mg), and AA (2.2 μL) in 49 mL of double distilled water, to which a solution of TBAm in ethanol (10 mg in 250 μl) was added. This solution is vacuum degassed while sonicating and then sparged with nitrogen. The pre-prepared beads (30 g) were added to this mixture followed by TEMED (12.5 μL) and APS in double distilled water (15 mg in 250 μL) to initiate polymerization, all under nitrogen. The 100 mL bottle was gently agitated for 1 hour at room temperature.The beads were filtered using 11 μm paper and washed with water (8 × 30 mL) at ambient temperature to remove the impurities, unreacted monomers, and low-affinity nanoMIPs. To elute high affinity nanoMIPs, the beads were heated in water (40 mL) to 60 °C, then filtered again and washed with 60 °C water in aliquots until approximately 100 mL of solution bearing the nanoparticles was collected. All nanoMIP solutions were stored at 4 °C.
The same solid-phase approach and method, for the production of nanoMIPs, was used for all the different SARMs nanoMIPs.
Dynamic light scattering
Particle size at 25 °C (effective hydrodynamic diameters (dh) was measured using a Brookhaven NanoBrook Omni spectrometer using Particle Solutions v 2.6) with n = 5 using this solution.SPR analysis
A 3 mL aliquot of the aqueous nanoparticle solution was dried and weighed, then resuspended as needed allowing for a concentration (in μg mL−1) of the initial solution to be calculated.Affinity and specificity of the imprinted nanoparticles for the different targets were studied using a Reichert 2 SPR system (Reichert Technologies, Buffalo, USA) with attached autosampler.
To immobilise the nanoMIPs, a carboxymethyl dextran hydrogel coated Au chip was preconditioned in PBS for 10 minutes then PBST both at 10 μL min−1 within the SPR. Then 1 mL of aqueous EDC/NHS solution (40 mg EDC and 10 mg NHS respectively) was passed over the chip (6 minutes at 10 μL min−1). Once prepared, 1 mL of 300 μg mL−1 of nanoMIPs in PBST (with 10 mM sodium acetate), was passed over the left channel (working channel) of the chip for 1 minute. Then quenching solution (1 M ethanolamine, pH 8.5) was added over both channels for 8 minutes; followed by a continuous flow of PBST at 10 μL min−1 All injections were taken from a stable baseline.
An effective rebinding method was developed prior to this work27 enabling kinetics of rebinding to be measured. Briefly this was a 2 minute association, 5 minute dissociation and a 1 minute regeneration cycle (regeneration buffer 10 mM glycine–HCl, pH 2) followed by a final stabilisation cycle (PBST for 1 minute). PBST was used throughout with the association ranges of analyte between 4–64 nM; alongside a blank association to baseline zero. In all cases, this was carried out in least triplicate.
Signals from reference channel were subtracted from signals from the working channel to give specific binding. The SPR responses were fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Langmuir fit bio-interaction (BI) model using the Reichert TraceDrawer software. Association rate constants (ka), dissociation rate constants (kd), and maximum binding (Bmax) were fitted globally, whereas the BI signal was fitted locally. Equilibrium dissociation constants (KD) were calculated by kd/ka.
1 Langmuir fit bio-interaction (BI) model using the Reichert TraceDrawer software. Association rate constants (ka), dissociation rate constants (kd), and maximum binding (Bmax) were fitted globally, whereas the BI signal was fitted locally. Equilibrium dissociation constants (KD) were calculated by kd/ka.
FBS was then spiked with the association ranges of analyte between 4–64 nM, alongside a blank association to baseline zero, and then injected to performed repeated SPR analysis, but in complex media.
Finally, calibration curves for each analyte were produced form this data taking n = 3 average and a theoretical limit of detection (LOD) calculated. Where signal saturation was observed (noted in more complex matrices), the linear section of the curve was used for this calculation.
Conclusions
Here, we have developed a series of molecularly imprinted nanoparticles for the specific recognition of the SARMs Andarine, Ligandrol, and RAD-140. These nanoMIPs were produced using a solid-phase synthesis method capable producing nanoMIP particles with high affinity (KD values of target interactions are in ×10−8 M range) and high selectivity (KD values of non-target interactions are in ×10−6 M range). The KD values observed in this study are consistent with previous studies and are equivalent to those of antibodies. Prior to the affinity testing of the SPR-based optical sensor, the nanoMIPs were characterised for quality and size, with the nanoMIPs found to be uniform. The nanoMIPs were immobilised onto a carboxymethyl dextran hydrogel-coated Au chip, which was activated using EDC and NHS, to allow for covalent coupling, these were then used to detect the target SARM molecules, displaying high affinity and high specificity. To demonstrate the ability of the nanoMIPs to select target SARMs molecules from a complex relevant media, FBS was spiked with the target SARMs. The nanoMIPs again, produced high affinity (KD values within the ×10−8 M range), consequently showing the potential for nanoMIPs to be used for the detection of SARMs from biological samples.As we have demonstrated in this paper, these simple to produce and cost effective nanoMIPs are able to offer affinities comparable to their biological counterparts and have the potential to lead to the development of new recognition materials that are able to offer suitable affinities for recognition in biological matrices. This work highlights the importance of developing matrix-specific protocols – a published figure for affinity of a material may not be true for all applications – it is therefore vital to explore performance on a case-by-case (or application-by-application) basis.
This paper does however, offer a suggestion at the potential of artificial recognition for anti-doping applications. We targeted serum based on the potential of this to be used in clinical setting to monitor drug use; and as the target molecule will be present in circulation. We are currently exploring the potential to detect these compounds (and their metabolites) in urine which will favour less invasive analysis. With these nanoMIPs we are also currently exploring their potential for application in ELISA format,33 but also as potential targeted clean-up for chromatographic applications.
Author contributions
AH was responsible for investigation and formal analysis. MS and RH were responsible for supervision and methodology. NT was responsible for methodology, writing (with MS) and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
MS and NT would like to thank EPSRC for financial support for this work (EP/S003339/1). RH and NT wish to thank the Partnership for Clean Competition (PCC) for support (2017R2000292G). AH wishes to thank Professor Anwar Baydoun and the Faculty of Health & Life Sciences graduate program for financial support.Notes and references
- S. Bhasin and R. Jasuja, Selective androgen receptor modulators as function promoting therapies, Curr. Opin. Clin. Nutr. Metab. Care, 2009, 12(3), 232–240 CrossRef CAS PubMed.
- I. V. Efimenko, D. Valancy, J. M. Dubin and R. Ramasamy, Adverse effects and potential benefits among selective androgen receptor modulators users: A cross-sectional survey, Int. J. Impotence Res., 2021 DOI:10.1038/s41443-021-00465-0.
- S. Ponnusamy, R. D. Sullivan, D. You, N. Zafar, C. He Yang and T. Thiyagarajan, et al., Androgen receptor agonists increase lean mass, improve cardiopulmonary functions and extend survival in preclinical models of duchenne muscular dystrophy, Hum. Mol. Genet., 2017, 26(13), 2526–2540 CrossRef CAS PubMed.
- World Anti-Doping Agency, The 2008 prohibited list international standard, The world Anti-Doping Code, 2007, pp. 1–11.
- P. Kintz, The forensic response after an adverse analytical finding (doping) involving a selective androgen receptor modulator (SARM) in human athlete, J. Pharm. Biomed. Anal., 2022, 207, 114433 CrossRef CAS PubMed.
- M. Thevis and W. Schänzer, Detection of SARMs in doping control analysis, Mol. Cell. Endocrinol., 2018, 464, 34–45 CrossRef CAS PubMed.
- M. Thevis, T. Kuuranne, K. Walpurgis, H. Geyer and W. Schänzer, Annual banned-substance review: Analytical approaches in human sports drug testing, Drug Test. Anal., 2016, 8(1), 7–29 CrossRef CAS PubMed.
- K. Bailey, T. Yazdi, U. Masharani, B. Tyrrell, A. Butch and F. Schaufele, Advantages and limitations of androgen receptor-based methods for detecting anabolic androgenic steroid abuse as performance enhancing drugs, PLoS One, 2016, 11(3), e0151860 CrossRef PubMed.
- C. Stacchini, F. Botrè, F. Comunità, X. de la Torre, A. P. Dima and M. Ricci, et al., Simultaneous detection of different chemical classes of selective androgen receptor modulators in urine by liquid chromatography-mass spectrometry-based techniques, J. Pharm. Biomed. Anal., 2021, 195, 113849 CrossRef CAS PubMed.
- S. Divari, E. Berio, P. Pregel, A. Sereno, L. Chiesa and R. Pavlovic, et al., Effects and detection of nandrosol and ractopamine administration in veal calves, Food Chem., 2017, 221, 706–713 CrossRef CAS PubMed.
- P. Kintz, L. Gheddar, A. Ameline and J. S. Raul, Identification of S22 (ostarine) in human nails and hair using LC-HRMS. application to two authentic cases, Drug Test. Anal., 2020, 12(10), 1508–1513 CrossRef CAS PubMed.
- E. Ventura, A. Gadaj, T. Buckley and M. H. Mooney, Development of a multi-residue high-throughput UHPLC–MS/MS method for routine monitoring of SARM compounds in equine and bovine blood, Drug Test. Anal., 2020, 12(9), 1373–1379 CrossRef CAS PubMed.
- T. F. H. Bovee, M. Thevis, A. R. M. Hamers, A. A. C. M. Peijnenburg, M. W. F. Nielen and W. G. E. J. Schoonen, SERMs and SARMs: Detection of their activities with yeast based bioassays, J. Steroid Biochem. Mol. Biol., 2010, 118(1), 85–92 CrossRef CAS PubMed.
- Y. Tang, X. Zeng and J. Liang, Surface plasmon resonance: An introduction to a surface spectroscopy technique, J. Chem. Educ., 2010, 87(7), 742–746 CrossRef CAS PubMed.
- S. Firdous, S. Anwar and R. Rafya, Development of surface plasmon resonance (SPR) biosensors for use in the diagnostics of malignant and infectious diseases, Laser Phys. Lett., 2018, 15(6), 65602 CrossRef.
- M. A. Morales and J. M. Halpern, Guide to selecting a biorecognition element for biosensors, Bioconjugate Chem., 2018, 29(10), 3231–3239 CrossRef CAS PubMed.
- P. S. Sharma, Z. Iskiero, A. Pietryzyk-Le, F. D'Souza and W. Kutner, Bioinspired intelligent molecularly imprinted polymers for chemosensing: A mini review, Electrochem. Commun., 2015, 50, 81–87 CrossRef CAS.
- J. Wackerlig and R. Schirhagl, Applications of molecularly imprinted polymer nanoparticles and their advances toward industrial use: A review, Anal. Chem., 2016, 88(1), 250–261 CrossRef CAS PubMed.
- N. W. Turner, C. W. Jeans, K. R. Brain, C. J. Allender, V. Hlady and D. W. Britt, From 3D to 2D: A review of the molecular imprinting of proteins, Biotechnol. Prog., 2006, 22, 1474–1489 CrossRef CAS PubMed.
- K. Mosbach and O. Ramstron, The emerging technique of molecular imprinting and its future impact on biotechnology, Nat. Biotechnol., 1996, 14, 163–170 CrossRef CAS.
- M. V. Sullivan, W. J. Stockburn, P. C. Hawes, T. Mercer and S. M. Reddy, Green synthesis as a simple and rapid route to protein modified magnetic nanoparticles for use in the development of a fluorometric molecularly imprinted polymer-based assay for detection of myoglobin, Nanotechnology, 2021, 32(9), 095502 CrossRef CAS PubMed.
- M. V. Sullivan, S. R. Dennison, G. Archontis, J. M. Hayes and S. M. Reddy, Towards rational design of selective molecularly imprinted polymers (MIPs) for proteins: Computational and experimental studies of acrylamide based polymers for myoglobin, J. Phys. Chem. B, 2019, 123, 5432–5443 CrossRef CAS PubMed.
- A. Poma, A. Guerreiro, M. J. Whitcombe, E. V. Piletska, A. P. F. Turner and S. A. Piletsky, Solid-phase synthesis of molecular imprinted polymer nanoparticles with a reusable template – “plastic antibodies”, Adv. Funct. Mater., 2013, 23, 2821–2827 CrossRef CAS PubMed.
- F. Canfarotta, A. Poma, A. Guerreiro and S. A. Piletsky, Solid-phase synthesis of molecularly imprinted nanoparticles, Nat. Protoc., 2016, 11(3), 443–455 CrossRef CAS PubMed.
- S. Subrahmanyam, A. Guerreiro, A. Poma, E. Moczko, E. Piletska and S. Piletsky, Optimisation of experimental conditions for the synthesis of high affinity MIP nanoparticles, Eur. Polym. J., 2013, 49, 100–105 CrossRef CAS.
- M. V. Sullivan, F. Allabush, D. Bunka, A. Tolley, P. M. Mendes and J. H. R. Tucker, et al., Hybrid aptamer-molecularly imprinted polymer (AptaMIP) nanoparticles selective for the antibiotic moxifloxacin, Polym. Chem., 2021, 12, 4405 RSC.
- M. V. Sullivan, O. Clay, M. P. Moazami, J. K. Watts and N. W. Turner, Hybrid aptamer-molecularly imprinted polymer (aptaMIP) nanoparticles from protein recognition-A trypsin model, Macromol. Biosci., 2021, 21(5), e2100002 CrossRef PubMed.
- N. Cennamo, G. D’Agostino, M. Pesavento and L. Zeni, High selectivity and sensitivity sensor based on MIP and SPR in tapered plastic optical fibers for the detection of l-nicotine, Sens. Actuators, B, 2014, 191, 529–536 CrossRef CAS.
- X. Xu, X. Tian, L. Cai, Z. Xu, H. Lei and H. Wang, et al., Molecularly imprinted polymer based surface plasmon resonance sensors for detection of sudan dyes, Anal. Methods, 2014, 6(11), 3751–3757 RSC.
- A. H. M. Safaryan, A. M. Smith, T. S. Bedwell, E. V. Piletska, F. Canfarotta and S. A. Piletsky, Optimisation of the preservation conditions for molecularly imprinted polymer nanoparticles specific for trypsin, Nanoscale Adv., 2019, 1(9), 379–3714 RSC.
- M. V. Sullivan, A. Henderson, R. A. Hand and N. W. Turner, A molecularly imprinted polymer nanoparticle-based surface plasmon resonance sensor platform for antibiotic detection in river water and milk, Anal. Bioanal. Chem., 2022, 414(12), 3687–3696 CrossRef CAS PubMed.
- M. Thevis, M. Kohler, N. Schlörer, G. Fußhöller and W. Schänzer, Screening for two selective androgen receptor modulators using gas chromatography-mass spectrometry in doping control analysis, Eur. J. Mass Spectrom., 2008, 14(3), 153–161 CrossRef CAS PubMed.
- S. S. Piletsky, A. E. G. Cass, E. V. Piletska, J. Czulak and S. A. Piletsky, A novel assay format as an alternative to ELISA: MINA test for biotin, ChemNanoMat, 2018, 4(12), 1214–1222 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb00270a |
| This journal is © The Royal Society of Chemistry 2022 |