Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Passerini chemistries for synthesis of polymer pro-drug and polymersome drug delivery nanoparticles†
Alessandra
Travanut
a,
Patrícia F.
Monteiro
a,
Sean
Smith
b,
Steven M.
Howdle
 b,
Anna M.
Grabowska
c,
Barrie
Kellam
b,
Anna M.
Grabowska
c,
Barrie
Kellam
 a,
Michael A. R.
Meier
a,
Michael A. R.
Meier
 d and
Cameron
Alexander
d and
Cameron
Alexander
 *a
*a
aSchool of Pharmacy University of Nottingham, Nottingham NG7RD, UK. E-mail: Cameron.alexander@nottingham.ac.uk
bSchool of Chemistry University of Nottingham, Nottingham NG7RD, UK
cSchool of Medicine, University of Nottingham, Nottingham NG7RD, UK
dKarlsruhe Institute of Technology, Institute of Organic Chemistry (IOC) and Institute of Biological and Chemical Systems, Functional Molecular Systems (IBCS-FMS), Straße am Forum 7, Building 30.48 76131, Karlsruhe, Germany. E-mail: m.a.r.meier@kit.edu
First published on 19th April 2022
Abstract
New materials chemistries are urgently needed to overcome the limitations of existing biomedical materials in terms of preparation, functionality and versatility, and also in regards to their compatibility with biological environments. Here, we show that Passerini reactions are especially suited for the preparation of drug delivery materials, as with relatively few steps, polymers can be synthesized with functionality installed enabling drug conjugation and encapsulation, self-assembly into micellar or vesicular architectures, and with facile attachment triggerable chemistries. The polymers can be made with a variety of building blocks and assemble into nanoparticles, which are rapidly internalized in triple negative breast cancer (TNBC) cells. In addition, the polymers transport drug molecules efficiently through 3D cell cultures, and when designed with chemistries allowing pH-mediated release, exhibit greater efficacy against TNBC cells compared to the parent drug.
Introduction
Polymers for therapeutic delivery are required to meet stringent criteria,1,2 including diverse functionalities to allow attachment and/or encapsulation of drugs and targeting agents, an intrinsically degradable backbone, and should be synthetically accessible in as few steps as possible.3,4 In addition, for systemic administration, it is advantageous if the polymers can assemble into nanoscale particles with a hydrophilic exterior, in order to circulate in the bloodstream and avoid early excretion.5–7 However, most existing polymer drug delivery systems in the clinic are derived from a relatively small range of lactides, glycolides and lactones.8 These chemistries are sub-optimal in terms of the ultimate function of biomedical polymer structures, with problems of poor compatibility with candidate drug compounds and low functional group content without complex and costly synthesis. In addition, for formulations that are administered systemically, the existing stock of materials are difficult to modify to alter their rate of biodegradation.9 In recent years, the Passerini multicomponent reaction (Passerini-MCR), which combines a carboxylic acid, an aldehyde and an isocyanide into a α-acyloxycarboxamide product in a straightforward and atom economic procedure,10–12 is emerging as an alternative route to biomedical materials. The Passerini reaction has been widely investigated in organic chemistry, but also offers a powerful route to functional polymers, as it can provide in a single step the synthesis of complex, multifunctional and highly defined macromolecular architectures.13 Passerini reactions also bring important advantages from a translational pharmaceutical perspective.14 These include facile and scalable chemistry, the potential for multiple functionalities (e.g. hydrogen bonding, or reactive linkers) in one structure from one reaction, easy routes to varied polymer architectures15 and predictable degradation under various biological conditions.16–18 In turn, these factors enable tuning of drug compatibility, extent of drug loading or encapsulation, and modulation of biodistribution. The intrinsic biodegradability of the Passerini ester-amide product (α-acyloxycarboxamides) extends the utility of this reaction for many further applications beyond drug delivery. We have previously shown a simple approach for the Passerini-MCR synthesis of a new biodegradable amphiphilic diblock copolymer, which self-assembled into vesicles that were well-tolerated by numerous cell lines and long-circulating in vivo.13 Here, we exploit the Passerini-MCR functionality to deliver an anti-cancer drug by two different routes from the same inherent polymer materials platform. In the first example, we use the self-assembly properties of the Passerini polymer amphiphile to form polymersomes, which can be used to encapsulate the drug for a more sustained release profile. In the second case, we utilize the high functional group content in the polyester–polyamide backbone to conjugate the anticancer drug via a pH-responsive linker for enhanced intracellular release. We chose to exemplify this approach with doxorubicin, which is one of the mainstay treatments for TNBC, an aggressive subtype of breast cancer and a leading cause of cancer deaths.19 TNBC is characterized by the absence of human epidermal growth factor receptor 2 and the lack of progesterone and estrogen receptors,20,21 it also affects young patients and those in certain population groups, and has high metastatic potential. Unfortunately, TNBC has poor clinical outcomes,22 with only 30–40% of chemotherapy-treated patients in early stage TNBC showing positive responses to therapies.23 Doxorubicin as a free drug has severe side-effects including dose-limiting cardiotoxicity, and hence the encapsulation of doxorubicin in a number of carriers has been carried out as a way to enhance therapeutic index. Liposomal doxorubicin is used successfully to treat a number of cancers, but for TNBC, in which combination therapies are typically employed, there is a need for carrier materials which might be amenable to both drug encapsulation and drug conjugation strategies. Here we have employed Passerini chemistries to explore both routes for doxorubicin delivery, and report the results of in vitro efficacy studies with TNBC cells in both 2D and 3D culture.Materials and methods
Materials
The following chemicals were used as received: trimethyl orthoformate (≥99%, Aldrich), thionyl chloride (≥97%, Aldrich), diisopropylamine (≥99%, Aldrich), phosphorus(V) oxidychloride (≥99%, Aldrich), 10-undecenal (≥90%, Aldrich), 3-mercaptopropionic acid (≥99%, Aldrich), 2-2-dimethoxy-2-phenylacetophenone (DMPA, (≥99%, Aldrich), tert-butyl isocyanide (98%, Aldrich), O-methyl-O′-succinylpolyethylene glycol 2′000 (Aldrich), triethylamine (≥99%, Aldrich), 3-buten-1-ol (≥98%, Aldrich), dimethyl sulfide (≥99%, Aldrich), N,N′-dicyclohexylcarbodiimide (99%, Aldrich), N-hydroxysuccinimide (≥97%, Aldrich), 1,5,7-triazabicyclo[4.4.0]dec-5-ene (Aldrich),4-(Dimethylamino)pyridine (≥99%, Aldrich), silica gel 60 (0.035–0.070, Aldrich), chloroform-d (CDCl3, 99.96% D, 0.03% v/v TMS, Aldrich), deuterium oxide (99.9% D, Aldrich), Triton X-100 (Aldrich), Sephadex® LH20, phosphate buffered saline (PBS) (Dulbecco's, Aldrich). TrypLE™ Express Enzyme (1X) no phenol red (GibcoTM), Hoechst 33342, CellMask™ Green plasma membrane stain was purchased from Thermo Fisher Scientific. Cyanine5 (Cy5) ammine fluorescent dye was purchased from Lumiprobe. CellTiter-Fluor™ Cell Viability Assay (Promega, Madison WI), L-glutamine (Aldrich), RPMI-1640 media and fetal bovine serum (FBS) were obtained from Life Technologies (Carlsbad, CA, USA). All solvents used were of technical grade.Measurements
Thin-layer chromatography (TLC) identification of reactants and products was performed on silica-gel-coated aluminum foil (Aldrich, silica gel 60, F 254 with fluorescence indicator). Compounds were visualized by permanganate stain solution (mixture of potassium permanganate, potassium carbonate and sodium hydroxide in water).Nuclear Magnetic Resonance (NMR) spectra were recorded using a Bruker DPX400 UltraShield™ Spectrometer at 400 MHz (1H) and 101 MHz (13C). All 1H NMR spectra were reported in ppm relative to the solvent signal for chloroform-d at 7.26 ppm. All 13C NMR spectra were reported in ppm relative to the central line of the triplet for chloroform-d at 77.16 ppm. The spectra produced were analysed with MestReNova 12.0.4 (Mestrelab Research S.L.).
ESI-TOF Mass Spectrometry measurements were carried out with ESI† positive ionization mode using a Bruker microTOFII mass spectrometer employed with flow injection sample induction with electrospray ionization (ESI).
Size exclusion chromatography (SEC) was performed on an Agilent 1260 infinity series HPLC with an online vacuum degasser, an Agilent online differential refractometer (DRI) and a Wyatt Heleos II Multi-Angle Light Scattering detector (MALS). Two Agilent 5 μm PLgel Mixed E columns (7.5 × 300 mm) were connected in series with an Agilent PLgel 5 μm guard column (7.5 × 50 mm). The column oven, DRI and MALS temperature were set to 35 °C and the flow rate was set to 1 mL per minute. Each sample was dissolved in HPLC grade tetrahydrofuran (THF), at a concentration of 1 mg mL−1. DRI calibration was performed using 30 kDa and 200 kDa linear polystyrene narrow dispersity standards dissolved in THF at a concentration of 1.0 mg mL−1.
Particle size measurements were recorded using a Malvern Zetasizer Nano Series (Malvern, UK) instrument supported by Zetasizer Software 7.11, where all the measurements were carried out at a scattering angle of 90° and a temperature of 25 °C and the results were recorded as average of 10 runs ± SD.
Flow cytometry analyses were performed on Coulter FC-500 cytometer (Beckman Coulter) and Kaluza Analysis 2.1 software was used to perform the analyses. Absorbance and fluorescence readings of cells in 96 well plates were performed by using a TECAN Spark 10 M plate reader.
Synthetic procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0% to 80%
0% to 80%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20%). Methyl 12-isocyanododecanoate (3) was collected as a yellow oil 2.79 g, 11.5 mmol, yield 80.4% – Rf: 0.34 EtOAc 20% – hexane 80%.
20%). Methyl 12-isocyanododecanoate (3) was collected as a yellow oil 2.79 g, 11.5 mmol, yield 80.4% – Rf: 0.34 EtOAc 20% – hexane 80%.
13C NMR (101 MHz, CDCl3) δ 174.30 Ch, 155.71 Cm, 51.46 Cg, 41.66 Ca, 34.12 Cf, 29.44-29.15 Cd, 28.72 Cb, 26.35 Ce, 24.97 Cc.
ESI-MS of [C14H25NO2] (M+Na)+ = calc. 262.19 found 262.17.
1H NMR (400 MHz, CDCl3) δ/ppm: δ 5.87 (dd, 1H, J = 16.9, 10.1, 6.8 Hz, CHb), 5.09 (dt, 2H, J = 19.9, 10.1 Hz, CHa), 4.10 (t, 2H, J = 6.7 Hz, CHd), 3.39–3.30 (m, 2H, CHn), 2.42–2.21 (m, 4H, CHce), 1.75–1.56 (m, 4H, CHmf), 1.40 (dd, 2H, J = 14.2, 6.8 Hz, CHh), 1.26 (s,12H, CHg).
13C NMR (101 MHz, CDCl3) δ 173.86 Cp, 155.38 Co, 134.96 Cb, 117.26 Ca, 63.25 Cd, 41.60 Cn, 34.27 Ce, 33.08 Cc, 29.37–29.18 Cg, 28.65 Ch, 26.27 Cf, 24.92 Cm ESI-MS of [C17H29NO2] (M + H)+ = calc. 280.22 found 280.22; (M + Na)+ = calc. 302.22 found 302.20.
Synthesis of AB-type monomer (5)
10-Undecenal (1.71 g, 10.0 mmol) and DMPA (134 mg, 0.50 mmol) were placed in a quartz tube and dissolved in 4.0 mL of tetrahydrofuran (THF). Then, the tube was purged with argon for 10 min and afterward the solution was exposed to UV light (365 nm) for 30 s. Subsequently, 3-mercaptopropionic acid (1.17 g, 11.0 mmol) was slowly added. The reaction mixture was stirred for 2 h under UV irradiation. Then, the solvent was evaporated under reduced pressure and the crude product was purified by silica gel column chromatography (n-hexane/ethyl acetate = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to yield a white solid (1.78 g, 65%). Rf = 0.30 (dichloromethane/methanol = 20
1) to yield a white solid (1.78 g, 65%). Rf = 0.30 (dichloromethane/methanol = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).
1).
1H NMR (400 MHz, CDCl3) δ/ppm: δ 9.76 (s, 1H, CHa), 2.80- 2.75 (t, 2H, J = 6.2 Hz, CHh) 2.70–2.61 (t, 2H, J = 6.2 Hz, CHi), 2.61–2.46 (m, 2H, CHg), 2.42–2.39 (m, 2H, CHc) 1.67–1.55 (m, 4H, CHdf), 1.35–1.27 (m, 12H, CHe).
13C NMR (101 MHz, CDCl3) δ 203.24 Ca, 177.49 Cn, 44.04 Cc, 34.74 Cg, 32.34 Ci, 29.64 Cf, 29.45, 29.27–28.94 Cg, 26.77 Cd, 22.21 Ch.
ESI-MS of [C14H26O3S] (M−H)− = calc. 273.16 found 273.15; (M + H)+ = calc. 275.16 found 275.16; (M + Na)+ = calc. 297.14 found 297.09.
Synthesis of P1 – Passerini-3CR polymerization
The synthesis was adapted from a previous route.13 To a vigorously stirred solution of O-methyl-O′-succinyl polyethylene glycol 2 kDa (0.25 g, 0.118 mmol) in 0.02 M DCM, AB-type monomer (5) (0.970 g, 3.54 mmol) was added slowly and solubilized. Finally, tert-butyl isocyanide (1.50 g, 18 mmol) was added dropwise. After stirring for three days at room temperature, the polymer was precipitated from ice-cold diethyl ether and Passerini diblock copolymer P1 was obtained as a white solid (1.152 g, 87%) and characterized by 1H NMR, 13C NMR and SEC in THF.1H NMR (400 MHz, CDCl3) δ/ppm: δ 6.10 (m, 25H, NHO), 5.06 (m, 26H, CHi), 4.21 (m, 2H, CHd), 3.78–3.66 (m, 176 H, CHb), 3.38 (s, 3H, CHa), 3.19–3.15 (m, 2H, CHg), 2.85–2.76 (m, 54H, CHfhv), 2.70 (t, J = 6.6 Hz, 48H, CHZ), 2.57 (t, J = 6.6 Hz, 48H, CHW), 1.94–1.76 (m, 52H, CHrs), 1.65–1.51 (m, 51H, CHu), 1.35–1.19 (m, 598H, CHqt).
13C NMR (101 MHz, CDCl3) δ 170.73 Ce, 169.11 Cm, 74.70 Ci, 70.7–70.6 Cbd, 51.61 Ca, 51.55 Cp, 34.89 Cf, 32.27 Ch, 31.92 Cv, 29.62 Ct, 29.55 Ct, 29.45 Ct, 29.30 Cq, 28.9 7 Cs, 28.82 Cu, 27.18 Cs, 24.82 Cg.
M
n,THEO: 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 g mol−1. Mn,SEC(THF): 16
300 g mol−1. Mn,SEC(THF): 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 g mol−1, Đ 1.55 (polystyrene equivalent values).
300 g mol−1, Đ 1.55 (polystyrene equivalent values).
Synthesis of P2 – Passerini-3CR polymerization
The synthesis was adapted from a previous route.13 To a vigorously stirred solution of O-methyl-O′-succinyl polyethylene glycol 2 kDa (0.076 g, 0.036 mmol) in DCM 0.02 M, AB bifunctional monomer (5) (0.051 g, 0.18 mmol) was added slowly and solubilized. Finally, but-3-en-1-yl 12-isocyanododecanoate (4) (0.260 g, 0.93 mmol) was added dropwise. After stirring for three days at room temperature, the polymer was precipitated from ice-cold diethyl ether and Passerini diblock copolymer P2 was obtained as a white solid (0.150 g, 0.030 mmol, yield 83%) and was characterized by 1H NMR, FT-IR and SEC in THF.1H NMR (400 MHz, CDCl3) δ 6.54(s, 5H, Hk), 5.83–5.74 (m, 5H, Ht), 4.28–4.24 (m, 5H, Hi), 4.15–4.10 (m, 10H, Hr), 3.71–3.64 (s, 126H, Hb), 3.38 (s, 3H, Ha), 3.26–3.16 (s, 10H, Hn), 2.93–2.53 (m, 29H, Hzwfg), 2.40–2.37 (m, 10H, Hs), 2.31–2.25 (m, 10H, Hp), 1.90–1.47 (m, 85H, Hv), 1.37–1.19 (s, 116H, Ho).
13C NMR (101 MHz, CDCl3) δ173.97 Cq, 173.67 Cz, 170.76 Cm, 169.81 Ch, 134.17 Ct, 117.26 Cu, 74.17 Ci, 72.06 Cd, 70.69 Cb, 63.38 Cr, 59.16 Ca, 39.39 Cn, 34.73 Cp, 34.45 Cv, 33.24 Cw, 31.99 Cv, 29.65–29.56 Cvy, 29.36 Co, 29.27 Cf, 29.04 Cg, 27.36 Co, 27.02 Co, 25.11 Cv.
P2 oxidation to P3via ozonolysis
P2 (0.100 g, 0.020 mmol) was dissolved in DCM (25 mL). The solution was cooled to −78 °C with stirring and then the flask was attached to an ozone generator equipped with an oxygen cylinder. Molecular oxygen was bubbled through the solution for 5 minutes, then the ozone generator was switched on, and the ozone/oxygen mixture was bubbled through the solution maintaining −78 °C for 1 h. After a persistent metallic blue color developed, the ozone generator was switched off, and molecular oxygen was bubbled through the reaction mixture for 5 minutes. Dimethyl sulfide (1 mL, 13 mmol) was then added over a 20 minutes period with stirring at −78 °C, turning the mixture deep yellow. The resulting mixture was stirred at −78 °C for a further 1 h, then was allowed to warm to room temperature and left stirring for a further 1 h. The reaction mixture was dried under vacuum and diluted with 3 mL of acetone. The Passerini diblock copolymer P3 was collected by precipitation in cold diethyl ether and obtained as a white solid (0.095 g, 0.018 mmol, yield 93%) characterized by 1H NMR, FT-IR and SEC in THF.1H NMR (400 MHz, CDCl3) δ 9.78 (s, 5H, Ht), 6.72–6.53 (s, 5H, Hk), 5.23–5.15 (m, 5H, Hi), 4.43–4.40 (m, 10H, Hr), 4.26–4.20 (m, 2H, Hd), 3.68–3.59 (s, 172H, Hb), 3.38 (s, 3H, Ha), 3.28–3.16 (m, 10H, Hj), 3.04–2.96 (m, 10H, Hn), 2.78–2.75 (m, 10H, Hs), 2.63–2.62 (m, 10H, Hw), 2.31–2.26 (m, 10H, Hp), 2.09–1.79 (m, 14H, Hyfg), 1.42–1.19 (m, 135H, Hvo).
13C NMR (101 MHz, CDCl3) δ = 211.41 Cu, 175.81 Cq, 173.81 Cz, 173.72 Cm, 165.70 Ch, 72.06 Ci, 70.90 Cd, 70.69 Cb, 62.78 Cr, 59.38 Ca, 54.23 Cj, 53.89 Cy, 39.58 Cn, 34.31 Cp, 34.14 Cv, 32.73 Cw, 31.60 Cv, 29.53–29.02 Cvyofg, 26.91 Co, 26.86 Co, 24.88 Cv.
Synthesis of P1-Cy5 and P3-Cy5
P1 (18.34 mg, 1.53 μmol) was solubilized in 0.5 mL of dry dimethylformamide (DMF). N,N′-Dicyclohexylcarbodiimide (0.78 mg, 3.82 μmol) and N-hydroxysuccinimide (0.26 mg, 2.29 μmol) were added to the mixture. The reaction mixture was stirred at room temperature for 12 h and then Cy5 amine (1 mg, 1.53 μmol) was dissolved in 0.1 mL of dry DMF and subsequently added. The reaction was left for 72 h. The crude mixture was diluted with 0.6 mL of acetone and the polymer was collected by precipitation into cold diethyl ether. The powder was dried, the polymer was solubilized in 0.5 mL of THF, diluted with 1 mL deionized water and then it was directly dialyzed against deionized water for 48 h (3.5 kDa cut-off, ThermoScientific). P1-Cy5 was collected after freeze drying (17.5 mg, yield 95.41%). The Cy5 conjugation yield was determined by UV/Vis spectroscopy. The Cy5 conjugation yield was calculated by absorbance spectroscopy: P1-Cy5-labelled polymer was solubilized in 40 μL of DMF and diluted with water to 400 μL of water, the absorbance was recorded at 646 nm with a Tecan Spark 10M plate reader in a 96 well plate. A solution of water/10% DMF was used as a blank. The Cy5 molar content was quantified by using the calibration curve in Fig. S12 (ESI†) and the Cy5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer molar ratio was calculated. The Cy5
polymer molar ratio was calculated. The Cy5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P1 polymer molar ratio was 1
P1 polymer molar ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and the conjugation yield was found to be 33%.
2 and the conjugation yield was found to be 33%.
P3 (5 mg, 0.97 μmol) was solubilized in the minimum amount of dry DMF. N,N′-Dicyclohexylcarbodiimide (0.5 mg, 2.42 μmol) and N-Hydroxysuccinimide (0.167 mg, 1.45 μmol) were added. The reaction mixture was stirred at room temperature for 12 h and then Cy5 amine (0.5 mg, 0.76 μmol) was solubilized in the minimum amount of dry DMF and then added. The reaction was left for 72 h. The crude was diluted with acetone and the polymer was collected by precipitation in cold diethyl ether. The powder was dried, the polymer was solubilized in 0.5 mL of THF, diluted with 1 mL DI water and then it was directly dialyzed against DI water for 48 h (3.5 kDa cut-off, ThermoScientific). P3-Cy5-labelled polymer was collected after freeze drying (4.9 mg; yield 98%). The Cy5 conjugation yield was calculated by absorbance spectroscopy: P3-Cy5-labelled polymer was solubilized in 40 μL of DMF and diluted with water to 400 μL of water, the absorbance was recorded at 646 nm with Tecan Spark 10M plate reader in a 96 well plate. A solution of water 10% DMF was used as blank. The Cy5 molar content was quantified by using the calibration curve in Fig. S12 (ESI†) and the Cy5:polymer molar ratio was calculated. Approximately 8% of the polymer chains were found to be functionalized with a Cy5 molecule through amidation leading to 3.53 ± 0.05 wt% and 10% of conjugation yield.
Synthesis of P4-Dox
The synthesis was adapted from a published route.25 Doxorubicin-HCl (0.128 g, 0.02 mmol) and triethylamine (0.04 g, 0.04 mmol) were solubilized in 0.2 mL of dimethyl sulfoxide (DMSO) and added to a solution of P3 (0.05 g, 0.009 mmol) in 0.2 mL of DMSO under stirring for 7 h at room temperature. The crude was diluted with acetone and precipitated in cold diethyl ether. The powder was dried, solubilized in chloroform and the polymer-doxorubicin conjugate P4-Dox (0.022 g, 0.002 mmol, 32% yield) was collected and dried under vacuum after size exclusion purification through Sephadex LH20.13C NMR (126 MHz, CDCl3) δ 216.64 Co, 186.18 Cp, 173.78 Ck, 173.21 Ck, 172.21 Ck, 171.42 Ck, 170.18 Cf, 170.08 Cf, 169.77 Cc, 156.00 Cm, 144.66 Cn, 127.41 Cn, 114.53 Cn, 113.66 Cn, 81.04 Cu, 75.31 Cu, 74.48 Ce, 69.04–65.98 Cb, 64.31 Ct, 61.82 Cr, 59.76 Cr, 59.19 Ca, 53.88 Cw, 40.99 Cs, 34.29 Cz, 31.94 Cu, 31.83 Cv, 31.71 Cv, 29.70 Cv, 29.38 Cq, 29.23 Cq, 28.61 Cv, 28.47 Cv, 28.23 Cv, 27.11 Cv, 26.81 Cv, 24.90 Cv, 24.75 Cv, 22.82 Cv, 22.55 Cv, 22.00 Cv, 15.45 Cj, 15.39 Cv, 8.80 Cv.
Formulation and characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio. The organic solvent was let evaporating overnight in the fume hood and the polymersomes size (z-average diameter) was measured using NanoZS equipment (Malvern, UK) at 25 °C (DLS).
1 volume ratio. The organic solvent was let evaporating overnight in the fume hood and the polymersomes size (z-average diameter) was measured using NanoZS equipment (Malvern, UK) at 25 °C (DLS).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio. The organic solvent was allowed to evaporate overnight under stirring. Then, the pH of the formulation was raised to 7.4 by adding ∼2 μL of NaOH 2 M. Doxorubicin hydrochloride solution in DMSO was added to the aqueous polymersomes solution (5 mg P1
1 volume ratio. The organic solvent was allowed to evaporate overnight under stirring. Then, the pH of the formulation was raised to 7.4 by adding ∼2 μL of NaOH 2 M. Doxorubicin hydrochloride solution in DMSO was added to the aqueous polymersomes solution (5 mg P1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 mg doxorubicin) leading to 30% v/v DMSO/PBS solution. Finally, the pH was raised to 8.2 by adding ∼0.5 μL of NaOH 2 M leading to a pH gradient of Δ = 4.2. The solution was left under stirring for 3 days in the dark. P1-Dox was directly dialyzed against PBS 150 mM for 24 h (3.5 kDa cut-off, ThermoScientific). The particle size (z-average diameter) was measured as reported above.
2 mg doxorubicin) leading to 30% v/v DMSO/PBS solution. Finally, the pH was raised to 8.2 by adding ∼0.5 μL of NaOH 2 M leading to a pH gradient of Δ = 4.2. The solution was left under stirring for 3 days in the dark. P1-Dox was directly dialyzed against PBS 150 mM for 24 h (3.5 kDa cut-off, ThermoScientific). The particle size (z-average diameter) was measured as reported above.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 volume ratio. The organic solvent was removed by dialysis against PBS 150 mM for 24 h (3.5 kDa cut-off, ThermoScientific). The size (z-average diameter) was measured using NanoZS equipment (Malvern, UK) at 25 °C (DLS).
8 volume ratio. The organic solvent was removed by dialysis against PBS 150 mM for 24 h (3.5 kDa cut-off, ThermoScientific). The size (z-average diameter) was measured using NanoZS equipment (Malvern, UK) at 25 °C (DLS).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (t = 5 min), 40
20 (t = 5 min), 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 (t = 20 min), 80
60 (t = 20 min), 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (t = 22 min) and was applied over 30 min at a flow rate of 1 mL min−1. The fluorescence excitation and emission wavelengths were 485 and 590 nm respectively, the UV detector was set at 485 nm. The injection volume was 10 μL. The analysis was performed in triplicate.
20 (t = 22 min) and was applied over 30 min at a flow rate of 1 mL min−1. The fluorescence excitation and emission wavelengths were 485 and 590 nm respectively, the UV detector was set at 485 nm. The injection volume was 10 μL. The analysis was performed in triplicate.
The drug loading and encapsulation efficiency were calculated using the following equations:
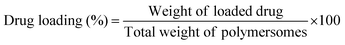 | (1) |
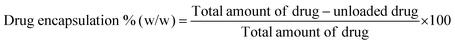 | (2) |
P1-Dox and P4-DoxIn Vitro drug release
Drug release studies were performed in vitro in PBS at 150 mM at pH 7.4 (blood pH)29 and sodium acetate buffer at 150 mM pH 5.0 containing 0.15 M NaCl30 (endosomal pH).31,32 A sample of 250 μL of P1-Dox or P4-Dox formulation was diluted to 500 μL with PBS or acetate buffer 150 mM and the pH was adjusted to 7.4 or 5. The solution was placed in a dialysis device (Slide-A-Lyzer™ mini dialysis device, 3.5 K MWCO, Thermo Scientific). The P1-Dox or P4-Dox solution was dialyzed against 1.7 mL of release media 150 mM at 37 °C and samples (0.75 mL) were taken at appropriate time points and replaced with 0.75 mL of fresh media to maintain sink conditions.33 The doxorubicin content of these samples was quantified by Agilent UV-Vis Spectrometer referring to the calibration curves.Calibration curves
Doxorubicin hydrochloride in DMSO 1 mg mL−1 was diluted with PBS 150 mM pH 7.4 leading to different concentrations. The procedure was repeated in triplicate, the samples were analysed by UV-Vis spectrophotometer and the absorbance at 500 nm was plotted against doxorubicin concentrations leading to the calibration curve reported. Doxorubicin hydrochloride 1 mg mL−1 in acetate buffer pH 5 150 mM was diluted with acetate buffer leading to different concentrations. The procedure was repeated in triplicate, the samples were analysed by UV-Vis spectrophotometer and the absorbance at 490 nm was plotted against doxorubicin concentrations leading to the calibration curve reported.Cell culture
The TNBC cell line MDA-MB-231 was acquired from the American Type Culture Collection (Manassas, VA). MDA-MB-231 cells were maintained in RPMI 1640 (Gibco BRL, USA) supplemented with 10% (v/v) FBS and 5% L-glutamine and incubated at 37 °C in 5% CO2.Effects of polymers on cellular metabolism
The CellTiter-Fluor cell viability assay was performed to determine the cytotoxicity of P1 and P3 copolymers against MDA-MB-231 cells following the protocol provided by the vendor (Promega, Madison WI).24 Cells were treated for 48 h with different concentrations of P1 and P3 copolymers.MDA-MB-231 cells were seeded into 96 well-plate at a density of 6 × 103 cells per well and incubated at 37 °C, 5% CO2 for 24 h. Afterwards, cells were exposed to different concentrations of P1 and P3 copolymers (in complete media - sterilized under 254 nm UV light for 30 minutes) for 48 h. Negative controls were prepared by treating the cells with 1% Triton X. Cells were then washed three times with complete media and incubated at 37 °C, 5% CO2 with a solution of media and CellTiter-Fluor Cell Viability Assay (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 1 h. 1 mg mL−1 solution of P1 and P3 copolymers in media was incubated with CellTiter-Fluor Cell in order to investigate any possible interactions. The fluorescence was then measured with Tecan Spark 10M (400nmEx/505nmEm). Experiments were made in replicates, repeated in different days and the protease activity (mean% ± SD) was reported compared to the controls and calculated as mentioned in eqn (3).
1) for 1 h. 1 mg mL−1 solution of P1 and P3 copolymers in media was incubated with CellTiter-Fluor Cell in order to investigate any possible interactions. The fluorescence was then measured with Tecan Spark 10M (400nmEx/505nmEm). Experiments were made in replicates, repeated in different days and the protease activity (mean% ± SD) was reported compared to the controls and calculated as mentioned in eqn (3).
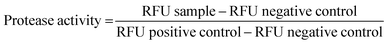 | (3) |
Preparation of 3D cancer cell spheroids
TNBC tumour spheroids were obtained using ultra-low attachment (ULA) 96-well round bottom plates (Corning, UK). MDA-MB-231 cells were seeded into ULA plates at a density of 4000 per well and cells were centrifuged at 200×g for 3 minutes after seeding to bring the cells closer together and aid in the formation of a single spheroid. Spheroids were then cultured for 4 days before final analysis or treatment.Evaluation of internalisation of P1-Cy5 and P3-Cy5 nanoparticles by cancer cells
Confocal microscopy
Cells were exposed to 25 μg mL−1P1-Cy5 or P3-Cy5 labelled nanoparticles in the dark at 37 °C in 5% CO2 for 4 h. Following incubation, cells were washed with PBS three times, incubated with CellMask Green plasma membrane stain at the concentration of 1 μL mL−1 (100 μL) for 10 minutes in the dark at 37 °C in 5% CO2 and washed with PBS three times. Cells were then incubated with Hoechst 33342 dye at concentration of 1 μg mL−1 (100 μL) for 30 minutes in the dark at room temperature and washed again with PBS (three times). Finally, cells were fixed with 4% formaldehyde (in PBS) for 10 min and again washed with PBS (three times). Cells were imaged with a Zeiss LSM 700 Confocal Laser Scanning Microscope. Zen 2.6 blue image Software was utilized for image processing.
Results
In order to exemplify the strategy for drug delivery materials, we chose to design Passerini polymers with functionality to enable self-assembly into nanoparticles and polymersome structures, and with conjugation or encapsulation of doxorubicin for targeting TNBC. This is because doxorubicin is a standard treatment for TNBC, which is an aggressive subtype of breast cancer and the leading cause of cancer deaths. TNBC is characterized by the absence of human epidermal growth factor receptor 2 and the lack of progesterone and estrogen receptors.20,21 TNBC affects young patients, has high metastatic potential and poor clinical outcomes.22 Only the 30–40% of the chemotherapy-treated patients in early stage TNBC show positive pathological responses.23The synthesis schemes are shown in Scheme 1. The Passerini diblock copolymers were synthesised in one-pot by reacting O-methyl-O′-succinyl polyethylene glycol 2 kDa (mPEG-COOH) as an irreversible chain transfer agent in a reaction involving AB-type monomer (5)34 and an excess of the isocyanide opportunely functionalized at the R group (Scheme 1A). The polymersome forming copolymer P1 was obtained in high yield (87%), with SEC data indicating a molar mass (Mn ∼ 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) and a molar mass distribution of Đ = 1.55. The molar mass by NMR was lower (∼11
000 g mol−1) and a molar mass distribution of Đ = 1.55. The molar mass by NMR was lower (∼11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), and the difference between this value and that from SEC may have been due to the different relaxation times, and hence measured integrals, for protons in the PEG block compared to the poly(ester) blocks. For the doxorubicin-polymeric prodrugs (Scheme 1C), a diblock copolymer P2 was obtained with high yield (83%) and high conversion of the AB monomer units. The alkene pendant groups of P2 were oxidatively cleaved to aldehyde groups via ozonolysis (P3). The peak t in the 1H NMR of the as-produced polymer (Fig. S9, ESI†) and the FT-IR spectra (Fig. S11, ESI†) showed complete conversion of the alkene groups and the concomitant oxidation of the thioether units into sulfones. Although the sulfone group is reducible and thus potentially biologically active, prior reports of polymers containing sulfones in their main chain have shown no acute adverse effects in cell lines.24,35 The Passerini copolymer chemistry was then exploited by conjugating the dye Cy5via amidation to the carboxylic end-groups leading to the fluorescently labeled P3-Cy5 (SI). Subsequently, the P3 side-chain aldehyde groups were conjugated with doxorubicin via an imine bond, in order to enable intracellular drug release. The 1H NMR spectrum of the polymer prodrug P4-Dox (Fig. S13, ESI†) showed complete conjugation of the drug due to the disappearance of the aldehyde protons, which was further confirmed by the imine signal at 1654 cm−1 in the FT-IR spectrum Fig. S11 (ESI†).
000 g mol−1), and the difference between this value and that from SEC may have been due to the different relaxation times, and hence measured integrals, for protons in the PEG block compared to the poly(ester) blocks. For the doxorubicin-polymeric prodrugs (Scheme 1C), a diblock copolymer P2 was obtained with high yield (83%) and high conversion of the AB monomer units. The alkene pendant groups of P2 were oxidatively cleaved to aldehyde groups via ozonolysis (P3). The peak t in the 1H NMR of the as-produced polymer (Fig. S9, ESI†) and the FT-IR spectra (Fig. S11, ESI†) showed complete conversion of the alkene groups and the concomitant oxidation of the thioether units into sulfones. Although the sulfone group is reducible and thus potentially biologically active, prior reports of polymers containing sulfones in their main chain have shown no acute adverse effects in cell lines.24,35 The Passerini copolymer chemistry was then exploited by conjugating the dye Cy5via amidation to the carboxylic end-groups leading to the fluorescently labeled P3-Cy5 (SI). Subsequently, the P3 side-chain aldehyde groups were conjugated with doxorubicin via an imine bond, in order to enable intracellular drug release. The 1H NMR spectrum of the polymer prodrug P4-Dox (Fig. S13, ESI†) showed complete conjugation of the drug due to the disappearance of the aldehyde protons, which was further confirmed by the imine signal at 1654 cm−1 in the FT-IR spectrum Fig. S11 (ESI†).
Nanoparticle formulations and TNBC cell culture studies
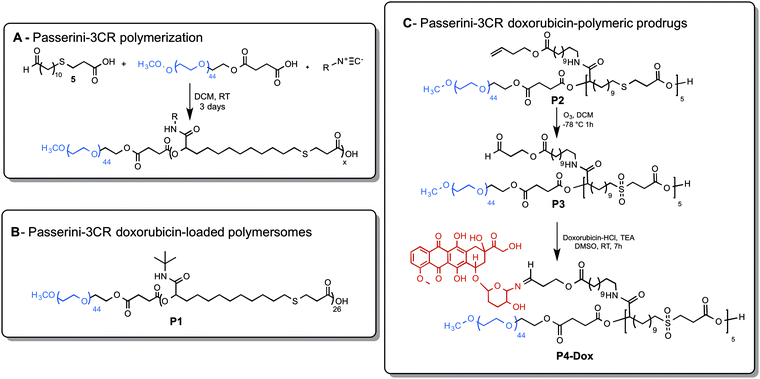
Polymers P1 and P4-Dox were found to self-assemble readily in buffer solutions, with mean diameters of ∼140 nm (P1) to ∼20 nm for P4-Dox). The dye-labelled polymer P1-Cy5 also self-assembled, but formed smaller particles (∼90 nm) and with a higher size dispersity than the parent polymer. As apparent from Fig. 1, a distinct polymersome membrane could be observed in the TEM micrographs of P1, whereas for P4-Dox, with a lower hydrophobic content, more conventional kinetically-trapped nanoparticulate structures were formed.The difference in self-assembly behaviour can be attributed to the different ratios of hydrophilic to hydrophobic blocks in the polymer structures. For P1 the degree of hydrophobicity based on block content13 was ∼70% (see Scheme 1B) based on SEC-derived mass or ∼55% based on NMR mass, whereas for P4-Dox the hydrophobic content was less than 30%. Accordingly, the packing parameters for P1 chains were in the expected region for vesicle/polymersome formation,36 whereas for P4-Dox, the hydrophilic : hydrophobic ratio indicated that more micellar-type structures should be formed. The overall diameters of the P4-Dox nanoparticles were larger than expected for ‘pure’ core-shell structures, in which the surface was only the PEG-layer and the core solely the hydrophobic regions, again in agreement with many prior studies of polymer nanoparticles.21
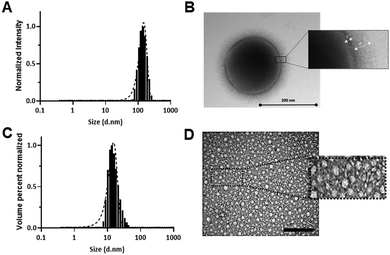
Tagging of the polymers with an amino-functional Cy5 label at the carboxyl terminus via NHS/DCC coupling allowed monitoring of their internalization in 2D monolayers and 3D spheroid cell cultures. MDA-MB-231 cells were chosen as a representative TNBC cell line, and entry of the polymers into the cells was evaluated by confocal microscopy and flow cytometry. Confocal images in 2D cultures of MDA-MB-231 cells (Fig. 2) showed that P1-Cy5 and P3-Cy5 nanoparticles were internalized by the cells.31
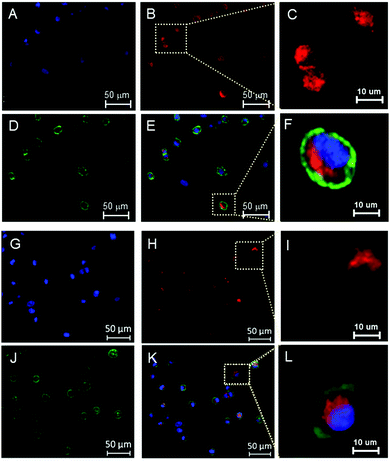
Transport of drug delivery carriers in vivo is known to correlate better with 3D cell cultures than with 2D monolayers, and thus the ability of the Cy5 labelled formulations to permeate throughout 3D spheroids was assayed. In these experiments, imaging was carried out after the cells had grown together for 4 days, such that the resultant spheroids were dense and close packed. Following an incubation time for the polymers of 16 h, the dye-labelled polymers in the cells were imaged by confocal microscopy and quantified by flow cytometry.
As can be seen in Fig. 3A–F and as reported in the ESI† (Fig S29, S30), the nanoparticles penetrated deep into the spheroids and were efficiently internalized by the tumour cells. The uptake of the polymers was quantified by flow cytometry and found to be concentration-dependent (Fig. 3G and H). These data were in accord with our previous studies of polymer nanoparticle penetration into TNBC spheroids.37,38
Effects of polymers with/without doxorubicin in breast cells
Following the demonstration of cell entry for both formulations into TNBC cells we evaluated their efficacy in drug delivery assays. Initially, P1 and P3 copolymers were dosed at increasing levels in 2D monolayers of TNBC (MDA-MB-231). The P1 polymers were well-tolerated up to 0.5 mg mL−1 whereas the P3 polymers exhibited signs of toxicity at 100 μg mL−1, which we attribute to the free aldehyde function in the latter (Fig. S23, ESI†). The P1 empty polymersomes were then loaded with doxorubicin by adapting the remote transmembrane pH gradient method reported for the formulation of liposomes by Li et al.26 The polymersomes were first assembled in 150 mM citrate buffer at pH 4 and then the pH gradient was created by adding 2 μL of NaOH 2M and raising the external pH to 7.4. Doxorubicin was dissolved in DMSO, which acted also to plasticise the polymer membrane, and added to the polymersomes aqueous solution leading to an overall 30% (v/v) and to increase the pH gradient from 3.43 to 4.2. As reported by Li et al.,26 the protonated doxorubicin can aggregate with the citrate anions forming insoluble fibrous structures, which are in equilibrium with the soluble drug molecules. Therefore, doxorubicin was expected to permeate throughout the polymeric bilayer and be trapped in the vesicular core as citrate complexes. P4-Dox prodrug nanoparticles were formulated as the self-assembled micellar-like nanoparticles formed in aqueous buffer solutions. The doxorubicin loaded nanoparticle drug loading was calculated by HPLC with values for P1-Dox and P4-Dox found to be 16 ± 3 wt% and 36 ± 2 wt%, respectively. Interestingly, the polymer:drug molecular ratio for P1-Dox polymersomes was found to be higher (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) than that reported by Ahmed et al.39P4-Dox drug loading was found to be higher than for P1-Dox as expected for a polymeric-prodrug formulation.32,40
4) than that reported by Ahmed et al.39P4-Dox drug loading was found to be higher than for P1-Dox as expected for a polymeric-prodrug formulation.32,40
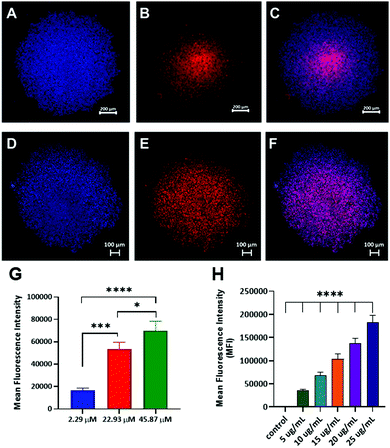
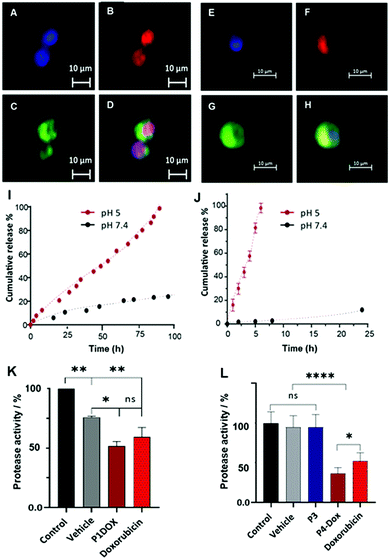
Preliminary microscopy assays (Fig. 4) indicated that free doxorubicin (Fig. 4A–D) rapidly entered MDA-MB-231 cells and reached the nucleus after 4 h, whereas P4-Dox polymers showed a different signal localization for doxorubicin (Fig. 4E–H). We therefore evaluated the release profiles for doxorubicin from the formulations over longer timescales and, as reported in Fig. 4(I and J), the drug cargoes in both P1-Dox and P4-Dox formulations were largely retained at ambient plasma pH,29 but were released under pH conditions mimicking endo/lysosomal intracellular processing.30–32 For the P1-Dox formulation, retention of the drug was expected to be reduced as the interior pH of the vesicles became acidic, and as shown in Fig. 4I, release of doxorubicin at pH 5 was faster than at pH 7.4. The overall time for full release was ∼100 h, whereas only 20% of drug was released at pH 7.4 from the vesicles over the same time period. For the P4-Dox prodrug nanoparticles, which were designed to liberate doxorubicin through the pH-responsive hydrolysis of the imine-linker, complete release of drug occurred within 6 hours (Fig. 4J), with <5% of drug released from the same polymer at pH 7.4. The ∼80% release of doxorubicin from P4-Dox under acidic conditions in 4 h (Fig. 4J) suggested that the drug should be liberated from the polymers if processed into digestive acidic intracellular compartments over this timescale.
We therefore decided to evaluate cytotoxicity of the polymers over a longer period in our efficacy assays. The data for 2D monolayers of MDA-MB-231 cells treated with P1-Dox polymersomes and P4-Dox prodrug nanoparticles in comparison to the free drug over 48 h is shown in Fig. 4(K and L). In these assays both P1-Dox nanoparticles and P4-Dox prodrug nanoparticles induced cell death, with P4-Dox showing a significantly enhanced cytotoxic effect compared to the free drug. While both formulations might have been expected to bypass membrane-bound transporters, which can act to pump free drug out of the cells, the greater efficacy of P4-Dox was most likely due to the greater overall release of doxorubicin over the 48 h period of incubation. The therapeutic potential of these materials will largely depend on their accumulation and retention time in tumours, and this may be problematic in TNBC as it is known that the EPR effect has limited relevance to many human cancers. However, the fact that it was possible to introduce the self-assembly properties and drug conjugation links through Passerini-related chemistries, and that these in turn enabled tuning of the drug release profiles, suggests that the materials may have promise if their biodistribution can be optimised.
Overall, both the P1-Dox polymersomes and the P4-Dox formulations were found to exhibit slow release of the drug cargoes under normal physiological conditions, and to accelerate release under acidic conditions. We believe these results indicate that these chemistries could be important in developing ‘platform’ delivery formulations, built on a standard Passerini materials scaffold but with drug release performance modulated by control of assembly behaviour.
Conclusions
We have shown a simple and efficient route for tunable amphiphilic diblock copolymers synthesis via Passerini-3CR and their formulation into doxorubicin-loaded polymersomes and prodrug nanoparticles. The copolymers were found to be well-tolerated by TNBC cells in the absence of a drug cargo. The Cy5 labelled formulations were internalized in 2D monolayers and 3D spheroids of TNBC and the uptake was found to be concentration-dependent. Additionally, P1-Dox and P4-Dox nanoparticles demonstrated pH-dependent drug release, with slow liberation of the cargo at plasma pH-values and accelerated release under pH conditions similar to those of intracellular compartments. P4-Dox prodrug nanoparticles showed both rapid drug release and improved toxicity against MDA-MB-231 cells, compared to P1-Dox and to the free drug. Our results suggest that the Passerini-3CR can provide a synthetically straightforward and flexible route to the synthesis of responsive drug delivery systems.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Engineering and Physical Sciences Research Council (Grant No. EP/N006615/1, EP/N03371X/1, EP/H005625/1, and EP/L013835/1). This work was also funded by the Royal Society (Wolfson Research Merit Award WM150086) to C.A. The authors thank the Nanoscale and Microscale Research Centre (nmRC) for providing access to instrumentation, Prof. Robert A. Stockman for providing access to the ozone generator, Douglas Crackett and Paul Cooling for expert technical assistance and Carol Turrill for outstanding administrative support.References
- W. Poon, B. R. Kingston, B. Ouyang, W. Ngo and W. C. W. Chan, Nat. Nanotechnol., 2020, 15, 819–829 CrossRef CAS PubMed.
- M. B. Ashford, R. M. England and N. Akhtar, Adv. Ther., 2021, 4, 2000285 CrossRef CAS.
- R. S. Riley, C. H. June, R. Langer and M. J. Mitchell, Nat. Rev. Drug Discovery, 2019, 18, 175–196 CrossRef CAS PubMed.
- W. S. Saw, T. Anasamy, Y. Y. Foo, Y. C. Kwa, C. S. Kue, C. H. Yeong, L. V. Kiew, H. B. Lee and L. Y. Chung, Adv. Ther., 2021, 4, 2000206 CrossRef CAS.
- M. Machtakova, H. Therien-Aubin and K. Landfester, Chem. Soc. Rev., 2022, 51, 128–152, 10.1039/d1cs00686j.
- V. M. Weiss, H. Lucas, T. Mueller, P. Chytil, T. Etrych, T. Naolou, J. Kressler and K. Mäder, Macromol. Biosci., 2018, 18 Search PubMed.
- J. Wang, S. Yuan, Y. Zhang, W. Wu, Y. Hu and X. Jiang, Biomater. Sci., 2016, 4, 1351–1360 RSC.
- C. Oerlemans, W. Bult, M. Bos, G. Storm, J. F. W. Nijsen and W. E. Hennink, Pharm. Res., 2010, 27, 2569–2589 CrossRef CAS PubMed.
- H. Seyednejad, A. H. Ghassemi, C. F. van Nostrum, T. Vermonden and W. E. Hennink, J. Controlled Release, 2011, 152, 168–176 CrossRef CAS PubMed.
- M. Passerini and L. Simone, Gazz. Chim. Ital., 1921, 51, 126–129 CAS.
- B. Alkan, O. Daglar, S. Luleburgaz, B. Gungor, U. S. Gunay, G. Hizal, U. Tunca and H. Durmaz, Polym. Chem., 2022, 13, 258–266, 10.1039/D1PY01528A.
- L. Banfi, A. Basso, C. Lambruschini, L. Moni and R. Riva, Chem. Sci., 2021, 12, 15445–15472 RSC.
- A. Travanut, P. F. Monteiro, S. Oelmann, S. M. Howdle, A. M. Grabowska, P. A. Clarke, A. A. Ritchie, M. A. R. Meier and C. Alexander, Macromol. Rapid Commun., 2021, 42, e2000321 CrossRef PubMed.
- M. Meier, Green Chem., 2014, 16, 1672 RSC.
- S. C. Solleder and M. A. R. Meier, Angew. Chem., Int. Ed., 2014, 53, 711–714 CrossRef CAS PubMed.
- M. Zhao, N. Liu, R.-H. Zhao, P.-F. Zhang, S.-N. Li, Y. Yue and K.-L. Deng, ACS Appl. Bio Mater., 2019, 2, 1714–1723 CrossRef CAS PubMed.
- B. T. Tuten, L. De Keer, S. Wiedbrauk, P. H. M. Van Steenberge, D. R. D'Hooge and C. Barner-Kowollik, Angew. Chem., Int. Ed., 2019, 58, 5672–5676 CrossRef CAS PubMed.
- Y.-Z. Wang, X.-X. Deng, L. Li, Z.-L. Li, F.-S. Du and Z.-C. Li, Polym. Chem., 2013, 4, 444–448 RSC.
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed.
- F. C. Geyer, F. Pareja, B. Weigelt, E. Rakha, I. O. Ellis, S. J. Schnitt and J. S. Reis-Filho, Am. J. Pathol., 2017, 187, 2139–2151 CrossRef CAS PubMed.
- P. F. Monteiro, M. Gulfam, C. J. Monteiro, A. Travanut, T. F. Abelha, A. K. Pearce, C. Jerome, A. M. Grabowska, P. A. Clarke, H. M. Collins, D. M. Heery, P. Gershkovich and C. Alexander, J. Controlled Release, 2020, 323, 549–564 CrossRef CAS PubMed.
- R. Dent, M. Trudeau, K. I. Pritchard, W. M. Hanna, H. K. Kahn, C. A. Sawka, L. A. Lickley, E. Rawlinson, P. Sun and S. A. Narod, Clin. Cancer Res., 2007, 13, 4429 CrossRef PubMed.
- G. Bianchini, J. M. Balko, I. A. Mayer, M. E. Sanders and L. Gianni, Nat. Rev. Clin. Oncol., 2016, 13, 674–690 CrossRef CAS PubMed.
- S. Oelmann, A. Travanut, D. Barther, M. Romero, S. M. Howdle, C. Alexander and M. A. R. Meier, Biomacromolecules, 2019, 20, 90–101 CrossRef CAS PubMed.
- N. Xiao, H. Liang and J. Lu, Soft Matter, 2011, 7, 10834–10840 RSC.
- X. Li, D. J. Hirsh, D. Cabral-Lilly, A. Zirkel, S. M. Gruner, A. S. Janoff and W. R. Perkins, Biochim. Biophys. Acta, Biomembr., 1998, 1415, 23–40 CrossRef CAS.
- A. Choucair, P. Lim Soo and A. Eisenberg, Langmuir, 2005, 21, 9308–9313 CrossRef CAS PubMed.
- R. R. Larson, M. B. Khazaeli and H. K. Dillon, Appl. Occup. Environ. Hyg., 2003, 18, 109–119 Search PubMed.
- J. A. Anderson, S. Lamichhane, T. Remund, P. Kelly and G. Mani, Acta Biomater., 2016, 29, 333–351 CrossRef CAS PubMed.
- V. Knorr, V. Russ, L. Allmendinger, M. Ogris and E. Wagner, Bioconjugate Chem., 2008, 19, 1625–1634 CrossRef CAS PubMed.
- C. Battistella and H.-A. Klok, Macromol. Biosci., 2017, 17, 1700022 CrossRef PubMed.
- Y. Zhang, C. Yang, W. Wang, J. Liu, Q. Liu, F. Huang, L. Chu, H. Gao, C. Li, D. Kong, Q. Liu and J. Liu, Sci. Rep., 2016, 6 Search PubMed.
- M. Gulfam, T. Matini, P. F. Monteiro, R. Riva, H. Collins, K. Spriggs, S. M. Howdle, C. Jerome and C. Alexander, Biomater. Sci., 2017, 5, 532–550 RSC.
- S. Oelmann, S. C. Solleder and M. A. R. Meier, Polym. Chem., 2016, 7, 1857–1860 RSC.
- D. Jeanmaire, J. Laliturai, A. Almalik, P. Carampin, D. A. Richard, E. Lallana, R. Evans, R. E. P. Winpenny and N. Tirelli, Polym. Chem., 2014, 5, 1393–1404 RSC.
- T. P. Smart, O. O. Mykhaylyk, A. J. Ryan and G. Battaglia, Soft Matter, 2009, 5, 3607–3610 RSC.
- V. Taresco, T. F. Abelha, R. J. Cavanagh, C. E. Vasey, A. B. Anane-Adjei, A. K. Pearce, P. F. Monteiro, K. A. Spriggs, P. Clarke, A. Ritchie, S. Martin, R. Rahman, A. M. Grabowska, M. B. Ashford and C. Alexander, Adv. Ther., 2020, 4 Search PubMed.
- H. Lu, R. H. Utama, U. Kitiyotsawat, K. Babiuch, Y. Jiang and M. H. Stenzel, Biomater. Sci., 2015, 3, 1085–1095 RSC.
- F. Ahmed, R. I. Pakunlu, G. Srinivas, A. Brannan, F. Bates, M. L. Klein, T. Minko and D. E. Discher, Mol. Pharmaceutics, 2006, 3, 340–350 CrossRef CAS PubMed.
- Y. Bae, N. Nishiyama, S. Fukushima, H. Koyama, M. Yasuhiro and K. Kataoka, Bioconjugate Chem., 2005, 16, 122–130 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb00045h |
| This journal is © The Royal Society of Chemistry 2022 |