Zwitterionic peptide-functionalized highly dispersed carbon nanotubes for efficient wastewater treatment†
Jie
Huang‡
 ,
Xiaojie
Sui‡
,
Haishan
Qi
*,
Xiang
Lan
,
Simin
Liu
and
Lei
Zhang
,
Xiaojie
Sui‡
,
Haishan
Qi
*,
Xiang
Lan
,
Simin
Liu
and
Lei
Zhang
 *
*
Department of Biochemical Engineering, School of Chemical Engineering and Technology, Frontier Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (MOE), Tianjin University, Tianjin 300350, P. R. China. E-mail: hsqi@tju.edu.cn; lei_zhang@tju.edu.cn
First published on 31st December 2021
Abstract
Multi-walled carbon nanotubes (MWCNTs) have displayed great potential as catalyst carriers due to their nanoscale structure and large specific surface area. However, their hydrophobicity and poor dispersibility in water restrict their applications in aqueous environments. Herein, the dispersibility of MWCNTs was significantly enhanced with a chimeric protein MPKE which consisted of a zwitterionic peptide unit and a mussel adhesive protein unit. The MPKE could be easily attached to MWCNTs (MPKE-MWCNTs) by a simple stirring process due to the versatile adhesion ability of mussel adhesive unit. As expected, the MPKE-MWCNTs displayed outstanding dispersibility in water (>7 months), as well as in alkaline solutions (pH = 12) and organic solvents (DMSO and ethanol) due to the hydrophilicity of the zwitterionic peptide unit. Moreover, the MPKE-MWCNTs were used as silver nanoparticle carriers for the reduction of 4-nitrophenol in wastewater, with the normalized rate constant knor up to 32.9 s−1 mmol−1. Meanwhile, they also exhibited excellent biocompatibility and antibacterial activity, which were favorable for wastewater treatment. This work provides a facile strategy for MWCNT modification, functionalization and applications in aqueous environments.
1. Introduction
Multi-walled carbon nanotubes (MWCNTs) have been considered as promising vehicles for metal nanoparticles, drugs, DNA aptamers, peptides and proteins.1–4 Due to their nanostructure, large specific surface area, electrical conductivity, etc., MWCNTs have shown great potential in biosensors, drug delivery, catalytic reactions, wastewater treatment, etc.5–8 However, the hydrophobicity of carbon materials makes them tend to agglomerate, significantly restricting their applications. For example, the poor dispersibility of MWCNTs can reduce the efficiency of the loaded catalysts.9 In addition, the aggregation of MWCNTs significantly affects their mechanical and electronic properties.10 Besides, a considerable number of studies have shown that MWCNTs exhibit cytotoxicity and genotoxicity.11,12To date, great efforts have been made to improve the dispersibility of MWCNTs (Table 1), including mechanical grinding, ultrasonication, chemical modification, etc.13,14 Mechanical grinding and ultrasonication are easy to operate but with short-term dispersion stability. For instance, MWCNTs maintained stable dispersibility for only 7 days after acid treatment and ball milling.13 The dispersibility of MWCNTs was stabilized in Milli-Q water for only 24 hours via high energy ultrasonication.15 In contrast, the chemical modification strategy endows MWCNTs with long-term dispersibility. For example, hyperbranched polylysine was used to modify MWCNTs through Michael addition, endowing MWCNTs with excellent dispersibility in an aqueous environment for 20 days.16 A novel star-like surfactant was designed to modify MWCNTs via hydrophobic polypropylene glycol backbones, endowing MWCNTs with good dispersibility in water for more than 30 days.17 These methods could efficiently enhance the dispersibility of MWCNTs in aqueous solutions and expand their applications. However, there are still some limitations, including strict reaction conditions, complex preparation processes, utilization of contaminated reagents.16 Therefore, developing a facile method to improve the dispersibility and biocompatibility of MWCNTs is significant.
Recently, peptide-engineered nanomaterials have attracted significant attention. By tailoring the amino acid sequences of peptides, peptide-functionalized nanomaterials can achieve improved homogeneous stability, biocompatibility, bioactivity and target binding capacity, thus advancing their applications in various fields.18–20 Particularly, due to the excellent hydrophilicity and biocompatibility, zwitterionic peptides have shown great potential for modification of nanomaterials. It is because their positive and negative groups can form a strong hydration layer via electrostatic interactions. Zwitterionic peptides have been applied in the modification of various materials, such as gold surfaces, conducting polymers, glass, mica and polytetrafluoroethylene (PTFE).21–24 Zwitterionic polymers were also utilized to modify carbon nanotubes for enhancing their hydrophilicity, such as poly(carboxybetaine methacrylate) (PCBMA) and poly(sulfobetaine methacrylate) (PSBMA).25,26 It is noteworthy that the modification methods of most peptides or polymers commonly require elaborate design, which depends on the nanomaterial substrates. In nature, marine organisms such as mussels, tube worms or barnacles can secrete adhesive proteins to adhere onto various substrates. It has been reported that mussel adhesive proteins (MAPs) were able to form various interactions with the substrates, including hydrogen bonds, metal-coordination, π–π/π-cation interactions, etc.27,28 Due to their versatile adhesion ability, MAPs have been considered as an adhesion platform for material functionalization.
In this work, we propose a facile method to enhance the dispersibility and biocompatibility of MWCNTs with a MPKE protein which consists of a zwitterionic peptide unit and a mussel-inspired adhesive protein unit (Fig. 1). The zwitterionic peptide unit contains 20 pairs of alternating lysine and glutamic acid (KE) peptides and the mussel-inspired adhesive protein unit contains 44 Dopa residues.23,29 MPKE can firmly modify MWCNTs due to the adhesion ability of Dopa residues.30 In addition, the zwitterionic peptides endowed MPKE-MWCNTs with enhanced dispersibility and biocompatibility. Then, the MPKE-MWCNTs were used as carriers to load silver nanoparticles (AgNPs) for reduction of 4-nitrophenol (4-NP) and antibacterial activity in wastewater treatment. The fabricated AgNPs@MPKE-MWCNTs exhibited high catalytic efficiency with a normalized rate constant (knor) of 32.9 s−1 mmol−1, which is beneficial for the wastewater treatment.31
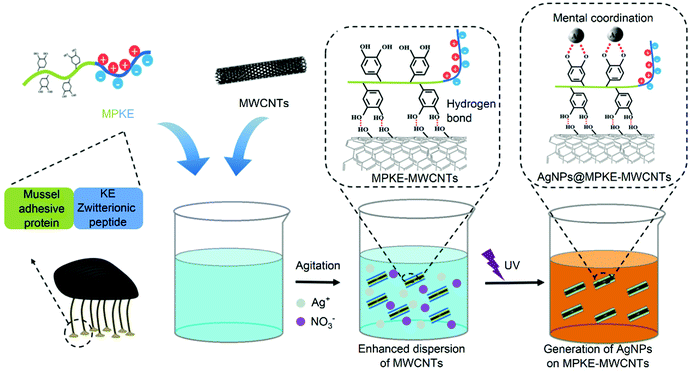 | ||
| Fig. 1 Schematic representation of the modification process of MWCNTs by MPKE and in situ loading of AgNPs onto MPKE-MWCNTs for reduction of 4-NP in wastewater. | ||
2. Materials and methods
2.1 Materials
Short hydroxyl functionalized multi-walled carbon nanotubes (MWCNTs, 30–50 nm outer diameter and 0.5–2 μm length) were provided by Chengdu Organic Chemicals Co., Ltd. Yeast extract and tryptone were obtained from OXOID Co., Ltd. The BCA protein assay kit was purchased from Cwbio. Nitrotetrazolium blue chloride (NBT) was purchased from Heowns Co., Ltd. Imidazole, Na2HPO4, NaH2PO4 and NaCl were obtained from Sangon Biotech Co., Ltd. Phosphate buffer saline (PBS), Isopropyl-β-D-thiogalactopyranoside (IPTG), kanamycin, chloramphenicol, dialysis bag MD25 (3500 MWCO) and sheep red blood cells (RBCs) were purchased from Solarbio Science & Technology Co., Ltd. The live/dead staining kit was purchased from Thermo Fisher Scientific.2.2 Production of the MPKE protein
MPKE was produced as previously reported.23 Briefly, E. coli was cultured with IPTG to induce MPKE proteins. After collecting bacteria through centrifugation, the precipitates were resuspended in the binding buffer (20 mM sodium phosphate, 0.5 M NaCl and 20 mM imidazole, pH 7.0) and crushed by ultrasonication. The supernatants were filtered using a 0.22 μm polyethersulfone membrane and then purified by Ni-affinity chromatography using the AKTA Pure L1 chromatography system. The target protein was eluted by elution buffer (20 mM sodium phosphate, 0.5 M NaCl and 500 mM imidazole, pH 7.0). Then, it was desalted by ultrafiltration and stored at 4 °C. All samples were analysed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). And the NBT staining kit was employed to determine the Dopa group.2.3 Preparation process of MPKE-MWCNTs
MPKE was dissolved in water solutions with 5% (v/v) acetic acid. Afterward, MWCNTs were added to the above solutions and the final concentrations of MPKE and MWCNTs were 0.5 and 0.3 mg mL−1, respectively. After stirring at room temperature overnight, the samples were collected by centrifugation and washed with pure water three times. Then, the samples were stored at 4 °C for further research.2.4 Characterization of the MPKE-MWCNTs
The microscopic morphology of samples was observed using a TEM (JEM-F200, JEOL) at an accelerating voltage of 200 kV. Besides, Fourier transform infra-red (FTIR) spectra were obtained using an ALPHA II (BRUKER, Germany) with a scan range of 400 to 4000 cm−1. The water contact angle test was conducted to characterize the hydrophilicity of the samples. The contact angle (θs) was measured using an optical θ meter (JC2000DM, Powereach). The volume of water was 5 μL and five parallel tests were conducted to determine the average contact angle. In addition, the samples were dissolved in water solutions and the absorbance was measured using a microplate reader (Infinite M200 Pro, Tecan). The size distribution and the zeta potential of samples were measured using a dynamic light scattering nanoparticle analyzer (ZS90, Malvern). The dispersion stability experiment was conducted via ultrasonication. The MPKE-MWCNT dispersion was treated by 100 W ultrasonication for 30 min. Then, the photographs and absorbance were recorded to determine the dispersibility of MPKE-MWCNTs.2.5 Antifouling assay
The FITC-BSA kit was employed to measure the protein adsorption. 500 μL of the samples were mixed with 1 mL of FITC-BSA solution (0.1 mg mL−1) and stirred overnight away from light (37 °C, 300 rpm). Afterward, the samples were washed with PBS solutions three times to remove the excessive FITC-BSA and observed using a fluorescence microscope (Nikon Eclipse Ti–S). The fluorescence intensity was analysed using ImageJ software.2.6 Hemolysis assay
The sheep RBCs were washed using PBS solutions three times before use and collected by centrifugation. Then, 200 μL of fresh RBCs were added to 800 μL of the sample solutions and incubated at 37 °C for 1 h. Equal amounts of RBCs were added to 800 μL of pure water or 0.9% NaCl as the negative control (100% hemolysis rate) and positive control (0% hemolysis rate), respectively. The samples were centrifuged at 1000 rpm for 10 min and 100 μL of the supernatant was added to a 96-well microplate. The absorbance at 541 nm was measured using a multifunctional microplate reader. The hemolysis rate (HR) was calculated by the following formula:| HR = (AS − AN)/(AP − AN) |
2.7 Cytotoxicity assay
The cytotoxicity assay was performed on NIH/3T3 cells using a previous method,32 with minor modifications. NIH/3T3 cells were seeded at a density of 6 × 103 cells per well onto 96-well plates and cultured in DMEM medium with 10% FBS overnight for attachment. Afterward, the cells were further cultured in DMEM medium containing 10% FBS and 40 μg mL−1 MPKE-MWCNTs for 24 h. The Calcein AM/PI reagent mixture solution was added to the above solutions and stained away from light at 37 °C for 30 min. The live and dead cells were observed and counted using an inverted microscope.2.8 Preparation and characterization of AgNPs@MPKE-MWCNTs
MWCNTs, MPKE and AgNO3 were mixed with final concentrations of 0.3, 0.5 and 0.3 mg mL−1 respectively. Then, the mixture was stirred at room temperature overnight and further exposed to UV irradiation (365 nm) for 4 h to reduce and load AgNPs onto MPKE-MWCNTs. The samples were collected by centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 2 min, 4 °C) and washed using pure water three times.
000 rpm, 2 min, 4 °C) and washed using pure water three times.
| Materials | Methods of improving dispersibility | Dispersion stability | pH range | Other solvents | Ref. |
|---|---|---|---|---|---|
| MWCNTs | Zwitterionic peptide containing protein; agitation | 7 Months | 1–12 | DMSO, Ethanol | This work |
| MWCNTs | Acid/alkaline treatment; ball milling | 1 Month | — | — | 13 |
| MWCNTs | Ultrasound-assisted acid etching and γ-irradiation | Several weeks | — | — | 33 |
| MWCNTs | Acid/alkaline treatment; high energy ultrasonication | 24 Hours | — | — | 15 |
| MWCNTs | Hyperbranched polylysine (HBPL); Michael addition | 20 Days | 3–11 | — | 16 |
| MWCNTs | Novel surfactant with a star-like molecular structure and terminated sulfonate; ultrasonication | 30 Days | — | — | 17 |
| MWCNTs | 4,4′-Di (n-tetradecyl) diphenylmethane disulfate salt (DSDM); ultrasonication | 5 Months | — | — | 34 |
| MWCNTs/SWCNTs | The BHT oxidation in THF; solvent exchange with THF-stabilized CNT dispersion | 6 Weeks | — | THF | 35 |
| MWCNTs | 1,3,6,8-Tetra(oct-1-yn-1-yl) pyrene (TOPy) and 1,3,6,8-tetra(dodec-1-yn-1-yl) pyrene (TDPy); ultrasonication | A few days | — | THF | 36 |
| MWCNTs | Shellac (SL) bio-resin; magnetic stirring | 14 Days | — | — | 37 |
| MWCNTs | S-Layer proteins, 0.1% Triton X 100; ultrasonication | 6 Months | — | — | 38 |
The microscopic morphology of the samples was observed using a SEM (Apreo S LoVac, FEI Co, Ltd) with an accelerating voltage of 5 kV and a TEM (JEM-2100F, JEOL) with an accelerating voltage of 200 kV. The elemental composition of the samples was obtained by Energy dispersive spectroscopy (Octane Elect EDS System, EDAX). The UV-Vis spectrum was obtained using a microplate reader (Infinite M200 Pro, Tecan). The AgNP loading capacity was measured using an IPC-OES (Agilent 7700X & Agilent 7800, Agilent Technologies, Inc.)
2.9 Reduction of 4-NP by AgNPs@MPKE-MWCNTs
A typical method for the reduction of 4-NP by NaBH4 was used to measure the catalytic efficiency of AgNPs@MPKE-MWCNTs.39 4-NP solution (10 mM, 600 μL), cold NaBH4 solution (0.5 M, 800 μL) and deionized water (2.6 mL) were mixed. Then, AgNPs@MPKE-MWCNT solution (0.3 mg mL−1, 200 μL) was added to the above solution. The UV-vis spectrum ranging from 300 to 600 nm of the mixture was recorded at a determined time interval.The kinetic parameters of the reaction satisfy the following pseudo first-order kinetic reaction equation:39
| ln (Ct/C0) = ln (At/A0) = kappt |
2.10 Antibacterial activity assays
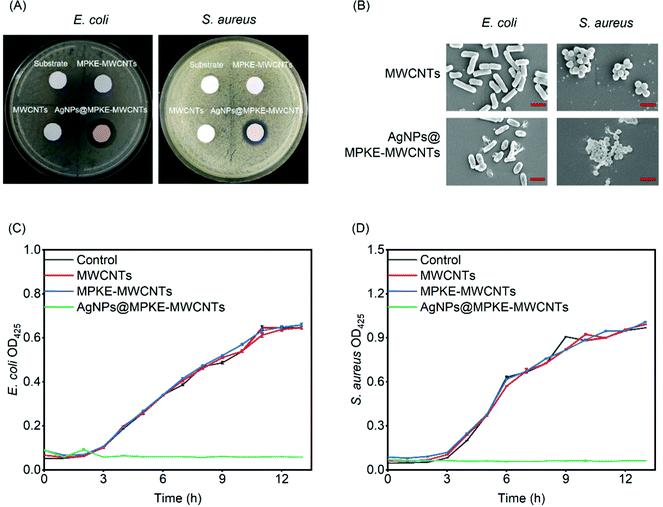
The antibacterial activity of the AgNPs@MPKE-MWCNTs was characterized by an inhibition zone method. The sample discs (diameter = 1 cm) were cocultured with Gram-negative E. coli and Gram-positive S. aureus on solid medium. Bacteriostatic circles were observed and measured after incubation at 37 °C overnight.E. coli or S. aureus (OD600 = 1.0, 5 μL) cells were seeded in 3 mL of liquid LB medium containing 25 μg mL−1 AgNPs@MPKE-MWCNTs. The absorbance of the culture medium at 600 nm was measured to determine the growth rate curves of the cells. The cocultured cells were fixed with glutaraldehyde and dried with ethanol. Then the bacterial morphology was observed by SEM (Apreo S LoVac, FEI Co, Ltd).
3. Results and discussion
3.1 Preparation and characterization of MPKE-MWCNTs
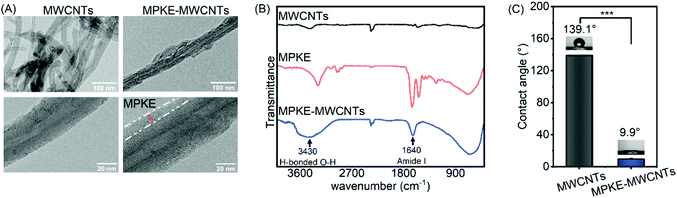
MPKE was produced by E. coli BL21(DE3) cells, which were constructed in our previous work.23 SDS-PAGE analysis indicated that the molecular weight was consistent with the theoretical value, and NBT staining confirmed the presence of Dopa residues (Fig. S1, ESI†). These results demonstrated the successful expression of the MPKE protein. Then, MPKE modified MWCNTs were prepared by simple mixing and stirring. As shown in Fig. 2A, the diameter of pristine MWCNTs was 39 nm and that of modified MWCNTs was 56 nm, indicating the existence of MPKE coating. The FTIR spectra confirmed the presence of the amide I group of MPKE at 1640 cm−1 and the formation of hydrogen bonds between MPKE and MWCNTs at 3430 cm−1 (Fig. 2B).9 These results indicated that MPKE could be coated on MWCNTs via hydrogen bonds. Moreover, the change of surface hydrophilicity was confirmed by measuring the contact angle. As shown in Fig. 2C, the MWCNTs were hydrophobic with a water contact angle of 139.1°, consistent with previous reports.40 After modification, the water contact angle decreased to 9.9°. These results demonstrated that the MPKE could be easily anchored onto MWCNTs and improved their hydrophilicity.3.2 Dispersion stability of MPKE-MWCNTs
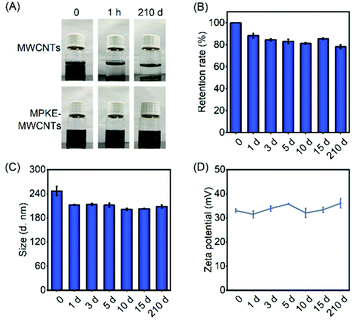
Long-term dispersion stability of carbon nanomaterials is critical to their applications in aqueous environments. As shown in Fig. 3A, pristine MWCNTs exhibited poor dispersibility, which gathered and precipitated within 1 hour. In stark contrast, MPKE-MWCNTs maintained excellent dispersibility for more than 7 months in an aqueous environment without visible aggregation (Table 1). The supernatant absorbance of the samples showed 78.3% retention of their initial ones after being stored for 7 months (Fig. 3B), demonstrating long-term dispersibility in water. Meanwhile, the dynamic light scattering (DLS) test was used to measure the particle size and zeta potential of MPKE-MWCNTs, which also exhibited insignificant change (Fig. 3C and D). Besides, after ultrasonication treatment, the MPKE-MWCNTs could still retain 90% of dispersibility compared to the untreated samples, indicating the strong attachment of MPKE onto MWCNTs (Fig. S2, ESI†). Moreover, even in alkaline solutions (pH = 12) and organic solvents (DMSO and ethanol) (Fig. S3, ESI†), the MPKE-MWCNTs also showed excellent dispersibility. The improved dispersity were mainly attributed to the hydrophilic zwitterionic peptides of MPKE, which could expose to the surface of MPKE-MWCNTs and generate a hydration layer.233.3 Biocompatibility tests
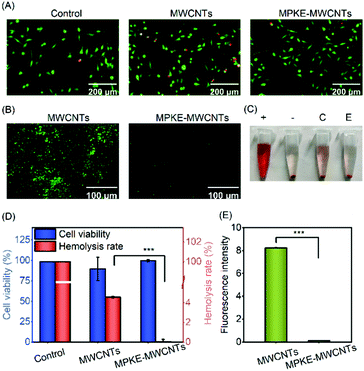
Pristine MWCNTs can lead to potential health and safety problems. For instance, MWCNTs can cause a significant shape change of erythrocytes and even hemolysis at high concentrations.41 Besides, MWCNTs may induce oxidative stress in mitochondria.42 Therefore, it is significant to evaluate the biocompatibility of modified MWCNTs. As shown in Fig. 4A and D, compared to the control samples, the MWCNT group showed significantly lower cell viability. However, no significant difference in cell viability could be observed between the control group and the MPKE-MWCNT group. In addition, the MPKE-MWCNTs also displayed excellent hemocompatibility with only 0.045% hemolysis rate (Fig. 4C and D), which was significantly lower than the clinical limitation (5% hemolysis rate). Furthermore, the anti-protein adsorption performance was measured using the FITC-BSA kit. As shown in Fig. 4B and E, the MPKE-MWCNTs showed excellent anti-protein absorption performance with a fluorescence intensity of only 0.13, while that of MWCNTs was 8.24. These results verified that MPKE-MWCNTs possessed excellent biocompatibility.3.4 AgNP loading and catalytic reduction of 4-NP
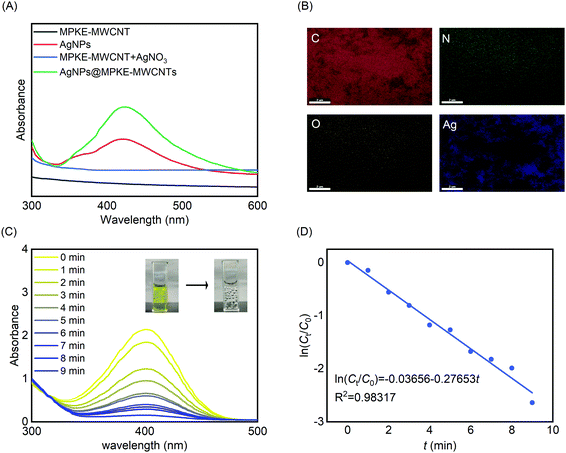
Because of their large specific surface area, MWCNTs were usually used as carriers of catalysts, such as Pa, Pd, Au, and Ag.43–46 Herein, we prepared AgNPs@MPKE-MWCNTs via in situ redox reactions and investigated their application for reduction of 4-NP in wastewater. UV-Vis spectra were used to confirm the generation of AgNPs. As shown in Fig. 5A, AgNPs@MPKE-MWCNTs exhibited an absorption peak at around 425 nm due to the plasmon resonance of AgNPs.47 As shown in Fig. S4 and S5A (ESI†), AgNPs with a particle size of 5–13 nm were generated on MPKE-MWCNTs, which was smaller than that of AgNPs in previous reports (Table 2). Besides, the TEM images showed clear lattice fringes with a 0.237 nm interplanar distance, corresponding to the (111) plane of silver nanoparticles.59,60 From EDS tests, it could be seen that the generated AgNPs were uniformly distributed onto MPKE-MWCNTs (Fig. 5B and Fig. S5B, ESI†). According to the ICP-OES tests, the content of Ag of AgNPs@MPKE-MWCNTs was 20.2 wt%. Compared with other AgNPs in Table 2, the AgNPs@MPKE-MWCNTs exhibited outstanding catalytic efficiency. In these studies, the reduction of 4-NP was used as a model reaction for determining the catalytic activity and wastewater treatment potential of AgNPs@MPKE-MWCNTs. The UV-vis absorption maximum of the mixture of 4-NP and NaBH4 was at 400 nm. After the addition of AgNPs@MPKE-MWCNTs, the reaction mixture gradually graded from yellow to colourless. In the meantime, the absorbance at 400 nm of the mixture decreased with the reaction time (Fig. 5C). Next, the reaction kinetics were investigated (Fig. 5D) and compared with those in other studies. It could be seen that AgNPs@MPKE-MWCNTs exhibited higher catalytic activity and knor (normalized rate constant) could reach 32.9 s−1 mmol−1. It was possibly because of the excellent dispersibility of MPKE-MWCNTs as well as the smaller particle size of the loaded AgNPs, which led to a larger specific surface area and more exposed atoms on the surface as potential catalytic points.61| Catalyst name | Catalyst used (mmol) | k app (s−1) | k nor (s−1 mmol−1) | Particle size (nm) | Ref. |
|---|---|---|---|---|---|
| AgNPs@MPKE-MWCNTs | 1.41 × 10−4 | 4.62 × 10−3 | 32.9 | 5–13 | This work |
| AgNPs | 9.26 × 10−3 | 2.84 × 10−3 | 0.307 | ∼8 | 48 |
| Ag@RF@Fe3O4 | 3.82 × 10−3 | 3.78 × 10−2 | 9.90 | — | 49 |
| Ag/Al2O3 | 3.50 × 10−3 | — | — | <20 | 50 |
| Ag/ZnO | 5.55 × 10−4 | 3.97 × 10−3 | 7.15 | 35–40 | 51 |
| Ag/ZnO/CaCO3 | 8.11 × 10−4 | 7.78 × 10−3 | 9.59 | — | 52 |
| Ag/TP | 5.5 × 10−2 | 5.27 × 10−3 | 0.0958 | 57–85 | 53 |
| Ag@NFC | 1.85 × 10−1 | 4.66 × 10−2 | 21.6 | <30 | 54 |
| Ag/PAM/PPy/GO | 1.59 × 10−3 | 3.38 × 10−2 | 21.3 | — | 55 |
| RGO–Ag | 1.11 × 10−2 | 3.03 × 10−3 | 27.3 | — | 56 |
| GO-PNVP-Ag | 4.63 × 10−3 | 1.90 × 10−2 | 4.10 | — | 57 |
| Ag−rGO | 1.48 × 10−2 | 4.40 × 10−2 | 2.97 | — | 58 |
3.5 Antibacterial performance of AgNPs@MPKE-MWCNTs
The number of bacteria is one of the water quality detection indicators and a large number of bacteria can cause disease or form biofilms to affect the stability of water equipment.62,63 Previous studies have demonstrated that AgNPs exhibited slow but long-term antibacterial performance with no harmful by-products at low concentrations.64 Herein, E. coli and S. aureus were used as model Gram-negative and positive bacteria respectively to evaluate the antibacterial performance of AgNPs@MPKE-MWCNTs. As shown in Fig. 6A, the control samples showed no bacteriostatic circles, whereas AgNPs@MPKE-MWCNTs exhibited obvious bacteriostatic circles and the diameters for E. coli and S. aureus were 1.83 and 1.45 cm, respectively. This result demonstrated the antibacterial activity of AgNPs@MPKE-MWCNTs. Besides, the SEM images indicated that the bacteria incubated with AgNPs@MPKE-MWCNTs had collapsed surfaces and inclusions outflowing (Fig. 6B), which may be ascribed to the damage effects of AgNPs on the biomolecules and cell membranes.65Moreover, growth curves of bacteria were also measured. Fig. 6C and D showed the growth curves of E. coli and S. aureus respectively. AgNPs@MPKE-MWCNTs could completely inhibit the growth of both Gram-negative and positive bacteria. The antibacterial mechanisms may be attributed to the multifunctional capability of AgNPs, including DNA degradation, protein oxidation, ROS production, lipid peroxidation, ATP consumption, biomolecular destruction and membrane interaction.65,66 Besides, due to the smaller nanoparticle size (5–13 nm), the AgNPs@MPKE-MWCNTs may enter cells more easily to kill the bacteria.67,68 It should be noted that AgNPs@MPKE-MWCNT samples still exhibited efficient bacteriostatic function after 2 months of storage (Fig. S6, ESI†). Therefore, these results demonstrated that the AgNPs@MPKE-MWCNTs displayed antibacterial activity, which is important to wastewater treatment.
4. Conclusions
In this work, a novel modification strategy was developed to improve the dispersibility of MWCNTs with excellent biocompatibility. With the adhesion of Dopa units, MPKE could simply and firmly modify MWCNTs. As for the zwitterionic peptide unit, the formed hydration layer endows MPKE-MWCNTs with outstanding hydrophilicity. Therefore, the MPKE-MWCNTs exhibited long-term dispersion stability for at least 7 months and could also be stabilized in other environments, including alkaline solution, DMSO and ethanol. In addition, compared with pristine MWCNTs, MPKE-MWCNTs possessed excellent biocompatibility. Furthermore, MPKE-MWCNTs could be applied as catalyst carriers of AgNPs for reduction of 4-NP in wastewater with a high catalytic activity (knor = 32.9 s−1 mmol−1) and bacterial inhibition. The feasible and effective modification of MPKE promotes the application of MWCNTs in aqueous environments.Author contributions
Jie Huang and Xiaojie Sui: conceptualization, investigation, methodology, software, validation, writing – original draft, data curation, resources, and formal analysis. Haishan Qi: conceptualization, investigation, methodology, software, validation, writing – original draft, supervision, writing – review & editing, and funding acquisition. Xiang Lan and Simin Liu: data curation, validation, resources, software, and formal analysis. Lei Zhang: conceptualization, supervision, writing – review & editing, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financially supported by the National Key R&D Program of China (2021YFC2100800), the National Natural Science Foundation of China (21606165, 21621004 and 21422605), the Program of Introducing Talents of Discipline to Universities (BP0618007), and the Open Funding Project of the National Key Laboratory of Biochemical Engineering.References
- R. Aghajari and A. Azadbakht, Anal. Biochem., 2018, 547, 57–65 CrossRef CAS PubMed.
- Y. Yan, R. Wang, Y. Hu, R. Sun, T. Song, X. Shi and S. Yin, Drug Delivery, 2018, 25, 1607–1616 CrossRef CAS.
- V. Gedi, C. K. Song, G. B. Kim, J. O. Lee, E. Oh, B. S. Shin, M. Jung, J. Shim, H. Lee and Y.-P. Kim, Sens. Actuators, B, 2018, 256, 89–97 CrossRef CAS.
- Q. Chen, Q. Wang, Y.-C. Liu, T. Wu, Y. Kang, J. D. Moore and K. E. Gubbins, J. Chem. Phys., 2009, 131, 015101 CrossRef PubMed.
- N. I. Khan, A. G. Maddaus and E. Song, Biosensors, 2018, 8, 7 CrossRef PubMed.
- V. Gedi, C. K. Song, G. B. Kim, J. O. Lee, E. Oh, B. S. Shin, M. Jung, J. Shim, H. Lee and Y.-P. Kim, Sens. Actuators, B, 2018, 256, 89–97 CrossRef CAS.
- Y. Qinzheng, L. Dianliang, L. Xiaoliang, G. Tiantian, Z. Xuedong, Z. Xinxin and W. Wenlong, Bioelectrochemistry, 2021, 142, 107941 CrossRef.
- K. M. Lee, C. P. P. Wong, T. L. Tan and C. W. Lai, Mater. Sci. Eng., B, 2018, 236-237, 61–69 CrossRef CAS.
- Y. C. Jung, H. Muramatsu, K. Fujisawa, J. H. Kim, T. Hayashi, Y. A. Kim, M. Endo, M. Terrones and M. S. Dresselhaus, Small, 2011, 7, 3292–3297 CrossRef CAS.
- G. Sakellariou, D. Priftis and D. Baskaran, Chem. Soc. Rev., 2013, 42, 677–704 RSC.
- L. Zhou, H. J. Forman, Y. Ge and J. Lunec, Toxicol. In Vitro, 2017, 42, 292–298 CrossRef CAS PubMed.
- D. Mohanta, S. Patnaik, S. Sood and N. Das, J. Pharm. Anal., 2019, 9, 293–300 CrossRef PubMed.
- B. Tserengombo, H. Jeong, E. Dolgor, A. Delgado and S. Kim, Nanomaterials, 2021, 11, 1323 CrossRef CAS PubMed.
- C. M. Park, Y. A. J. Al-Hamadani, J. Heo, N. Her, K. H. Chu, M. Jang, S. Lee and Y. Yoon, Ultrason. Sonochem., 2017, 39, 750–757 CrossRef CAS PubMed.
- Z. Wu and S. Mitra, J. Nanopart. Res., 2014, 16, 2563 CrossRef.
- H. Lu, L. Zou, Y. Xu and Y. V. Li, J. Appl. Polym. Sci., 2018, 135, 46249 CrossRef.
- M. Qiao, Q. Ran and S. Wu, Appl. Surf. Sci., 2018, 433, 975–982 CrossRef CAS.
- Y. Wang, K. Xia, L. Wang, M. Wu, X. Sang, K. Wan, X. Zhang, X. Liu and G. Wei, Small, 2021, 17, 2005578 CrossRef CAS PubMed.
- G. Wei, Z. Su, N. P. Reynolds, P. Arosio, I. W. Hamley, E. Gazit and R. Mezzenga, Chem. Soc. Rev., 2017, 46, 4661–4708 RSC.
- Z. Su, H. Shen, H. Wang, J. Wang, J. Li, G. U. Nienhaus, L. Shang and G. Wei, Adv. Funct. Mater., 2015, 25, 5472–5478 CrossRef CAS.
- C. Zhang, J. Lu, Y. Hou, W. Xiong, K. Sheng and H. Lu, ACS Appl. Mater. Interfaces, 2018, 10, 17463–17470 CrossRef CAS.
- J. Wang, D. Wang and N. Hui, Bioelectrochemistry, 2020, 136, 107595 CrossRef CAS PubMed.
- H. Qi, W. Zheng, X. Zhou, C. Zhang and L. Zhang, Chem. Commun., 2018, 54, 11328–11331 RSC.
- W. Zheng, M. Liu, H. Qi, C. Wen, C. Zhang, J. Mi, X. Zhou, L. Zhang and D. Fan, J. Colloid Interface Sci., 2020, 576, 68–78 CrossRef CAS PubMed.
- A. Surapathi, H.-y. Chen, E. Marand, J. Karl Johnson and Z. Sedlakova, J. Membr. Sci., 2013, 429, 88–94 CrossRef CAS.
- X. Zhao, L. Cheng, R. Wang, N. Jia, L. Liu and C. Gao, J. Membr. Sci., 2019, 589, 117257 CrossRef CAS.
- Q. Guo, J. Chen, J. Wang, H. Zeng and J. Yu, Nanoscale, 2020, 12, 1307–1324 RSC.
- K. Zhang, F. Zhang, Y. Song, J.-B. Fan and S. Wang, Chin. J. Chem., 2017, 35, 811–820 CrossRef CAS.
- H. Qi, W. Zheng, C. Zhang, X. Zhou and L. Zhang, ACS Appl. Mater. Interfaces, 2019, 11, 12846–12853 CrossRef CAS.
- P. K. Forooshani and B. P. Lee, J. Polym. Sci., Part A: Polym. Chem., 2017, 55, 9–33 CrossRef PubMed.
- Y. Li, P. Zhang, Z. Ouyang, M. Zhang, Z. Lin, J. Li, Z. Su and G. Wei, Adv. Funct. Mater., 2016, 26, 2122–2134 CrossRef CAS.
- Z. Wang, R. X. Zhang, C. Zhang, C. Dai, X. Ju and R. He, J. Agric. Food Chem., 2019, 67, 887–894 CrossRef CAS PubMed.
- S. Bibi, T. Yasin, M. Nawaz and G. J. Price, Mater. Chem. Phys., 2018, 207, 23–29 CrossRef CAS; S. Bibi, T. Yasin, M. Nawaz and G. J. Price, Mater. Chem. Phys., 2018, 207, 23–29 CrossRef.
- J. Hou, W. Du, F. Meng, C. Zhao and X. Du, J. Colloid Interface Sci., 2018, 512, 750–757 CrossRef CAS.
- J. P. V. Damasceno and A. J. G. Zarbin, Chem. Commun., 2019, 55, 5809–5812 RSC.
- S. B. Kandy, G. P. Simon, W. Cheng, J. Zank, K. Saito and A. R. Bhattacharyya, ACS Omega, 2019, 4, 6647–6659 CrossRef PubMed.
- A. K. M. M. Alam, M. D. H. Beg, R. M. Yunus, M. R. Islam and Q. T. H. Shubhra, Colloid Interface Sci. Commun., 2021, 42, 100395 CrossRef CAS.
- A. Breitwieser, U. B. Sleytr and D. Pum, Nanomaterials, 2021, 11, 1346 CrossRef CAS PubMed.
- M. H. Rashid and T. K. Mandal, Adv. Funct. Mater., 2008, 18, 2261–2271 CrossRef CAS.
- A. P. Evseev, E. A. Vorobyeva, Y. V. Balakshin, K. D. Kushkina, A. V. Stepanov, V. S. Chernysh, N. G. Chechenin and A. A. Shemukhin, Surf. Interfaces, 2021, 23, 100955 CrossRef CAS.
- Y. Heo, C.-A. Li, D. Kim and S. Shin, Clin. Hemorheol. Microcirc., 2017, 65, 49–56 CAS.
- G. Visalli, A. Facciola, M. Curro, P. Lagana, V. La Fauci, D. Iannazzo, A. Pistone and A. Di Pietro, Int. J. Environ. Res. Public Health, 2019, 16, 792 CrossRef CAS PubMed.
- Y. Ya, S. Huiping, Y. Pingping, K. Shi-Zhao and M. Jin, Appl. Surf. Sci., 2011, 257, 3620–3626 CrossRef.
- Y. Haitang, L. Bingyue, C. Rongjing, X. Ruimin and L. Shanhu, J. Nanopart. Res., 2017, 19, 354 CrossRef.
- H. Wei, B. Peiyan, S. Changshuai, W. Jin and W. Erkang, J. Mater. Chem. A, 2016, 4, 4485–4489 RSC.
- S. U. B. Ramakrishna, D. S. Reddy, S. S. Kumar and V. Himabindu, Int. J. Hydrogen Energy, 2016, 41, 20447–20454 CrossRef CAS.
- S. Tang, S. Vongehr and X. Meng, J. Phys. Chem. C, 2010, 114, 977–982 CrossRef CAS.
- D. Ayodhya and G. Veerabhadram, Inorg. Nano-Met. Chem., 2020, 51, 1831–1841 Search PubMed.
- H.-L. Cao, C. Liu, F.-Y. Cai, X.-X. Qiao, A. B. Dichiara, C. Tian and J. Lu, Water Res., 2020, 179, 115882 CrossRef CAS PubMed.
- M. Nasrollahzadeh, Z. Issaabadi and S. M. Sajadi, J. Mater. Sci.: Mater. Electron., 2019, 30, 3847–3859 CrossRef CAS.
- S. A. Rasaki, C. Zhao, R. Wang, J. Wang, H. Jiang and M. Yang, Mater. Res. Bull., 2019, 119, 110536 CrossRef CAS.
- L. Chang, Y. Feng, B. Wang, X. Huang, D.-P. Yang and Y. Lu, RSC Adv., 2019, 9, 41336–41344 RSC.
- M. Ismail, M. I. Khan, S. B. Khan, K. Akhtar, M. A. Khan and A. M. Asiri, J. Mol. Liq., 2018, 268, 87–101 CrossRef CAS.
- H. Heidari, J. Cluster Sci., 2018, 29, 475–481 CrossRef CAS.
- H. Mao, C. Ji, M. Liu, Z. Cao, D. Sun, Z. Xing, X. Chen, Y. Zhang and X.-M. Song, Appl. Surf. Sci., 2018, 434, 522–533 CrossRef CAS.
- S. Ganguly, P. Das, M. Bose, T. K. Das, S. Mondal, A. K. Das and N. C. Das, Ultrason. Sonochem., 2017, 39, 577–588 CrossRef CAS PubMed.
- S. Singh, R. K. Gundampati, K. Mitra, K. Ramesh, M. V. Jagannadham, N. Misra and B. Ray, RSC Adv., 2015, 5, 81994–82004 RSC.
- M. K. Joshi, H. R. Pant, H. J. Kim, J. H. Kim and C. S. Kim, Colloids Surf., A, 2014, 446, 102–108 CrossRef CAS.
- Y. Zhu, J. Zhang, J. Song, J. Yang, T. Xu, C. Pan and L. Zhang, J. Mater. Chem. B, 2017, 5, 8451–8458 RSC.
- M. S. Tamboli, M. V. Kulkarni, R. H. Patil, W. N. Gade, S. C. Navale and B. B. Kale, Colloids Surf., B, 2012, 92, 35–41 CrossRef CAS PubMed.
- H. Zhang, X. Li and G. Chen, J. Mater. Chem., 2009, 19, 8223–8231 RSC.
- D. van der Kooij, G. L. Bakker, R. Italiaander, H. R. Veenendaal and B. A. Wullings, Appl. Environ. Microbiol., 2017, 83, e02737 Search PubMed.
- J. W. Park, K.-Y. Park, Y. Na, S. Park, S. Kim, J. H. Kweon and S. K. Maeng, Water Res., 2019, 165, 115025 CrossRef CAS PubMed.
- K. Chamakura, R. Perez-Ballestero, Z. Luo, S. Bashir and J. Liu, Colloids Surf., B, 2011, 84, 88–96 CrossRef CAS PubMed.
- O. T. Fanoro and O. S. Oluwafemi, Pharmaceutics, 2020, 12, 1044 CrossRef CAS PubMed.
- S.-L. Loo, W. B. Krantz, A. G. Fane, Y. Gao, T.-T. Lim and X. Hu, Environ. Sci. Technol., 2015, 49, 2310–2318 CrossRef CAS PubMed.
- T. Harifi and M. Montazer, Ind. Eng. Chem. Res., 2014, 53, 1119–1129 CrossRef CAS.
- Y. Chen, Y. Ding and J. Zheng, Chem. Eng. J., 2020, 379, 122268 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1tb02348a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |