Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Hydrogelation tunability of bioinspired short peptides†
Sara
La Manna
a,
Daniele
Florio
a,
Valeria
Panzetta
 bcd,
Valentina
Roviello
c,
Paolo Antonio
Netti
bcd,
Concetta
Di Natale
bcd,
Valentina
Roviello
c,
Paolo Antonio
Netti
bcd,
Concetta
Di Natale
 bcd and
Daniela
Marasco
bcd and
Daniela
Marasco
 *a
*a
aDepartment of Pharmacy, University of Naples “Federico II”, 80131, Naples, Italy. E-mail: daniela.marasco@unina.it
bInterdisciplinary Research Centre on Biomaterials (CRIB), University of Naples “Federico II”, 80125, Naples, Italy
cDepartment of Ingegneria Chimica del Materiali e della Produzione Industriale (DICMAPI), University of Naples “Federico II”, 80125, Naples, Italy
dIstituto Italiano di Tecnologia, Largo Barsanti e Matteucci 53, 80125, Naples, Italy
First published on 24th October 2022
Abstract
Supramolecular assemblies of short peptides are experiencing a stimulating flowering. Herein, we report a novel class of bioinspired pentapeptides, not bearing Phe, that form hydrogels with fibrillar structures. The inherent sequence comes from the fragment 269–273 of nucleophosmin 1 protein, that is normally involved in liquid–liquid phase separation processes into the nucleolus. By means of rheology, spectroscopy, and scanning microscopy the crucial roles of the extremities in the modulation of the mechanical properties of hydrogels were elucidated. Three of four peptide showed a typical shear-thinning profile and a self-assembly into hierarchical nanostructures fibers and two of them resulted biocompatible in MCF7 cells. The presence of an amide group at C-terminal extremity caused the fastest aggregation and the major content of structured intermediates during gelling process. The tunable mechanical and structural features of this class of hydrogels render derived supramolecular systems versatile and suitable for future biomedical applications.
Introduction
Short peptide-based hydrogels are used in supramolecular technologies to build new smart light-responsive1 and biocompatible2 materials as well as delivery systems of therapeutics or vaccines.3 These structures can be programmed to self-organize into various forms: nanofibers, nanotubes, nanoribbons, or hydrogels, acquiring the ability to cross cell membranes and other challenging biological barriers.4–6 They are also widely used as starting materials in regenerative medicine7 in wound repair,8 for stem cell culture niche and differentiation9 and in cancer personalized treatments.10,11The hydrogelation mechanism, consists in the formation of ordered structures regulated by different forces: hydrogen, hydrophobic, electrostatic, van der Waals and aromatic bonds.12,13 These interactions allow the formation of hierarchical nano/micro fibrillar structures that then interweave into α-helices, β-sheets, hairpins and turns.14
The de novo design of supramolecular peptide-systems has been largely investigated,15 probing that even minor modifications may affect self-assembly in unpredictable ways, for example the C-terminal amidation accelerated the gelation kinetics of several Fmoc-Phe derivatives,16,17 with respect to a carboxylic acid termini.18 Many short peptides (<8 amino acids) self-assemble providing different nanostructures on the basis of the amphiphilicity and self-complementarity of sequences.19,20 The ability of phenylalanine residue to aid amyloid oligomerization process through π–π stacking is widely reported21 and in the building of microstructures, one of the most employed template is the homo-dipeptide Phe–Phe.22,23
Studies related to it demonstrated slight changes in the size and hydrophobicity of the N-terminal residue of the X-Phe dipeptide (X = Ile, Val, etc.) significantly affected the self-assembly process.24 Examples of short-peptide hydrogels without phenylalanine are however available: a pioneering study reported on the catalytic activities of designed heptapeptides bearing alternatively hydrophobic (L), cationic (R and K) and Zn2+ coordinating His residues.25 More recently designed bioinks-lysine-containing sequences revealed able to form three-dimensional hydrogels.26 In them, a hydrophobic N-terminal stretch, constituted by three, four and five residues, acts as a “template” for the self-assembly, while the variation of a cationic C-terminal residue (mainly Lys) regulated the stiffness and yield stress of hydrogels.27 In short sequences, not only the identity of single residues but also the overall charge exerts a crucial role, especially in dependence of N-and/or C-terminal chemical groups:28,29
Nature is an amazing source of chemical and structural diversities: proteins contain small fragments, sometimes hidden in the protein architecture, with unexpected features that are exploitable to develop innovative peptide-based molecules with unique characteristics.30 Following this approach, recently, we reported on the hydrogel formation of an amyloidogenic hexapeptide protein fragment (268–273, FINYVK) of Nucleophosmin 1 (NPM1), located in the second helix of the wild-type (wt) three helix bundle of the C-terminal domain (CTD).31 NPM1 is a homopentameric nucleolar protein mainly involved in chaperon activity32,33 and nucleolar organization through liquid–liquid phase separation (LLPS) phenomena.34,35NPM1 resulted the most mutated gene in a subtype of Acute Myeloid Leukaemia (AML) providing mutations within the third helix of the CTD.36,37 Many SAR investigations focused on the conformational consequences of AML mutations provided experimental evidences that protein regions including different mutations are able to self-aggregate in an amyloid way often producing cytotoxic fibers.38–51
Herein, to obtain hydrogels driven from different interactions we further modified the original hexapeptide by focusing on a five-residues fragment 269–273 (INYVK), deleting the N-terminal Phe residue. We investigated the extremity charge effects on the self-recognition mechanism of four different sequences with N- and C-termini free, or both acetylated/amidated or alternated (Table 1).
By means of different solution spectroscopies, as fluorescence and circular dichroism, we analyzed pentapeptides’ ability to aggregate, and the conformational intermediates involved in gelling process. The morphology of deriving structures was investigated through scanning electron microscopy as well as the viscoelastic properties of hydrogels by rheological analysis while biocompatibility of single pentapeptides was tested in MCF7 cancer cells.
Results and discussion
Peptides were synthesized following standard procedures, purified and identified through LC-MS (Fig. S1, ESI†).Fluorescence studies
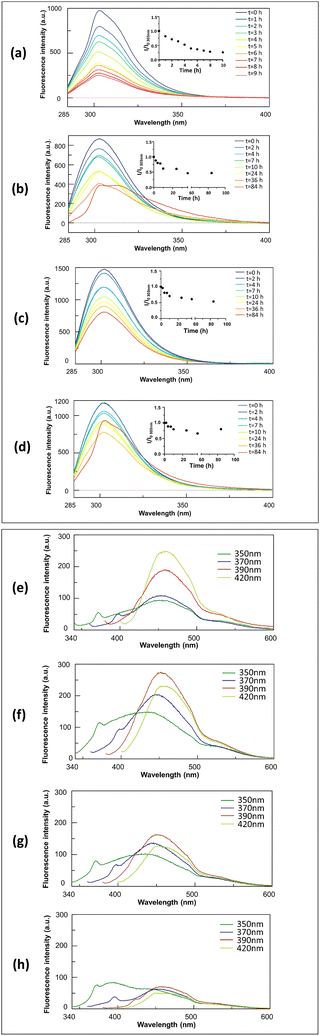
The role of the charges at the C- and N-termini of the fragment 269–273 of NPM1 in the hydrogelation process was firstly investigated through the evaluation of the Critical Aggregation Concentration (CAC) of each sequence performing ANS titrations. In them, the variations of ANS fluorescence signal at increasing amount of peptides were detected. As shown in Fig. 1, all peptides showed fluorescence changes providing CAC values in the high micromolar range (Table 1), except the fully unprotected sequence, H-INYVK-OH, which did not exhibit any variation.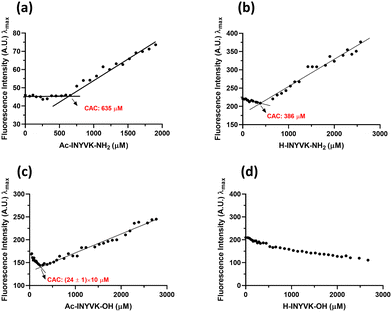 | ||
| Fig. 1 CAC values estimation. ANS fluorescence intensity at λmaxversus peptide concentration (a) Ac-INYVK-NH2, (b) H-INYVK-NH2, (c) Ac-INYVK-OH, (d) H-INYVK-OH. | ||
In particular, the highest CAC value (∼640 μM) was provided by the fully protected variant, Ac-INYVK-NH2, while sequences partially protected presented intermediate CACs (∼400 and 250 μM). The zwitterionic form of the sequence H-INYVK-OH negatively affects the self-aggregation as also confirmed by fluorescence results of tyrosine emission shown in Fig. 2, upper panel. In them, by reporting the fraction I/I0 as function of time (insets of Fig. 2(a)–(d)), the greatest variation was exhibited by acetylated/amidated sequence with a decrease of ∼80%, in a time interval of 11 h (Fig. 2(a)). Conversely, the fully deprotected sequence showed slight signal variation, ∼30% despite very long analysis, 80 h (Fig. 2(d)). As expected, partially protected sequences presented an intermediate behaviour providing variations of I/I0 of ∼50% and both reached saturation later with respect to the fully protected peptide (Fig. 2(b) and (c)).
These data were confirmed by the comparison of t1/2 values (Table 1): these are the times at which the normalized Tyr fluorescence intensity (I/I0) reached its maximum value/2.
The ability of pentapeptides to generate autofluorescence during self-aggregation47 was evaluated by monitoring emission spectra at different wavelengths of excitation during time. As expected, at t0 of aggregation, no significative fluorescence signals were observed for all four pentapeptides (Fig. S2, ESI†) but after 6 days of stirring, interestingly, three pentapeptides exhibited marked autofluorescence as reported in Fig. 2, lower panel. For the fully protected sequence the blue fluorescence emission is the most sensitive to the aggregation with a maximum at ∼455 nm, upon excitation at 420 nm (Fig. 2(e)). The two partially protected peptides exhibited greater intensities upon excitations at 390 nm (Fig. 2(f) and (g)) while the fully unprotected sequence, again, did not show significative intrinsic fluorescence (Fig. 2(h)). The amyloid character of the aggregation of the fully protected peptide was confirmed by ThT-assay reported in Fig. S3 (ESI†). In it, Ac-INYVK-NH2 was the only sequence to exhibit an increase of fluorescence signal over time and a t1/2 value comparable with that estimated with Tyr emission assay (Table 1).
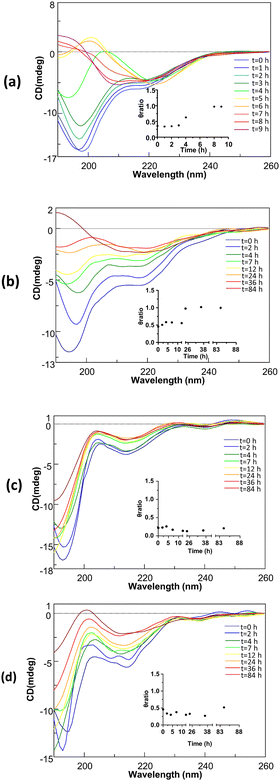
Conformational analysis through circular dichroism
Conformational variations during aggregation were analyzed by CD spectroscopy52 and the overlays of spectra at indicated times are reported in Fig. 3.At t = 0 all peptides presented spectra indicating a prevalent random coil state, mostly for C-terminal acid variants (Fig. 3(c) and (d)), as suggested by the absolute minimum at λ < 200 nm even if a small relative minimum around 218 nm can suggest a certain contribution of β-structure, again minor for acidic sequence. However, time evolutions CD profiles of four sequences appeared quite different and the time-dependent profiles of θratio, reported as insets of Fig. 3(a) and (b), help in the analysis of conformational transitions. Ac-INYVK-NH2, exhibited a strong tendency to aggregate within the first 3 hours of analysis as indicated by the progressive decrease of Cotton effect. This behavior is followed by different conformational contents that, in the interval 4–7 h seemed overwhelmed by β-structure (for the prevalence of a minimum at 219 nm) and by helices in the interval 8–9 hours (for the two minima at 219 and 198 nm and one positive band at 195 nm) (Fig. 3(a)). A similar behavior was exhibited by amidated sequence H-INYVK-Ac (Fig. 3(b)) for which the β-transition is much less evident and occurred in longer times (84 h). Also, acidic sequences showed a progressive reduction of CD signal, but for them conformational transitions appeared less pronounced especially for the acetylated variant (Fig. 3(c) and (d)).
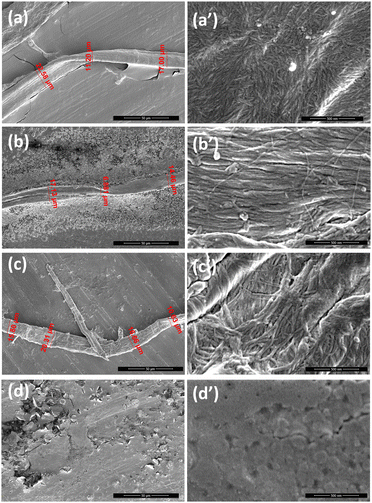
Scanning electron microscopy (SEM) analysis
The morphology of aggregates deriving from our pentapeptides was analyzed by SEM. Images were registered at t = 0 (Fig. S4, ESI†) and after 6 days of aggregation (Fig. 4 and Fig. S5, ESI†). For freshly prepared samples, only Ac-INYVK-NH2 and H-INYVK-NH2 demonstrated the presence of fibers (Fig. S4, ESI†). Indeed, the full protected peptide provided a fiber with an average diameter of 13 μm (Table 1) constituted by a network of fibers better visible at 500 nm of magnification (Fig. S4a′′′, ESI†). Conversely, fibers of only amidate peptide, even if with a similar size, did not appear yet fully mature, since immersed in the stub. After 6 days, both amidate sequences exhibited numerous twisted fibers longer with respect to those observed at t = 0. After 6 days, also Ac-INYVK-OH peptide formed fibers (Fig. 4(b) and Fig. S5b, ESI†), with diameters and lengths similar to the others (Table 1). In line with previous data, no fiber formation was detected for the full free peptide, even after long time of incubation (Fig. 4(d) and Fig. S5d, ESI†).Hydrogel formation
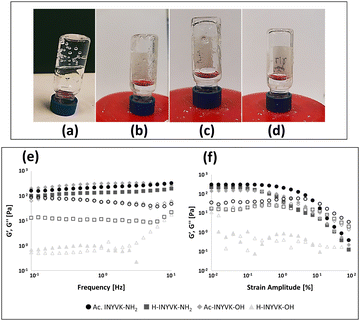
The heating–cooling53 method was employed to form hydrogels as assessed by 3D self-supporting hydrogels and inverted tube test54 (Fig. 5, upper panel).For Ac-INYVK-NH2 the presence of a stable gel was observable throughout the solution (Fig. 5(a)), for H-INYVK-NH2 a suspension containing small white precipitates was observable (Fig. 5(b)) while Ac-INYVK-OH presented a non-homogeneous, opaque hydrogel (Fig. 5(c)). As expected, the free pentapeptide, did not exhibit any gel (Fig. 5(d)).
Rheological analysis
Rheological studies were performed on all pentapeptides, prepared in glass vials, subjected to heating–cooling process and transferred to the rheometer for the analysis. Dynamic frequency and strain sweep experiments were performed at 0.1% and 1 Hz, at three temperatures, and three out of four sequences (except fully unprotected peptide), exhibited a linear viscoelastic (LVE) behavior (Fig. 5 lower panel and Fig. S6, ESI†). At 10 and 20 °C (Fig. S6a, b, ESI† and Table 2), frequency sweeps profiles for H-INYVK-NH2 presented viscoelastic moduli one order of magnitude higher than Ac-INYVK-NH2 and Ac-INYVK-OH systems, and two order of magnitude higher to those of H-INYVK-OH. This behaviour could be due to the presence of one protonable group at the N-terminal extremity of only amidated sequence, that likely favored non covalent crosslinks in inter-molecular self-assembly, causing an immediate increase of storage and loss moduli values (Table 1).55| Sequence | G′@1 Hz (Pa) | G′′@1 Hz (Pa) | ||||
|---|---|---|---|---|---|---|
| Temperature (°C) | ||||||
| 10 | 20 | 30 | 10 | 20 | 30 | |
| Ac-INYVK-NH2 | 5.3 | 25.3 | 227.6 | 0.84 | 3.5 | 56.6 |
| H-INYVK-NH2 | 50.6 | 163.0 | 136.0 | 13.6 | 16.3 | 11.5 |
| Ac-INYVK-OH | 5.0 | 22.1 | 317.1 | 0.9 | 4.8 | 58.2 |
| H-INYVK-OH | 0.1 | 1.0 | 0.8 | 0.1 | 0.6 | 0.8 |
In frequency profiles at 30 °C (Fig. 5(e) and Table 2), the viscoelastic moduli increased for all gels, except for the free pentapeptide. For acetylated sequences storage moduli G′ were larger than loss moduli G′′, indicating a characteristic viscoelasticity of the gel, no crossovers were detectable within the 0.1–10 Hz frequency range and G′ did not show a frequency dependence. These features suggested the presence of strong junctions among macromolecular chains responsible for the emergence of a viscoelastic plateau.56 On the contrary, the H-INYVK-OH sequence behaved as a gel within the range 10−1 to 100 Hz (G′–G′′) and as a liquid for higher frequencies (Fig. 5(e) and Fig. S6a, b, ESI†).
On the other hand, strain sweeps profiles (Fig. 5(f)) indicated that high values of strains determined changes in the gel structures causing a decrease of the measured moduli likely due to the rupture of bonds and/or entanglements. As expected, the yield strain decreased by increasing temperature (Fig. 5(f) and Fig. S6c, d, ESI†).
Similar experiments carried out at 37 °C for Ac-INYVK-NH2 (Fig. S7, ESI†), exhibited storage and loss moduli values almost overlapped to those at 30 °C.
In addition, the viscosity curves at 20 °C (Fig. S8, ESI†) indicated that all sequences had a shear-thinning behaviour, typical of macromolecules, except the free sequence that, since unable to polymerize efficiently, exhibited a very low viscosity (1–4 mPa s, water viscosity at 20 °C ∼1 mPa s) close to the sensitivity limit of the rheometer.
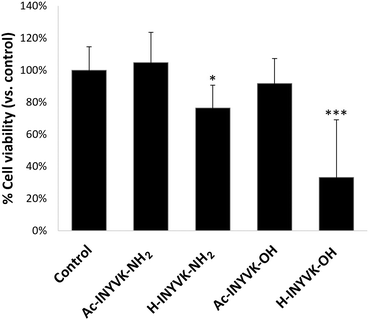
Cytotoxicity
Cell compatibility of hydrogels was investigated in cancer cells for their potential application as delivery systems of anticancer drugs.57 Cell toxicity experiments were performed using human MCF7 breast cancer cells incubated with all peptides at a final concentration of 0.5 mg mL−1 in cell culture medium. Results obtained from MTT assays (Fig. 6) revealed that both amidated pentapeptides exhibited a good compatibility with a cell viability close to 100%. Conversely, the free amine sequences negatively affected the cell viability, in particular the fully unprotected peptide H-INYVK-OH, at a greater extend. This behavior could be ascribed to unspecific effects of protonable/deprotonable extremities that can induce not selective interactions within the cell and cause its death.58Experimental
Peptide synthesis and purification
Pentapeptides were synthesized as C-terminal acids or amides on Wang (substitution 0.5 mmol g−1) and Rink-amide (substitution 0.78 mmol g−1) resins, respectively, using 9-fluorenylmethoxycarbonyl/tert-butyl (Fmoc/tBu) as already reported.31 Briefly, Fmoc deprotections were carried out using a 40% (v/v) piperidine solution in DMF (reaction time: 10 min). The coupling reactions were carried out by treatment with a solution of the Fmoc-amino acid (5 equiv.) and (HBTU)/(HOBt)/(DIEA) (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) in DMF. All couplings were performed for 20 minutes and repeated twice. When required, the N-terminal primary amino group was acetylated by treatment with a solution at 20% of acetic anhydride and 5% DIEA in DMF. All sequences were removed from the resin by treatment with a TFA
10) in DMF. All couplings were performed for 20 minutes and repeated twice. When required, the N-terminal primary amino group was acetylated by treatment with a solution at 20% of acetic anhydride and 5% DIEA in DMF. All sequences were removed from the resin by treatment with a TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS
TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (90
H2O (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 v/v/v) mixture for 90 min at room temperature. Then, they were precipitated in cold ether, dissolved in a water/acetonitrile (1
5 v/v/v) mixture for 90 min at room temperature. Then, they were precipitated in cold ether, dissolved in a water/acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) mixture, and lyophilized.
1 v/v) mixture, and lyophilized.
Crude products were identified through LC-MS analysis and purified by RP-HPLC, applying a linear gradient of 0.1% TFA CH3CN in 0.1% TFA water from 5–70% over 13 min with a semipreparative 2.2 × 5 cm C18 column at a flow rate of 15 mL min−1. Pure peptides were analyzed through LC-MS (Fig. S1, ESI†), treated with (HFIP) 1,1,1,3,3,3-hexafluoro-2-propanol, lyophilized and stored at −20 °C until use.
Hydrogel formation
Hydrogels were prepared following the heating–cooling method.59 In detail, solutions were prepared by dissolving freeze-dried peptide samples (2 mg mL−1, 2.5 mM) in 20 mM borate buffer (pH 8.7) and the peptide, Ac-INYVK-OH, in 20 mM acetate buffer (pH 4.0).Homogeneous solutions were obtained by sonication at 25 °C until they were visibly clear, then they were immersed in a thermoblock at 90 °C for 30 minutes and then cooled at room temperature for 2 hours. The formation of gels was assessed via tube-inverting method60 reported in Fig. 5, upper panel.
Fluorescence assays
Florescence assays were performed on a spectrofluorometer Jasco FP 8300 with a cell of 10 mm path-length quartz cuvette. Tyrosine, ThT and intrinsic fluorescence signals were measured on samples at 400 μM in borate (pH 8.7) or acetate (pH 4.0) buffers (10 mM for Tyr and intrinsic fluorescence and 50 mM for ThT), at 25 °C under magnetic stirring. λexc: 275 nm, λemiss: 303 nm for Tyr emission, λexc: 440 nm, λemiss: 483 nm for ThT assay while, for intrinsic fluorescence, excitation wavelengths of acquisition were 350, 370, 390 and 420 nm. Critical aggregation concentration (CAC) values were determined by fluorescence titration of the dye 8-anilino-1-naphthalene sulfonic acid ammonium salt (ANS) (λexc = 350 nm; λemiss: 491–480 nm). Small aliquots (2 μL) of each peptide at 2 mM were added at 250 μL of ANS 80 μM in borate or acetate buffer 20 mM.Circular dichroism
CD spectra were registered with a Jasco J-815 spectropolarimeter (JASCO, Tokyo, Japan), at 25 °C in the spectral range 190–260 nm and are averaged over two scans, with the subtraction of blanks. Peptides concentrations were 400 μM in 10 mM borate and acetate buffer using a 0.1 cm path-length quartz cuvette. The θ ratio was calculated as the ratio between the two CD signals at two minima in each spectrum at corresponding time. Concerning acid peptides, this value was obtained as the ratio between the minimum at ∼218 nm and that 195 nm while for amidated sequences the value was initially calculated as the ratio 219/198 nm, and, after the conformational transitions (t = 8, 9 h for Ac-INYVK-NH2, and t = 36, 84 h for H-INYVK-NH2), as 220/210 nm. For the fully protected sequence the absence of another minimum besides that at ∼222 nm, did not allow to evaluate this ratio at t = 5, 6, 7 h.SEM analysis
Samples, at two different times of aggregation (t = 0 and 6 days), were dropped on stub at 400 μM concentrations and introduced into chamber of field emission scanning (Nova NanoSem 450 FEI/ThermoFisher Scientific), to obtain Sem micrographs at 3.00 and 5.00 kV in high vacuum mode, with an Everhart Thornley Detector (ETD) and the Through the Lens Detector (TLD), as previously reported.61–65Rheological studies
Rheological measurements were performed on peptides subjected to heating–cooling procedure already described using a stress-controlled shear rheometer (Anton Paar MCR 502) equipped with 25 mm stainless steel parallel plate geometry tool and a Peltier heating system to control the temperature at 10, 20 or 30 °C. 500 μL of hydrogels were prepared as described above, placed on the rheometer stage and the geometry was lowered onto the sample to a gap height of 0.75 mm. Rheological studies included a dynamic strain sweep (0.1 to 100 strain%) collected at a constant frequency of 1 Hz, a dynamic frequency sweep (frequency range of 0.1 to 10 Hz) collected at 0.1% strain and a flow test obtained in a shear rate range of 10−2 to 102 s−1.66Cell culture and in vitro cytotoxicity assay
MCF7 cells were grown at 37 °C in a humidified atmosphere of 5% CO2 in Eagle's minimum essential medium (EMEM, Sigma-Aldrich) containing 10% fetal bovine serum (FBS), 100 μg mL−1L-glutamine and 100 U mL−1 penicillin/streptomycin. Cells were seeded in triplicates in 96-well plates at a density of 8000 cells per well and allowed to adhere overnight. Pre-formed gels (2 mg mL−1) were prepared as previously describes, diluted in cell culture medium at a final concentration of 0.5 mg mL−1 and added to the cells for 24 h. Control cells were incubated with borate buffer and acetate buffer diluted in cell culture medium at the same final concentration used for the pre-formed gels. After the incubation, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays was used according to the manufacturer's instructions. Briefly, 200 μL of MTT labelling reagent (final concentration 0.5 mg mL−1; Sigma-Aldrich) were added to each well for 3 h. Then, the supernatant was removed and substituted with 200 μL of isopropanol for 10 min at 37 °C, 5% CO2. The optical density of each well sample was determined at 550 nm determination at 495 nm using a microplate reader. A blank absorbance value of 0.058, obtained from wells without cells, but treated with MTT reagent, was subtracted from all the absorbance values. Then, the average absorbance value of cells treated with gels were normalized to those of control cells incubated with the same buffer and cell viability was expressed as a percentage of the corresponding control. Standard deviations of normalized absorbance values were calculated via error propagation.Conclusion
The application fields of peptide hydrogels closely depend on self-recognition processes and on the type of generated structures.67 In the present study, by applying a reductionist approach, we analyzed the gelling properties of the short 269–273 stretch of NPM1. We investigated the INYVK peptide in four different forms deriving from the partial absent, and complete amidation/acetylation at the extremities of the backbone. The most prone gelling sequence resulted the fully protected peptide that provided: the highest CAC value, the fastest kinetic of aggregation, the most hierarchical nanostructures bundled into twisted, ribbon-like fibers and the greatest value of ratio G′/G′′. Interestingly the gelling process is accompanied by a series of structural transitions including α-helical intermediates, related to the native structure of the protein fragment and Ac-H-INYVK-NH2 sequence provided good cellular compatibility. From our results, even just the presence of N-terminal acetylated group, that increased hydrophobicity, favored hydrogelation, even if at a lesser extent with not-homogeneous gel-states. Conversely, the fully deprotected sequence did not exhibit any feature of gel even if analyzed for long times suggesting that an amidated C-terminus is required for the gelation of the sequence. Indeed, the only amidated sequence stood out for the presence of more ordered helical structures and of nanostructures at early stages of aggregation assuming the greatest values of viscoelastic moduli with respect to the others. Overall data indicate that the absence of Phe269 in NPM1's fragment does not alter the ability of this stretch to self-assembly even if it delays the aggregation as evident by the comparison of ThT profiles of 269–273 (Fig. S3, ESI†) and its longer parent fragment 268–273.31 In the case of 269–273, the aromatic contribution to self-recognition process is due to Tyr271 side chain, exclusively. Also the gel features of the fully protected pentapeptide appeared different from the already investigated hexapeptide stretch: indeed, for Ac-FINYVK-NH2, similar rheological investigations indicated lower mechanical performance of the hydrogel (data not shown) corroborating our reductionist approach to investigate the sequence INYVK. However, to detail self-recognition mechanism of our sequences, solution and solid state NMR investigations would be required to experimentally follow the process but overall data allow to speculate that involved physical factors in self-assembly process are comparable with to those reported for similar systems.68,69 In conclusion we have presented a novel class of self-assembling peptides that exhibited tuneable and versatile conditions to become hydrogels.Author contributions
S. L. M. and D. F. contributed equally to this work. S. L. M. synthesized and characterized the peptides and performed CD and fluorescence experiments. V. R. made SEM experiments. V. P. performed rheological studies and cellular assays. D. M. designed the concept. D. M. and P. A. N. supervised the experiments. D. M., D. F. and C. D. N. wrote the manuscript. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Sara La Manna was supported by AIRC fellowship for Italy.References
- B. Pramanik and S. Ahmed, Gels, 2022, 8, 533 CrossRef CAS PubMed.
- J. Li, R. Xing, S. Bai and X. Yan, Soft Matter, 2019, 15, 1704–1715 RSC.
- A. Lakshmanan, S. Zhang and C. A. Hauser, Trends Biotechnol., 2012, 30, 155–165 CrossRef CAS PubMed.
- Z. Li, B. Cai, W. Yang and C.-L. Chen, Chem. Rev., 2021, 121, 14031–14087 CrossRef CAS PubMed.
- R. Jamaledin, C. Di Natale, V. Onesto, Z. B. Taraghdari, E. N. Zare, P. Makvandi, R. Vecchione and P. A. Netti, J. Clin. Med., 2020, 9, 542 CrossRef CAS PubMed.
- E. Lagreca, V. Onesto, C. Di Natale, S. La Manna, P. A. Netti and R. Vecchione, Prog. Biomater., 2020, 9, 153–174 CrossRef CAS PubMed.
- W. Y. Seow and C. A. Hauser, Mater. Today, 2014, 17, 381–388 CrossRef CAS.
- T. Guan, J. Li, C. Chen and Y. Liu, Adv. Sci., 2022, 9, 2104165 CrossRef CAS PubMed.
- K. S. Hellmund and B. Koksch, Front. Chem., 2019, 7, 172 CrossRef CAS PubMed.
- Z. Yang, H. Xu and X. Zhao, Adv. Sci., 2020, 7, 1903718 CrossRef CAS PubMed.
- T. Agarwal, G. M. Fortunato, S. Y. Hann, B. Ayan, K. Y. Vajanthri, D. Presutti, H. Cui, A. H. Chan, M. Costantini and V. Onesto, Mater. Sci. Eng., C, 2021, 124, 112057 CrossRef CAS PubMed.
- G. A. Braun, B. E. Ary, A. J. Dear, M. C. Rohn, A. M. Payson, D. S. Lee, R. C. Parry, C. Friedman, T. P. Knowles and S. Linse, Biomacromolecules, 2020, 21, 4781–4794 CrossRef CAS PubMed.
- I. W. Fu, C. B. Markegard, B. K. Chu and H. D. Nguyen, Adv. Healthcare Mater., 2013, 2, 1388–1400 CrossRef CAS PubMed.
- A. Dasgupta, J. H. Mondal and D. Das, RSC Adv., 2013, 3, 9117–9149 RSC.
- S. Das and D. Das, Front. Chem., 2021, 9, 770102 CrossRef CAS PubMed.
- D. M. Ryan, T. M. Doran and B. L. Nilsson, Chem. Commun., 2011, 47, 475–477 RSC.
- B. L. Abraham, W. Liyanage and B. L. Nilsson, Langmuir, 2019, 35, 14939–14948 CrossRef CAS PubMed.
- M. Gorlero, R. Wieczorek, K. Adamala, A. Giorgi, M. E. Schininà, P. Stano and P. L. Luisi, FEBS Lett., 2009, 583, 153–156 CrossRef CAS PubMed.
- A. Mahler, M. Reches, M. Rechter, S. Cohen and E. Gazit, Adv. Mater., 2006, 18, 1365–1370 CrossRef CAS.
- R. Das, B. Gayakvad, S. D. Shinde, J. Rani, A. Jain and B. Sahu, ACS Appl. Bio Mater., 2020, 3, 5474–5499 CrossRef CAS PubMed.
- W. P. Esler, C. Das and M. S. Wolfe, Bioorg. Med. Chem. Lett., 2004, 14, 1935–1938 CrossRef CAS PubMed.
- M. Reches and E. Gazit, Science, 2003, 300, 625–627 CAS.
- S. Marchesan, A. V. Vargiu and K. E. Styan, Molecules, 2015, 20, 19775–19788 CrossRef CAS PubMed.
- N. S. De Groot, T. Parella, F. X. Aviles, J. Vendrell and S. Ventura, Biophys. J., 2007, 92, 1732–1741 Search PubMed.
- C. Rufo, Y. Moroz, O. Moroz, J. Stöhr, T. Smith, X. Hu and W. DeGrado, Short peptides self-assemble to produce catalytic amyloids, Nat. Chem., 2014, 6, 303–309 CrossRef CAS PubMed.
- Y. Loo, A. Lakshmanan, M. Ni, L. L. Toh, S. Wang and C. A. Hauser, Nano Lett., 2015, 15, 6919–6925 CrossRef CAS PubMed.
- K. H. Chan, W. H. Lee, M. Ni, Y. Loo and C. A. E. Hauser, Sci. Rep., 2018, 8, 17127 CrossRef PubMed.
- M. Andreasen, K. K. Skeby, S. Zhang, E. H. Nielsen, L. H. Klausen, H. Frahm, G. Christiansen, T. Skrydstrup, M. Dong and B. Schiøtt, Biochemistry, 2014, 53, 6968–6980 CrossRef CAS PubMed.
- D. Roberts, C. Rochas, A. Saiani and A. Miller, Langmuir, 2012, 28, 16196–16206 CrossRef CAS PubMed.
- K. Seidi, M. H. Ayoubi-Joshaghani, M. Azizi, T. Javaheri, M. Jaymand, E. Alizadeh, T. J. Webster, A. A. Yazdi, M. Niazi and M. R. Hamblin, Nano Today, 2021, 39, 101157 CrossRef CAS.
- D. Florio, C. Di Natale, P. L. Scognamiglio, M. Leone, S. La Manna, S. Di Somma, P. A. Netti, A. M. Malfitano and D. Marasco, Bioorg. Chem., 2021, 114, 105047 CrossRef CAS PubMed.
- L. Federici and B. Falini, Protein Sci., 2013, 22, 545–556 CrossRef CAS PubMed.
- M. S. Lindstrom, Biochem. Res. Int., 2011, 2011, 195209 Search PubMed.
- M. C. Ferrolino, D. M. Mitrea, J. R. Michael and R. W. Kriwacki, Nat. Commun., 2018, 9, 5064 CrossRef PubMed.
- D. M. Mitrea, J. A. Cika, C. B. Stanley, A. Nourse, P. L. Onuchic, P. R. Banerjee, A. H. Phillips, C. G. Park, A. A. Deniz and R. W. Kriwacki, Nat. Commun., 2018, 9, 842 CrossRef PubMed.
- B. Falini, N. Bolli, A. Liso, M. P. Martelli, R. Mannucci, S. Pileri and I. Nicoletti, Leukemia, 2009, 23, 1731–1743 CrossRef CAS PubMed.
- C. G. Grummitt, F. M. Townsley, C. M. Johnson, A. J. Warren and M. Bycroft, J. Biol. Chem., 2008, 283, 23326–23332 CrossRef CAS PubMed.
- D. Florio, V. Roviello, S. La Manna, F. Napolitano, A. Maria Malfitano and D. Marasco, Bioorg. Chem., 2022, 127, 106001 CrossRef CAS PubMed.
- S. La Manna, D. Florio, C. Di Natale, P. L. Scognamiglio, T. Sibillano, P. A. Netti, C. Giannini and D. Marasco, Int. J. Biol. Macromol., 2021, 188, 207–214 CrossRef CAS PubMed.
- S. La Manna, D. Florio, C. Di Natale, F. Napolitano, A. M. Malfitano, P. A. Netti, I. De Benedictis and D. Marasco, Bioorg. Chem., 2021, 113, 104997 CrossRef CAS PubMed.
- D. Florio, C. Di Natale, P. L. Scognamiglio, M. Leone, S. La Manna, S. Di Somma, P. A. Netti, A. M. Malfitano and D. Marasco, Bioorg. Chem., 2021, 114, 105047 CrossRef CAS PubMed.
- D. Florio, M. Cuomo, I. Iacobucci, G. Ferraro, A. M. Mansour, M. Monti, A. Merlino and D. Marasco, Pharmaceuticals, 2020, 13, 171 CrossRef CAS PubMed.
- C. Di Natale, D. Florio, S. Di Somma, A. Di Matteo, L. Federici, P. A. Netti, G. Morelli, A. M. Malfitano and D. Marasco, Int. J. Biol. Macromol., 2020, 164, 3501–3507 CrossRef CAS PubMed.
- S. La Manna, P. L. Scognamiglio, V. Roviello, F. Borbone, D. Florio, C. Di Natale, A. Bigi, C. Cecchi, R. Cascella, C. Giannini, T. Sibillano, E. Novellino and D. Marasco, FEBS J., 2019, 286, 2311–2328 CrossRef CAS PubMed.
- S. La Manna, V. Roviello, P. L. Scognamiglio, C. Diaferia, C. Giannini, T. Sibillano, G. Morelli, E. Novellino and D. Marasco, Int. J. Biol. Macromol., 2019, 122, 517–525 CrossRef CAS PubMed.
- D. Florio, A. M. Malfitano, S. Di Somma, C. Mugge, W. Weigand, G. Ferraro, I. Iacobucci, M. Monti, G. Morelli, A. Merlino and D. Marasco, Int. J. Mol. Sci., 2019, 20, 829 CrossRef CAS PubMed.
- C. Di Natale, S. La Manna, A. M. Malfitano, S. Di Somma, D. Florio, P. L. Scognamiglio, E. Novellino, P. A. Netti and D. Marasco, Biochim. Biophys. Acta, Proteins Proteomics, 2019, 1867, 637–644 CrossRef CAS PubMed.
- A. De Santis, S. La Manna, I. R. Krauss, A. M. Malfitano, E. Novellino, L. Federici, A. De Cola, A. Di Matteo, G. D’Errico and D. Marasco, Biochim. Biophys. Acta, Gen. Subj., 2018, 1862, 967–978 CrossRef CAS PubMed.
- A. Russo, C. Diaferia, S. La Manna, C. Giannini, T. Sibillano, A. Accardo, G. Morelli, E. Novellino and D. Marasco, Biochim. Biophys. Acta, Proteins Proteomics, 2017, 1865, 176–185 CrossRef CAS PubMed.
- P. L. Scognamiglio, C. Di Natale, M. Leone, R. Cascella, C. Cecchi, L. Lirussi, G. Antoniali, D. Riccardi, G. Morelli, G. Tell, F. Chiti and D. Marasco, Oncotarget, 2016, 7, 59129–59143 CrossRef PubMed.
- C. Di Natale, P. L. Scognamiglio, R. Cascella, C. Cecchi, A. Russo, M. Leone, A. Penco, A. Relini, L. Federici, A. Di Matteo, F. Chiti, L. Vitagliano and D. Marasco, FASEB J., 2015, 29, 3689–3701 CrossRef CAS PubMed.
- C. Bortolini, L. H. Klausen, S. V. Hoffmann, N. C. Jones, D. Saadeh, Z. Wang, T. P. Knowles and M. Dong, ACS Nano, 2018, 12, 5408–5416 CrossRef CAS PubMed.
- H. Wang, Z. Yang and D. J. Adams, Mater. Today, 2012, 15, 500–507 CrossRef CAS.
- W. M. Dawson, F. J. Martin, G. G. Rhys, K. L. Shelley, R. L. Brady and D. N. Woolfson, Chem. Sci., 2021, 12, 6923–6928 RSC.
- O. Guaresti, S. Basasoro, K. González, A. Eceiza and N. Gabilondo, Eur. Polym. J., 2019, 119, 376–384 CrossRef CAS.
- R. A. Hule, R. P. Nagarkar, A. Altunbas, H. R. Ramay, M. C. Branco, J. P. Schneider and D. J. Pochan, Faraday Discuss., 2008, 139, 251–264 RSC ; discussion 309–225, 419–220.
- H. Huang, Y. Ding, X. S. Sun and T. A. Nguyen, PLoS One, 2013, 8, e59482 CrossRef CAS PubMed.
- B. Cheng, B. Thapa, K. Remant and P. Xu, J. Mater. Chem. B, 2015, 3, 25–29 RSC.
- Q. Li, G. Zhang, Y. Wu, Y. Wang, Y. Liang, X. Yang, W. Qi, R. Su and Z. He, J. Colloid Interface Sci., 2021, 583, 234–242 CrossRef CAS PubMed.
- J. Huang, C. L. Hastings, G. P. Duffy, H. M. Kelly, J. Raeburn, D. J. Adams and A. Heise, Biomacromolecules, 2013, 14, 200–206 CrossRef CAS PubMed.
- N. Bagheri, V. Mazzaracchio, S. Cinti, N. Colozza, C. Di Natale, P. A. Netti, M. Saraji, S. Roggero, D. Moscone and F. Arduini, Anal. Chem., 2021, 93, 5225–5233 CrossRef CAS PubMed.
- R. Jamaledin, R. Sartorius, C. Di Natale, R. Vecchione, P. De Berardinis and P. A. Netti, Microorganisms, 2020, 8, 650 CrossRef CAS PubMed.
- V. Onesto, C. Di Natale, M. Profeta, P. A. Netti and R. Vecchione, Prog. Biomater., 2020, 9, 203–217 CrossRef CAS PubMed.
- M. Profeta, C. Di Natale, E. Lagreca, V. Mollo, P. A. Netti and R. Vecchione, Pharmaceutics, 2021, 13, 1069 CrossRef CAS PubMed.
- C. Di Natale, V. De Gregorio, E. Lagreca, F. Mauro, B. Corrado, R. Vecchione and P. A. Netti, Front. Bioeng. Biotechnol., 2022, 10, 851893 CrossRef PubMed.
- C. Di Natale, E. Lagreca, V. Panzetta, M. Gallo, F. Passannanti, M. Vitale, S. Fusco, R. Vecchione, R. Nigro and P. Netti, Front. Bioeng. Biotechnol., 2021, 9, 660691 CrossRef PubMed.
- A. M. Garcia, M. Melchionna, O. Bellotto, S. Kralj, S. Semeraro, E. Parisi, D. Iglesias, P. D’Andrea, R. De Zorzi and A. V. Vargiu, ACS Nano, 2021, 15, 3015–3025 CrossRef CAS PubMed.
- S. R. Leonard, A. R. Cormier, X. Pang, M. I. Zimmerman, H.-X. Zhou and A. K. Paravastu, Biophys. J., 2013, 105, 222–230 CrossRef CAS PubMed.
- K. Nagy-Smith, E. Moore, J. Schneider and R. Tycko, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 9816–9821 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1. LC-MS analysis of all pentapeptides; Fig. S2. Overlay of emission spectra at t = 0 of aggregation; Fig. S3. ThT fluorescence profiles of four pentapeptides; Fig. S4. SEM micrographs of all pentapeptides at t = 0 of aggregation; Fig. S5. SEM micrographs of all pentapeptides after 6 days of aggregation; Fig. S6. Storage modulus G′ and loss modulus G′′ at 10 °C and 20 °C in dynamic frequency and amplitude sweeps; Fig. S7. Storage modulus G′ and loss modulus G′′ at 30 °C and 37 °C and Fig. S8. Viscosity curves at 20 °C of all pentapeptides. The following files are available free of charge. See DOI: https://doi.org/10.1039/d2sm01385a |
| This journal is © The Royal Society of Chemistry 2022 |