Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Investigating histidinylated highly branched poly(lysine) for siRNA delivery
Ali
Alazzo
ab,
Nurcan
Gumus
b,
Pratik
Gurnani
 b,
Snjezana
Stolnik
b,
Ruman
Rahman
b,
Snjezana
Stolnik
b,
Ruman
Rahman
 c,
Keith
Spriggs
b and
Cameron
Alexander
c,
Keith
Spriggs
b and
Cameron
Alexander
 *b
*b
aDepartment of Pharmaceutics, College of Pharmacy, University of Mosul, Mosul, Iraq
bDivision of Molecular Therapeutics and Formulation, School of Pharmacy, University of Nottingham, Nottingham NG7 2RD, UK. E-mail: cameron.alexander@nottingham.ac.uk
cBioDiscovery Institute, University of Nottingham, Nottingham NG7 2RD, UK
First published on 23rd November 2021
Abstract
The temporary silencing of disease-associated genes utilising short interfering RNA (siRNA) is a potent and selective route for addressing a wide range of life limiting disorders. However, the few clinically approved siRNA therapies rely on lipid based formulations, which although potent, provide limited chemical space to tune the stability, efficacy and tissue selectivity. In this study, we investigated the role of molar mass and histidinylation for poly(lysine) based non-viral vectors, synthesised through a fully aqueous thermal condensation polymerisation. Formulation and in vitro studies revealed that higher molar mass derivatives yielded smaller polyplexes attributed to a greater affinity for siRNA at lower N/P ratios yielding greater transfection efficiency, albeit with some cytotoxicity. Histidinylation had a negligible effect on formulation size, yet imparted a moderate improvement in biocompatibility, but did not provide any meaningful improvement over silencing efficiency compared to non-histidinylated derivatives. This was attributed to a greater degree of cellular internalisation for non-histidinylated analogues, which was enhanced with the higher molar mass material.
Introduction
RNA interference (RNAi) is an evolutionarily conserved pathway that enables post-transcriptional regulation of gene expression through cleavage and degradation of messenger RNA (mRNA) in a selective manner. This is accomplished by utilizing short (21–25 base pair) double-stranded non-coding RNA (dsRNA), such as siRNA (small interfering RNA) to transiently interfere with mRNA translation.1–4 In principle it is possible to target specifically any transcribed genes providing its sequence is known, therefore RNAi has attracted attention as a potent and selective therapeutic approach for a range of diseases particularly cancers, and those lacking ‘druggable’ targets,5 by downregulating expression of disease related proteins. However, the bioavailability of administered ‘naked’ siRNAs at their intracellular target, the cytosol, is limited by, instability against endogenous nucleases, poor cellular uptake and weak endosomal escape.6–9 Therefore, several non-viral delivery systems, including polymeric nanoparticles and liposomes have gained attention as synthetic carriers for genetic material, including siRNA, in order to overcome the limitations of unpackaged nucleic acids, and thus to enhance cell specificity and knockdown efficiency.4Polymeric delivery systems are typically designed around cationic moieties which can electrostatically condense negatively charged nucleic acids into small (<200 nm diameter) polyelectrolyte complexes, often referred to as polyplexes.10–13 In contrast to other formulation systems, the chemical versatility that can be derived for polymeric materials, for example through variations in molar mass,14 architecture,15 and monomer chemistries16 offers significant adaptability to tune physical properties including stimuli responsiveness,17 particle size, surface charge, and therapeutic loading capacity.18 In turn, these factors enable modulation of biological activities, such as biodistribution,19 cytocompatibility, cellular uptake20 and endosomal escape,21 allowing macromolecular gene delivery vectors to target specific tissue types or diseases. Of the many polymer–gene delivery systems reported, branched macromolecules have garnered significant attention due to their dense and globular architecture, larger surface area compared to linear polymers and controllable charge density which have been reported to enhance nucleic acid condensation and transfection efficiency.22,23 These properties are most evident from the widespread use of the branched polycation, poly(ethyleneimine), bPEI, which has demonstrated higher transfection efficiency over the linear analogue albeit with significant cytotoxicity concerns under certain biological settings.24 Nonetheless, this architectural blueprint has paved the way for an array of highly effective branched polycations derived from numerous material platforms, such as poly(β-aminoester)s,23 poly(amidoamine)s,25 poly(amino acids)/polypeptides,26 partially hydrolysed poly(oxazoline)s27 and poly(meth)acrylates/acrylamides.28
Although many of these delivery platforms offer high potency, future clinical use also depends on their scalability and straightforward manufacture, which ideally should occur under aqueous conditions to avoid potential toxicity from organic solvents, catalysts and side products. We and others have reported on the use of an aqueous atom-economical ‘one-pot’ thermal condensation polymerisation to prepare highly branched poly(lysine) (hb-polyK) and poly(lysine-co-histidine) (hb-polyKH2) utilising the ABB’ structure of lysine for branching.26,29–34 This approach offers a practical route to biodegradable cationic poly(amino acid)s, which in our previous studies exhibited high transfection efficiency for plasmid DNA cargo but have not yet been explored as siRNA vectors.26
Accordingly, in this work we investigate the activity of both hb-polyK and histidinylated hb-polyK to condense and deliver siRNA. To this end, the two different molecular mass variants of hb-polyK (13 and 33 kDa) and histidinylated hb-polyK (18 and 34 kDa); denoted by hb-polyK-13 kDa, hb-polyK-33 kDa, hb-polyKH2-18 kDa and hb-polykH2-34 kDa respectively, were used to condense siRNA and the resultant polyplexes were evaluated for their physico-chemical properties and biological function.
Materials and methods
Materials
Polymers hb-polyK-13 kDa, hb-polyK-33 kDa, hb-polyKH2-18 kDa, hb-polykH2-34 kDa copolymers and their labelled forms were synthesised and characterised as reported in our previous work.29 Luc-siRNA (target sequence: 5′-CCGCAAGAUCCGCGAGAUU-3′) and negative control (scrambled) siRNA were purchased from Eurogentec (Seraing, Liège, Belgium). Silencer™ Select GAPDH Positive Control siRNA and Cy3 labelled siRNA was obtained from Thermo Fisher Scientific (Loughborough, UK). GAPDH activity assay kits were received from Source Bioscience (Nottingham, UK), and RNase free water from Sigma-Aldrich (Gillingham, UK) The Luciferase assay kits were from Promega (Madison, WI, USA). All cell culture products including Dulbecco's modified Eagle's medium (DMEM), Roswell Park Memorial Institute media RPMI), foetal bovine serum (FBS), phosphate-buffered saline (PBS), L-glutamic acids were obtained from Sigma-Aldrich (Gillingham, UK). All the other materials were obtained from Sigma-Aldrich (Gillingham, UK) and used without further purification unless stated.Preparation of polyplexes
The siRNA polyplexes derived from hb-polyK-13 kDa, hb-polyK-33 kDa, hb-polyKH2-18 kDa and hb-polykH2-34 kDa were prepared at 40 μg ml−1 siRNA in HEPES buffer (10 mM in RNase free water, pH 7.4). The concentrations of the polymers were adjusted to be equivalent to the required nitrogen/phosphate (N/P) ratio. Polyplexes were formulated by mixing equal volumes of siRNA and the polymer solutions, vortexed for 10 s, and incubated for 30 min at room temperature before further use.Particle sizes and zeta potential measurements
The hydrodynamic radii of siRNA polyplexes were measured using a Viscotek 802 instrument supported by Omnisize 0.3 V software, where all the measurements were carried out at a scattering angle of 90° and a temperature of 25 °C and the results were recorded as average of 10 runs ± SD. For zeta potential, the prepared polyplexes were diluted to using a solution of 5 mM NaCl (1 ml) and the measurements were performed using a NanoZetasizer (Malvern, UK) and recorded as three readings of 10–20 runs each.Atomic force microscopy
Polymer/siRNA polyplexes prepared at N/P ratio of 10 in HEPES buffer (10 mM, pH 7.4) were examined by Atomic Force Microscope (AFM) imaging using Mica as a support disc. The specimens were prepared by depositing 100 μl of polyplexes (40 μg ml−1 of siRNA) on freshly cleaved Mica for 30 s, and then the excess of solution was removed. Samples were dried using dry filtered air for 1 min before being imaged in air using the Peak Force Tapping mode of Fast Scan AFM (Dimension ICON from Bruker). The cantilevers were Tap150A with a spring constant of 5 N m−1 operating at resonant frequencies of approximately 150 kHz. Images were analysed using Nanoscope Software version 7.3.Gel retardation assays (agarose gel electrophoresis)
The hb-polyK polymers were dissolved in HEPES buffer (10 mM, pH 7.4) and mixed with 0.5 μg siRNA at various N/P ratios. After incubating for 30 min at room temperature, the obtained sample, the mixture of the 6×-loading dye (5 μl) and siRNA polyplexes (25 μl), were loaded onto 1% (w/v) agarose gel that was prepared with 1% Tris–acetate–EDTA buffer solution and contains 2 μg ml−1 ethidium bromide. Electrophoresis was performed at 70 V for 60 min, after which the gel was imaged using a UV-transilluminator (Syngene, Germany).Ethidium bromide displacement assay
Ethidium bromide displacement was measured using a Tecan Microplate reader (Spark®). The test siRNA solution (40 μg ml−1 in HEPES, 10 mM, pH 7.4) was incubated with ethidium bromide (2 μg ml−1) for 15 minutes. Next, aliquots of 50 μl were mixed with 50 μl of polymer solutions at concentrations equivalent to the different N/P ratios. The samples (n = 4) were incubated for 30 minutes at the room temperature, and then the fluorescence intensity of siRNA–ethidium bromide intercalate was measured at a wavelength of 590 nm using an excitation wavelength of 520 nm.Cell culture
A549 (human adenocarcinoma alveolar basal epithelial, ATCC® CCL-185) cells stably expressing the firefly luciferase transgene denoted by Luc-A549, and H1299 (human non-small cell lung carcinoma, ATCC CRL-5803) were cultured in DMEM and RPMI, respectively, supplemented with 10% FBS and 1% L-glutamic acids under a humidified atmosphere containing 5% CO2. Routinely, the cells were cultured in a T75 flask and split when approximately 90% confluent using Trypsin/EDTA to detach the cells.Toxicity assays
The effects of siRNA polyplexes on target cell lines (Luc-A549 or H1299) were determined with two different methods; membrane leakage via the lactate dehydrogenase (LDH assay), and mitochondrial function (MTT assay). Both assays were performed by following the manufacturer's instructions. In brief, Luc-A549 or H1299 cells were seeded on 96-well plates at density of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 100 μl of culture medium for 24 hours. Subsequently, the culture media were replaced with the transfection medium (FBS free medium) containing siRNA polyplexes at different N/P ratios (final concentration of siRNA was 2 μg ml−1 in FBS-free media) and incubated for 4 hours. Thereafter, the treatment medium was collected for conducting LDH assays, replaced with fully supplemented medium, and the cells were incubated for a further 24 hours for the MTT assay.
000 cells per well in 100 μl of culture medium for 24 hours. Subsequently, the culture media were replaced with the transfection medium (FBS free medium) containing siRNA polyplexes at different N/P ratios (final concentration of siRNA was 2 μg ml−1 in FBS-free media) and incubated for 4 hours. Thereafter, the treatment medium was collected for conducting LDH assays, replaced with fully supplemented medium, and the cells were incubated for a further 24 hours for the MTT assay.
Lactate dehydrogenase (LDH) detection assay
At 4 h post-transfection, 50 μl aliquots of the treatment medium were mixed with 50 μl LDH reagent and the contents were kept at room temperature for 20–30 min before adding the stop solution (5 μl). The released LDH was detected by measuring the absorbance at 490 nm using a Tecan Microplate reader. The cells treated with Triton (0.2% in HEPES) and FBS free medium were used as controls and the percentage of released LDH was calculated using the following equation:| Released LDH (%) = S − C/T − C × 100 |
Metabolic activity (MTT) assay
To determine metabolic activity, after the 24 h post-transfection incubation, 25 μl of MTT reagent (5 mg ml−1 in PBS) was added to each well and the cells were incubated for 3 hours. DMSO (150 μl) was then added and mixed thoroughly before measuring the absorbance at 570 nm using a Tecan Microplate reader. The untreated cells and cells treated with Triton 0.2% were used as negative and positive controls. The metabolic activity of negative control was accepted as 100% whilst that of positive control 0%, and the percentage of metabolic activity of specimens was calculated using the following equation:| Metabolic activity (%) = S − T/C − T × 100 |
In vitro transfection study
The cells, LucA459 or H1299, were seeded into a 24-well plate at a density of 5 × 104 cell per well and incubated in 500 μl of fully supplemented medium, 24 h before transfection. Then, the cells were washed with PBS, covered with 500 μl of FBS free medium and treated with Luc-siRNA and GAPDH-siRNA polyplexes for Luc-A549 and H1299 cells respectively (at a concentration of 1 μg of siRNA per well). After 4 hours, the transfection medium was replaced with fully supplemented medium, and the cells were incubated for further 24 hours. Then, the cells were washed with PBS and lysed in reporter lysis buffer (Promega), 100 μl for each well. Luc-A549 lysate was used to detect the expression of luciferase using luciferase assay kit (Promega) and H1299 lysate to detect the expression of GAPDH using GAPDH activity assay kit (Source Bioscience), following the supplier's protocol. The results were normalised using total protein concentration of each sample, which was detected using a Coomassie Plus (Bradford) assay kit (following the supplier's protocol).Cellular uptake
Polyplexes were prepared using FITC-labelled polymers and Cy3 labelled siRNA. For confocal microscopy, the cells (Luc-A549 or H1299) were seeded into 8-well chamber slides (from Ibidi) at a seeding density of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in 300 μl fully supplemented growth medium. After 24 hours, the cells washed with PBS, covered with 300 μl of FBS free medium, and treated with the labelled siRNA polyplexes at concentration of 2 μg of siRNA per ml for 4 hours. Then, the cells were washed with PBS three times, fixed with 4% paraformaldehyde for 15 minutes, stained with DRAQ5 (20 μM in PBS for 5 minutes) or Hoechst (1 μg ml−1 in PBS for 10 minutes), and then covered with mounting medium. The samples were imaged using a Zeiss 510 Meta Confocal microscope. For flow cytometry, after the incubation of cells with labelled siRNA polyplexes for 4 hours, the cells were washed with PBS three times, and then detached from the plates using 500 μl of AccutaseTM solution for 15 minutes at 37 °C. The cells were collected in FACS tubes, mixed with 150 μl of Trypan blue (4%) and measured directly using a Beckman Coulter Astiros flow cytometer and data were analysed using Kaluza 1.5 software.
000 in 300 μl fully supplemented growth medium. After 24 hours, the cells washed with PBS, covered with 300 μl of FBS free medium, and treated with the labelled siRNA polyplexes at concentration of 2 μg of siRNA per ml for 4 hours. Then, the cells were washed with PBS three times, fixed with 4% paraformaldehyde for 15 minutes, stained with DRAQ5 (20 μM in PBS for 5 minutes) or Hoechst (1 μg ml−1 in PBS for 10 minutes), and then covered with mounting medium. The samples were imaged using a Zeiss 510 Meta Confocal microscope. For flow cytometry, after the incubation of cells with labelled siRNA polyplexes for 4 hours, the cells were washed with PBS three times, and then detached from the plates using 500 μl of AccutaseTM solution for 15 minutes at 37 °C. The cells were collected in FACS tubes, mixed with 150 μl of Trypan blue (4%) and measured directly using a Beckman Coulter Astiros flow cytometer and data were analysed using Kaluza 1.5 software.
Statistical analysis
All experiments in the study were performed at least three times as independently and the data are expressed as mean ± SD or SE (standard deviations or standard error, respectively). Statistical analyses of the results were evaluated by using One-way or Two-way ANOVA with a Tukey's multiple comparison based on the number of variables on graphs. Where * = significant (p < 0.05), ** = very significant (p < 0.01), *** = extremely significant (p < 0.001), and **** = extremely significant (p < 0.0001). GraphPad Prism 9.0 software was used for data analysis.Results
Variations of polyplex size and siRNA condensation with molar mass and histidinylation
In this study, we utilised four previously prepared highly branched lysine or lysine-co-histidine poly(amino acids) (hb-polyK-13 kDa, hb-polyK-33 kDa, hb-polyKH2-18 kDa and hb-polyKH2-34 kDa) synthesised at a low (∼13–18 kg mol−1) or high molar mass (∼35 kg mol−1) in order to evaluate the impact of molar mass and histidinylation on siRNA complexation and transfection efficiency. Polyplexes were prepared by mixing polymer and siRNA components at various N/P ratios, with nitrogen content calculated based on a fluorescamine assay performed in our previous study.29 The properties of these polymers are given in Table 1 and their structures and representation of polyplexes formed with siRNA are given in Scheme 1.| Polymer | K![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H ratioa H ratioa |
M n (kDa) | M w (kDa) | (Đ) | DBc | ANBd | Buffer capacitye | μmol of aminef |
|---|---|---|---|---|---|---|---|---|
| a Molar ratios of lysine to histidine. b Malls GPC molecular masses. c Degree of branching. d Average number of branches. e Buffer capacity at pH range of 5–7. f μmol of amine per mg of polymers (fluorescamine assay). | ||||||||
| hb-polyK-13 kDa | — | 13.4 | 22.7 | 1.7 | 0.34 | 0.24 | 35.2% | 5.3 |
| hb-polyK-33 kDa | — | 33.6 | 58.8 | 1.7 | 0.39 | 0.27 | 37.4% | 5.3 |
| hb-polyKH2-18 kDa | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
18.1 | 25.3 | 1.4 | 0.46 | 0.39 | 44.0% | 4.0 |
| hb-polyKH2-34 kDa | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
34.5 | 62.8 | 1.8 | 0.40 | 0.32 | 43.9% | 3.6 |
 | ||
| Scheme 1 Representative structures of hyperbranched poly(lysine) and poly(lysine-co-histidine) and complexes with siRNA. | ||
Initial evaluation of the formulations revealed stronger siRNA binding capacity for the non-histidinylated polymers, as evidenced by the maximum fluorescence reduction and full siRNA complexation being achieved at lower N/P ratios compared to the histidinylated polymers in the ethidium bromide displacement assay (Fig. 1A). In addition, the gel retardation data (Fig. 1B), suggested that the lysine-only polymers were better at retaining siRNA, and with the higher molar mass hb-polyK retarding siRNA fully at N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 2.
P 2.
Interestingly, although full siRNA complexation was achieved at lower N/P ratios for the higher molar mass polymers across both compositions, the same trend was not evident in the ethidium bromide displacement study. The importance of ‘matching’ charge distribution in polyelectrolyte interactions is reported in several studies,35–37 where ‘mismatching’ the charge between two opposite electrolytes leads to swollen and incompletely compacted complexes.
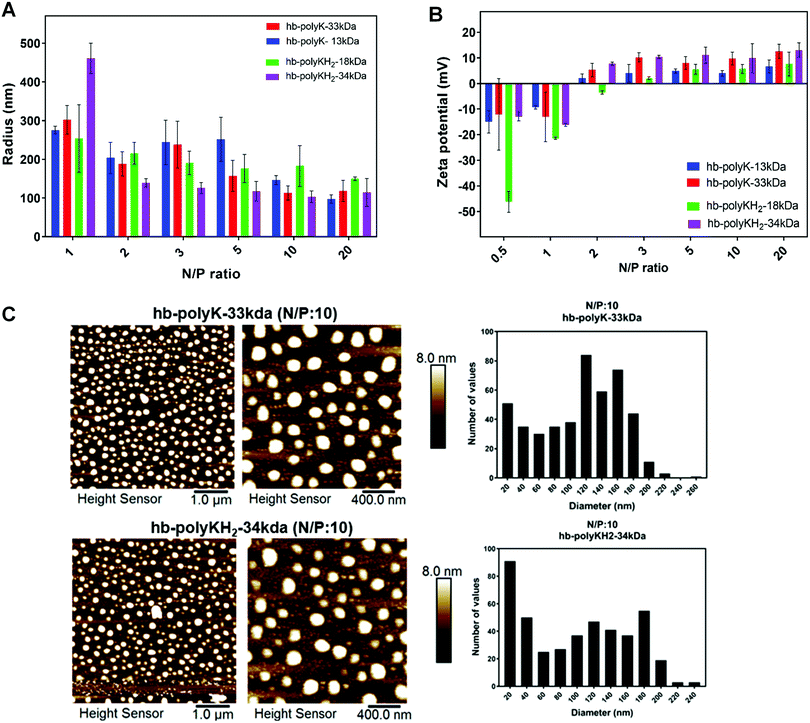
Particle sizes of polyplexes derived from all four branched polymers displayed an N/P ratio dependent trend, with hydrodynamic radii decreasing with increasing polymer concentrations (N/P ratio), culminating in all polymers condensing siRNA sufficiently at N/P = 20 to produce sub-200 nm radii particles (Fig. 2A). The inversion of zeta potential measurements from positive to negative occurred at ∼N/P = 2 for all polyplexes, with the minimum zeta potential (∼−15 mV) recorded at N/P = 0.5 which increased with N/P ratio, reaching a maximum at N/P = 20 (∼12 mV) (Fig. 2B).
This steady increase in zeta potential with N/P ratio likely contributes a greater electrostatic stabilisation of these complexes, evidenced by their smaller particle size. Zeta potentials of polyplexes derived from the high molar mass polymers were generally higher than that of their low molecular analogue, but no discernible difference was seen in the zeta potentials due to histidinylation. Although the electrostatic interactions are the main determining forces in polycation:nucleic acid complexes, some studies have shown that non-ionic interactions such as hydrogen bonds and hydrophobic interaction contribute significantly to these complexes.38–40 In the case of siRNA, the presence of the 2′ hydroxyl group of ribose provides potential hydrogen bond acceptors,41 which could explain the relative stability of siRNA polyplexes with nearly neutral zeta potentials.
Particle size and morphology were further validated through AFM measurements for polyplexes derived from all four polymers at N/P = 10 (Fig. 2C). Atomic force micrographs indicated that the polymers could compact the siRNA into small spherical nanoparticles with a relatively broad size distribution; the polyplex diameters varied between 40–200 nm. The smaller average size of siRNA polyplexes reported by AFM compared to DLS is attributed to the deposition and drying of polyplexes onto the Mica substrate compared to hydrated complexes under light scattering measurements. Furthermore, microscopy analyses show more clearly the number distributions of polyplexes, while the intensity DLS histograms are skewed towards larger particles. Overall, our formulation studies indicate that all four polymers display efficient siRNA binding and form relatively small (<200 nm radii polyplexes).
Evaluation of membrane damage and changes in metabolic activity in cells treated with polymer:siRNA complexes
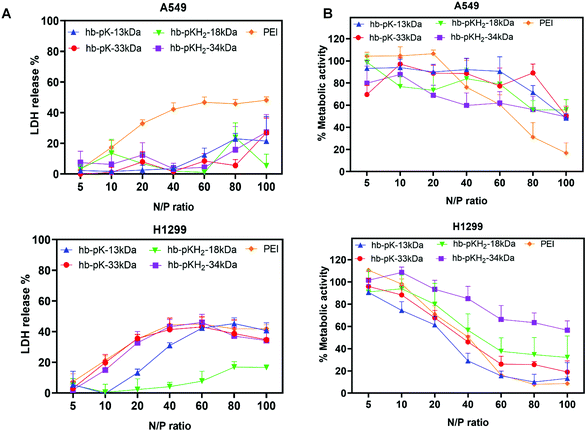
Polycations are known to disrupt cell membranes and induce toxicity. Accordingly, the effects of the complexes on cell membranes and metabolic activity were evaluated by monitoring lactate dehydrogenase release (LDH) and MTT conversion in two cell lines of importance in lung cancers, i.e. Luc-A549 and H1299 cells, treated with siRNA complexes derived from the four branched polycations and branched 25 kDa PEI as a control, all formulated with 2 μg ml−1 siRNA at N/P ratios 10 to 100.14To evaluate any acute cell membrane disruption, LDH release was monitored after the initial 4 hours’ exposure to the siRNA polyplexes (i.e., before polyplex removal and serum supplementation). In Luc-A549 cells, none of the branched polyplexes showed a severe negative influence on plasma membrane integrity at up to N/P ratio of 80, indicated by <20% LDH release, meanwhile PEI polyplexes displayed significantly higher (30–40%) LDH release at N/P ratio >10. In contrast, hb-polyK-13 kDa, hb-polyK-33 kDa, and hb-polyKH2-34 kDa polyplexes induced an increasing LDH release profile with higher N/P ratios in H1299 cells, comparable to that of PEI polyplexes (35% at N/P = 20). The polyplexes of hb-polyKH2-18 kDa showed the least impact on the integrity of H1299 membrane with negligible LDH release at N/P ratios of up to 80 (Fig. 3A). Overall, the membranes of H1299 cells were more sensitive to polyplex treatments than Luc-A549 cells, possibly attributed to differences in fluidity and composition between A549 and H1299 cell membranes. For instance, A549 cells produce large amounts of lecithin, which is a major contributor to phospholipid biosynthesis, thus may provide greater structural integrity in comparison with H1299.26,42,43 Interestingly, the most membrane active polyplexes displayed a plateau in LDH release at high N/P ratios, suggesting the interaction of those polyplexes (or the associated non-complex polycation) reached a maximum at N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ∼ 40, which may have been indicative of binding saturation at intracellular membranes.
P ∼ 40, which may have been indicative of binding saturation at intracellular membranes.
To examine cytotoxicity, metabolic activity of the cells was assessed by an MTT assay 24 h post-transfection (Fig. 3B).
In both cell lines the metabolic activity decreased with increased N/P ratios across all siRNA polyplex formulations; however this trend was not identical across the cell lines and was more pronounced in H1299 cells than A549 cells. In addition, H1299 cells displayed significantly lower metabolic activity when treated with polyplexes from all polymers except hb-polyKH2-34 kDa over N/P = 20 compared to A549 cells, with metabolic activities below 40% recorded for the non-histidinylated derivatives in this cell line, compared to that of PEI. The difference between A549 and H1299 may be attributed to the differences in membrane fluidity as reported previously.26,42,43 No substantial differences in metabolic activity were observed in cells treated with the high molecular weight polymers.
High molar mass derivatives display most pronounced gene silencing, with histidinylation negatively affecting knockdown efficiency
To assess the in vitro siRNA transfection efficiency of the hyperbranched polymers, the expression of Luciferase (firefly) and GAPDH genes was knocked down in Luc-A549 and H1299 cells using polyplexes prepared at N/P ratios of 5, 10 and 20. The knockdown efficiency was evaluated as a percentage relative to enzyme/protein activity in cells treated with polyplexes containing scrambled siRNA (Fig. 4).The most pronounced silencing in Luc-A549 cells was observed with polyplexes derived from the polymers with high molar mass (hb-polyK-33 kDa and hb-polyKH2-34 kDa), with the lysine-only analogue most effective at all N/P ratios (70%, 50% and 70%, 60% silencing for N/P = 10 and 20 respectively). Notably, the higher molar mass histidinylated derivative (hb-polyKH2-34 kDa) displayed marginally lower knockdown of luciferase expression than non-histidinylated analogues, suggesting minimal silencing improvement from histidinylation (25, 50 and 60% knockdown at N/P rations 5, 10 and 20 respectively). Polyplexes from the low molar mass polymers (hb-polyK-13 kDa and hb-polyKH2-18 kDa) did not show a statistically significant silencing effect on luciferase expression (Fig. 4A).
In H1299 cells, a similar trend of gene silencing was observed, where only the polyplexes of hb-polyK-33 kDa and hb-polyKH2-34 kDa achieved a significant reduction in GAPDH activity (35% and 25% at N/P of 10 and 20 respectively). Interestingly, the overall silencing efficiency was lower in H1299 cells compared to Luc-A549 across all treatments, including PEI, suggesting that this was either due to the efficiency of siRNA itself in recognition or availability to GAPDH encoding mRNA, or is cell dependent (Fig. 4B). According to these in vitro transfection results, siRNA delivered by polyplexes derived from the higher molar mass polymers (hb-polyK-33 kDa and hb-polyKH2-34 kDa) induced superior gene knockdown than lower molar mass derivatives, a structure–function trend which has been reported in many other studies.14,32
Flow cytometry and confocal microscopy analysis of polyplex internalisation in A549 and H1299 cells
To gain a greater insight into the silencing properties of the siRNA polyplexes and to probe potential mechanisms for the variations in knockdown efficiency, the internalisation of the polyplexes in A549 and H1299 cells was studied using Cy3-labelled siRNA and FITC-labelled polymers. The Cy3 signal (siRNA) from flow cytometry analysis of both cell lines treated with fluorescent polyplexes indicated the highest internalisation occurred with PEI, followed by hb-polyK-33 kDa across all tested N/P ratios, with all other complexes displaying low internalisation of the labelled siRNA (Fig. 5A, B and C, D). The increased relative cellular uptake of PEI and hb-polyK-33 kDa is likely a major contributor to the enhanced knockdown efficiencies observed above. Comparatively, the FITC signal (polymers) did not show an identical trend to the Cy3 signal, suggesting large proportions of free polymer not associated with the polyplex were also associating with the cell membranes or being internalised, particularly in the case of lysine-only analogues. Confocal micrographs of hb-polyK-33 kDa (lysine-only) and hb-polyKH2-34 kDa (histidine containing) displayed punctate Cy3 signal indicating polyplexes were likely internalised through endosomal uptake in both cell lines (Fig. 6). Interestingly, the images showed that there was poor colocalization between the signals of Cy3-labelled siRNA (red channel) and FITC-labelled polymers (green channel), suggesting a dissociation of polyplexes and release of the siRNA at the time of imaging (after 4 hours of treatment), or as suggested above, uptake of polymers not associated with the polyplexes. Similarly, in H1299 cells, polyplexes derived from high molar mass polymers were internalised significantly (2-fold) more than lower molar mass analogues based on the Cy3 (siRNA) median intensity (Fig. 6B) The FITC signal (polymer) again did not show the same fluorescence trend as the Cy3 (siRNA) signal, indicating uptake of uncomplexed polymer. It should be noted that fluorescein dyes are pH sensitive and their fluorescence can be quenched in the acidic environment of late lysosomes. Thus there may have been an underestimate of the amount of complexed polymer in these intracellular compartments. However, the inherent buffering by the polymers within the complexes and thus the modulation of local pH renders a full interpretation of polymer content in the endo/lysosomes difficult. Nevertheless, when taken together, the results indicated that the hyperbranched polymers of higher molar masses were able to deliver siRNA more efficiently than the lower molecular mass polymers and that histidinylation did not have a positive effect on transfection efficiency or cellular uptake.Discussion
When the results are evaluated together, it is clear that the high molar mass analogues exhibit the highest transfection efficiencies and cellular uptake, likely arising from a greater propensity to condense siRNA into smaller, and more colloidally stable polyplexes with positive zeta potentials at lower N/P ratios. This is in agreement with data reported in the literature, where higher molar mass polymers are required to condense the relatively small siRNA nucleic acids into stable polyplexes, which have low charge and high rigidity of siRNA relative to plasmid DNA or mRNA.8,44,45Interestingly, these results are in contrast to our previously reported findings for plasmid DNA polyplexes with these polymers, where low transfection efficiency was obtained with the polyplexes of the higher molecular mass polymers.29 This we attributed primarily to the formation of highly stable polyplexes, with poor unpacking to release their payload, and the fact that access of DNA to the nucleus is needed for effective mRNA and subsequent protein production. Accordingly, there is a fine balance between stable complexation and intracellular decomplexation, and has previously been noted to be very important for efficient transfection.46,47 Thus, the variations between plasmid DNA and siRNA in size and flexibility gave an advantage for the higher molecular mass polymers, in the case of siRNA, to form relatively stable polyplexes with good intracellular decomposition and siRNA release, as suggested by confocal images (which showed poor colocalisation between the signals of siRNA and the polymers). Similarly, Kwok et al.48 compared complexation of linear and branched PEI with both siRNA and DNA and correlated the results of transfection with the stability of the polyplexes in the presence of heparin. They found that the transfection of the more stable polyplexes of branched PEI was better than that for the polyplexes of the linear PEI in the case of siRNA. However, linear PEI, which showed better unpacking properties, achieved better transfection in the case of DNA polyplexes. Zhou et al.49 found that the gene silencing of polyamidoamine dendrimers was a function of the dendrimer generation, where the high generations produced more stable complexes. Also, it was reported that the improved stability of siRNA polyplexes provided with cysteine, which act as an inner crosslinker (using disulfide linkages), enhanced the transfection of those polyplexes significantly.
Furthermore, our results revealed that the lysine-only polymer (hb-polyK-33 kDa) has a better silencing efficiency in comparison with the histidine-containing polymer (hb-polyKH2-34 kDa). This could be explained by the effect of histidine on the structure of the polymer; the incorporation of histidine (an AB type monomer) during thermal polycondensation produced polymers of more dendritic units through the ‘capping’ of partially reacted lysine monomers, thus restricting the flexibility of these analogues.29 This, in combination with the small size and stiffness of siRNA, would affect the condensing ability of the histidinylated polymers, as shown clearly in ethidium bromide displacement and gel retardation. Also, the incorporation of histidine might affect the ability of the polymers to interact with, and permeabilise, the cellular membrane, particularly for the low molecular mass polymers, as indicated by LDH assays. All these factors, in turn, affected the cellular uptake and silencing efficiency of the polyplexes of the histidinylated polymers.
The weaker membrane interaction of the histidinylated polymers could also be expected to affect their ability to escape from the endosomal barrier. In this regard, the uptake study revealed (based on the signal of FITC-labelled polymers) that the histidinylated polymers, free or associated with polyplexes, accumulated to less extent inside the cells than the lysine-only polymers. This supports the assertion that endosomal escape might be affected in the case of histidinylated polyplexes, where the effect of free polymer on endosomal buffering was well reported.50,51
Conclusion
In this study we investigated the ability of highly branched poly(lysine) and poly(lysine-co-histidine) polycations to condense and deliver siRNA for potential gene silencing therapies. The physicochemical characterization indicated that the polymers of higher molar masses yielded smaller polyplexes with higher zeta potentials at lower N/P ratios than lower molar mass analogues, suggesting a greater affinity for siRNA. Consequently, higher molar mass polymers exhibited a higher degree of cellular uptake, leading to more potent silencing efficiency without significant cytotoxicity at N/P ratios where silencing was effective. This was likely due to the small size of these nucleic acids, imparting low overall charge and rigidity, which required polycations of relatively high molecular mass to condense them into stable nanoparticles. While there is always the caveat with hyperbranched polymers that polydisperse samples, for which in our case the Đ values ranged between 1.4–1.8, these findings provide important structure–property relationships for future designs of polymeric non-viral vectors for siRNA. Furthermore, the data highlight the necessity to design vectors for specific nucleic acid cargoes which retain vital features such as biodegradability, cytocompatibility, and manufacturability for further clinical assessment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Engineering and Physical Sciences Research Council (grant EP/H005625/1) and by the Department of Health and Social Care using UK Aid funding and is managed by the Engineering and Physical Sciences Research Council (EPSRC, grant number: EP/R013764/1). The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health and Social Care. This research was also funded through the Royal Society through a Wolfson Research Merit Award [WM150086], and the Higher Committee for Education Development in Iraq (scholarship to AA), and the Turkish Ministry of National Education (scholarship to NG). We also thank Douglas Crackett and Paul Cooling for expert technical assistance and Carol Turrill for outstanding administrative support.References
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967–977 CrossRef CAS PubMed.
- G. Ozcan, B. Ozpolat, R. L. Coleman, A. K. Sood and G. Lopez-Berestein, Adv. Drug Delivery Rev., 2015, 87, 108–119 CrossRef CAS.
- J. Park, J. Park, Y. Pei, J. Xu and Y. Yeo, Adv. Drug Delivery Rev., 2016, 104, 93–109 CrossRef CAS PubMed.
- G. Mahmoodi Chalbatani, H. Dana, E. Gharagouzloo, S. Grijalvo, R. Eritja, C. D. Logsdon, F. Memari, S. R. Miri, M. R. Rad and V. Marmari, Int. J. Nanomed., 2019, 14, 3111–3128 CrossRef.
- M. A. Subhan and V. P. Torchilin, Transl. Res., 2019, 214, 62–91 CrossRef CAS PubMed.
- T. Bus, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2018, 6, 6904–6918 RSC.
- T. C. Roberts, K. Ezzat, S. El Andaloussi and M. S. Weinberg, Methods in Molecular Biology, 2016, vol. 1364, pp. 291–310 Search PubMed.
- D. J. Gary, N. Puri and Y. Y. Won, J. Controlled Release, 2007, 121, 64–73 CrossRef CAS.
- C. Scholz and E. Wagner, J. Controlled Release, 2012, 161, 554–565 CrossRef CAS PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. Van Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- C. Tros de Ilarduya, Y. Sun and N. Düzgüneş, Eur. J. Pharm. Sci., 2010, 40, 159–170 CrossRef CAS PubMed.
- P. Gurnani, A. K. Blakney, J. Yeow, C. R. Bouton, R. J. Shattock, M. M. Stevens and C. Alexander, Polym. Chem., 2020, 11, 5861–5869 RSC.
- P. Gurnani, A. K. Blakney, R. Terracciano, J. E. Petch, A. J. Blok, C. R. Bouton, P. F. McKay, R. J. Shattock and C. Alexander, Biomacromolecules, 2020, 21, 3242–3253 CrossRef CAS PubMed.
- A. K. Blakney, Y. Zhu, P. F. McKay, C. R. Bouton, J. Yeow, J. Tang, K. Hu, K. Samnuan, C. L. Grigsby, R. J. Shattock and M. M. Stevens, ACS Nano, 2020, 14, 5711–5727 CrossRef CAS PubMed.
- F. Saviano, T. Lovato, A. Russo, G. Russo, C. R. Bouton, R. J. Shattock, C. Alexander, F. Quaglia, A. K. Blakney, P. Gurnani and C. Conte, J. Mater. Chem. B, 2020, 8, 4940–4949 RSC.
- J. J. Green, R. Langer and D. G. Anderson, Acc. Chem. Res., 2008, 41, 749–759 CrossRef CAS PubMed.
- C. Yu, L. Li, P. Hu, Y. Yang, W. Wei, X. Deng, L. Wang, F. R. Tay and J. Ma, Adv. Sci., 2021, 2100540 CrossRef CAS.
- D. Pezzoli, E. Giupponi, D. Mantovani and G. Candiani, Sci. Rep., 2017, 7, 44134 CrossRef PubMed.
- R. S. Burke and S. H. Pun, Bioconjugate Chem., 2008, 19, 693–704 CrossRef CAS.
- J. Shi, J. L. Choi, B. Chou, R. N. Johnson, J. G. Schellinger and S. H. Pun, ACS Nano, 2013, 7, 10612–10620 CrossRef CAS PubMed.
- S. A. Smith, L. I. Selby, A. P. R. Johnston and G. K. Such, Bioconjugate Chem., 2019, 30, 263–272 CrossRef CAS.
- A. B. Cook and S. Perrier, Adv. Funct. Mater., 2020, 30, 1901001 CrossRef CAS.
- D. Zhou, L. Cutlar, Y. Gao, W. Wang, J. O’Keeffe-Ahern, S. McMahon, B. Duarte, F. Larcher, B. J. Rodriguez, U. Greiser and W. Wang, Sci. Adv., 2016, 2, e1600102 CrossRef PubMed.
- Y. K. Sung and S. W. Kim, Biomater. Res., 2019, 23, 8 CrossRef CAS.
- B. Zhang, X. Ma, W. Murdoch, M. Radosz and Y. Shen, Biotechnol. Bioeng., 2013, 110, 990–998 CrossRef CAS PubMed.
- A. Alazzo, M. A. Al-Natour, K. Spriggs, S. Stolnik, A. Ghaemmaghami, D.-H. Kim and C. Alexander, Mol. Omics, 2019, 15, 77–87 RSC.
- A. B. Cook, R. Peltier, J. Zhang, P. Gurnani, J. Tanaka, J. A. Burns, R. Dallmann, M. Hartlieb and S. Perrier, Polym. Chem., 2019, 10, 1202–1212 RSC.
- B. Newland, H. Tai, Y. Zheng, D. Velasco, A. Di Luca, S. M. Howdle, C. Alexander, W. Wang and A. Pandit, Chem. Commun., 2010, 46, 4698–4700 RSC.
- A. Alazzo, T. Lovato, H. Collins, V. Taresco, S. Stolnik, M. Soliman, K. Spriggs and C. Alexander, J. Interdiscip. Nanomed., 2018, 3, 38–54 CrossRef CAS.
- M. Scholl, T. Q. Nguyen, B. Bruchmann and H.-A. Klok, Macromolecules, 2007, 40, 5726–5734 CrossRef CAS.
- M. Scholl, T. Q. Nguyen, B. Bruchmann and H.-A. Klok, J. Polym. Sci., Part A: Polym. Chem., 2007, 45, 5494–5508 CrossRef CAS.
- Z. Kadlecova, Y. Rajendra, M. Matasci, L. Baldi, D. L. Hacker, F. M. Wurm and H. A. Klok, J. Controlled Release, 2013, 169, 276–288 CrossRef CAS PubMed.
- Z. Kadlecova, Y. Rajendra, M. Matasci, D. Hacker, L. Baldi, F. M. Wurm and H.-A. Klok, Macromol. Biosci., 2012, 12, 794–804 CrossRef CAS PubMed.
- Z. Kadlecova, L. Baldi, D. Hacker, F. M. Wurm and H.-A. Klok, Biomacromolecules, 2012, 13, 3127–3137 CrossRef CAS PubMed.
- H. Dautzenberg and W. Jaeger, Macromol. Chem. Phys., 2002, 203, 2095–2102 CrossRef CAS.
- S. Danielsen, G. Maurstad and B. T. Stokke, Biopolymers, 2005, 77, 86–97 CrossRef CAS.
- V. Capel, D. Vllasaliu, P. Watts, P. A. Clarke, D. Luxton, A. M. Grabowska, G. Mantovani and S. Stolnik, Drug Delivery, 2018, 25, 644–653 CrossRef CAS.
- L. E. Prevette, T. E. Kodger, T. M. Reineke and M. L. Lynch, Langmuir, 2007, 23, 9773–9784 CrossRef CAS PubMed.
- L. M. Pei, M. Lavertu, F. M. Winnik and M. D. Buschmann, Biomacromolecules, 2009, 10, 1490–1499 CrossRef PubMed.
- M. H. Allen, M. D. Green, H. K. Getaneh, K. M. Miller and T. E. Long, Biomacromolecules, 2011, 12, 2243–2250 CrossRef CAS PubMed.
- S. T. Chou, K. Hom, D. Zhang, Q. Leng, L. J. Tricoli, J. M. Hustedt, A. Lee, M. J. Shapiro, J. Seog, J. D. Kahn and A. J. Mixson, Biomaterials, 2014, 35, 846–855 CrossRef CAS.
- R. Tedja, C. Marquis, M. Lim and R. Amal, J. Nanopart. Res., 2011, 13, 3801–3813 CrossRef CAS.
- M. Lieber, G. Todaro, B. Smith, A. Szakal and W. Nelson-Rees, Int. J. Cancer, 1976, 17, 62–70 CrossRef CAS.
- M. Alameh, M. Lavertu, N. Tran-Khanh, C. Y. Chang, F. Lesage, M. Bail, V. Darras, A. Chevrier and M. D. Buschmann, Biomacromolecules, 2018, 19, 112–131 CrossRef CAS PubMed.
- X. Liu, K. A. Howard, M. Dong, M. Andersen, U. L. Rahbek, M. G. Johnsen, O. C. Hansen, F. Besenbacher and J. Kjems, Biomaterials, 2007, 28, 1280–1288 CrossRef CAS PubMed.
- S. P. Strand, S. Lelu, N. K. Reitan, C. de Lange Davies, P. Artursson and K. M. Vårum, Biomaterials, 2010, 31, 975–987 CrossRef CAS PubMed.
- D. V. Schaffer, N. A. Fidelman, N. Dan and D. A. Lauffenburger, Biotechnol. Bioeng., 2000, 67, 598–606 CrossRef CAS PubMed.
- A. Kwok and S. L. Hart, Nanomedicine, 2011, 7, 210–219 CrossRef CAS.
- J. Zhou, J. Wu, N. Hafdi, J. P. Behr, P. Erbacher and L. Peng, Chem. Commun., 2006, 2362–2364 RSC.
- T. C. B. Klauber, R. V. Søndergaard, R. R. Sawant, V. P. Torchilin and T. L. Andresen, Acta Biomater., 2016, 35, 248–259 CrossRef CAS.
- J. Cai, Y. Yue, Y. Wang, Z. Jin, F. Jin and C. Wu, J. Controlled Release, 2016, 238, 71–79 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |