Open Access Article
Open Access ArticleNanomechanical analysis of SARS-CoV-2 variants and predictions of infectiousness and lethality†
Yiwen
Hu
ab and
Markus J.
Buehler
 *abc
*abc
aLaboratory for Atomistic and Molecular Mechanics (LAMM), Massachusetts Institute of Technology, 77 Massachusetts Ave., Cambridge, MA 02139, USA. E-mail: mbuehler@mit.edu; Tel: +1-617-452-2750
bDepartment of Mechanical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Ave., Cambridge, MA 02139, USA
cCenter for Computational Science and Engineering, Schwarzman College of Computing, Massachusetts Institute of Technology, 77 Massachusetts Ave., Cambridge, MA 02139, USA
First published on 20th July 2022
Abstract
As variants of the pathogen that causes COVID-19 spread around the world, estimates of infectiousness and lethality of newly emerging strains are important. Here we report a predictive model that associates molecular motions and vibrational patterns of the virus spike protein with infectiousness and lethality. The key finding is that most SARS-CoV-2 variants are predicted to be more infectious and less lethal compared to the original spike protein. However, lineage B.1.351 (Beta variant) is predicted to be less infectious and more lethal, and lineage B.1.1.7 (Alpha variant) is predicted to have both higher infectivity and lethality, showing the potential of the virus to mutate towards different performance regimes. The relatively more recent lineage B.1.617.2 (Delta variant), although contains a few key spike mutations other than D614G, behaves quite similar to the single D614G mutation in both vibrational and predicted epidemiological aspects, which might explain its rapid circulation given the prevalence of D614G. This work may provide a tool to estimate the epidemiological effects of new variants, and offer a pathway to screen mutations against high threat levels. Moreover, the nanomechanical approach, as a novel tool to predict virus-cell interactions, may further open up the door towards better understanding other viruses.
Introduction
As variants of the pathogen that causes COVID-19 spread around the world, estimates of infectiousness and lethality of newly emerging strains are important. Here we report, building off earlier work1 in which we correlated infectiousness and lethality of different types of coronavirus pathogens such as Middle East Respiratory Syndrome (MERS), Severe Acute Respiratory Syndrome (SARS) and Coronavirus Disease 2019 (COVID-19), a predictive model that associates molecular motions and vibrational patterns of the virus spike protein with infectiousness and lethality.First reported in China in 2019,2–4 the novel coronavirus disease, COVID-19, has spread rapidly around the world, resulting in more than 180 million global cases as of July 2021.5,6 SARS-CoV-2, the virus that causes COVID-19, can easily transmit from human to human, compared to previous two highly pathogenic human coronaviruses—namely, SARS-CoV and MERS-CoV. Similar to other human coronaviruses, SARS-CoV-2 enters human cells through the interaction between the receptor-binding domain (RBD) of its spike protein and the human cell receptor, and it shares the same human receptor angiotensin-converting enzyme 2 (ACE2) with SARS-CoV.7 Composed of an amino (N)-terminal S1 subunit and a carboxyl (C)-terminal S2 subunit, the spike protein of coronavirus bears functional significance in the infection process. While the S2 subunit participates more in the membrane fusion, the S1 subunit, consisting of an N-terminal domain (NTD), the receptor-binding domain (RBD) and two smaller intermediate subdomains, is responsible for recognizing and binding to the host cell receptor.8 Previous literature reveals a prerequisite conformational state for receptor binding where at least one RBD should be in upward position, which makes it receptor-accessible, otherwise there would be steric clashes hindering the binding process.9 In experimental work, this specific type of receptor-accessible state has been captured for MERS-CoV, SARS-CoV and SARS-CoV-2.7,8 Given the essential role that the spike protein plays in receptor recognition, viral fusion and cell entry,7,10,11 it is also a promising target for drug and vaccine development.
While vaccination progress has been progressing well in many countries across the world, an emergence of more transmissible SARS-CoV-2 variants has caused great concern.12,13 SARS-CoV-2 D614G variant emerged in late January or early February 2020 and has quickly become the most prevalent form in the global pandemic.14 Not only does the D614G variant adopts a range of more open conformations than the original virus, but it also has added interactions to prevent premature dissociation of the trimer and increase the number of spikes that can facilitate infection.15,16 A number of experimental and statistical research have also shown that D614G variant has enhanced infectivity and transmissibility compared to the wild-type virus.14,17,18 The lineage B.1.1.7 (Alpha variant), first reported in the UK in December 2020, is predicted to be 43 to 90% more transmissible than the predecessor lineage based on statistical and dynamic modeling approaches.12 Moreover, transmission of SARS-CoV-2 between human and animals has also been observed, with the most notable mink mutation reported in Denmark causing the culling of all farmed mink in the country.19 Experimental evidence reveals that while mink-specific mutations do not increase entry into human cells, they do cause partial evasion from neutralization by a therapeutic antibody and convalescent plasma/sera.20 A concerning finding is that, an experiment suggests that mRNA-vaccine activity against SARS-CoV-2 variants that encode E484K-, N501Y- or K417N/E484K/N501Y-mutant spike may be reduced by a small—but significant—margin.21
Some recent research focuses on understanding and predicting epidemiological behavior of SARS-CoV-2 and its variants using different indicators from biological or clinical aspect. By evaluating the binding energy of SARS-CoV-2 spike protein and the host cell receptor with a machine-learning model, researchers report that most likely future mutations will make SARS-CoV-2 more infectious.22 It has also been shown that the viral load can be considered as an independent predictor of the transmissibility and mortality of SARS-CoV-2.23,24 Furthermore, with machine learning technique for natural language processing, the generated semantic landscapes for SARS-CoV-2 are able to predict mutations that could potentially evade the immune system.25
The research reported here focuses on the spike protein of coronavirus, where it makes physical interaction with the human cell receptor ACE2 and then enters human body. We conduct a normal mode analysis (NMA) of different SARS-CoV-2 variants in receptor-accessible conformation with one upward RBD, analyze their vibrational behaviors, and utilize the correlation that we reported in earlier work1 to make predictions on the infectiousness and lethality (Fig. 1).
Compared to previous predictive models,22–25 our presented predictive model is the first one to make predictions based on pure nanomechanical properties, i.e. the average fluctuation level and mobility ratio of receptor-binding domain that we identified in previous work.1 This work may provide a tool to estimate the epidemiological effects of new variants, and offer a pathway to screen mutations against high threat levels. Moreover, the nanomechanical approach, as a novel tool to predict virus-cell interactions, may offer a tool to better understand other viruses.
Materials and methods
Data preparation
The beta-coronavirus spike proteins that we consider in this paper are all coronavirus spike proteins with one upward receptor-binding domain (RBD), and they are MERS-CoV S (PDB ID: 5X5F), SARS-CoV S (PDB ID: 6CRZ, 6CRW), SARS-CoV-2 S (PDB ID: 6Z97, 7DDN). All coronavirus spike protein structures considered above, accessible from the Protein Data Bank,26 contain complete RBD structure, so that we can evaluate the largest flexibility in the extended loop of upward RBD based on our previous results.1 In regards to SARS-CoV-2 variants, we use the SARS-CoV-2 spike protein structure with PDB ID 6Z97 as our base protein structure, and implement the spike mutations of different variants using PyMOL. A few mutations are not implemented due to missing regions in templet protein structure (Table 1). The atomistic models of all coronavirus spike proteins, including the wild-type and the mutants, are prepared with Visual Molecular Dynamics (VMD).27 The spike protein structures are immersed in a TIP3P water box with a minimum distance of 20 Å to the nearest protein atom, followed by ionization with Sodium and Chlorine atoms. Then, we carry out 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps of conjugate gradient energy minimization and equilibrate the system for 80 ns with temperature controlled at 300 K based on Nanoscale Molecular Dynamics (NAMD).28 The simulation is performed with CHARMM36 all-atom additive protein force field,29 and both electrostatic and van der Waals calculations for non-bonded interactions are truncated smoothly at cutoff distance 12 Å using smooth switching functions. Nosé-Hoover Langevin piston pressure control,30,31 together with Langevin dynamics, are employed to simulate the NPT ensemble. After equilibration, we compute the average root mean square derivations (RMSD) based on the last 20 ns equilibrium period and pick out 10 frames with the nearest RMSD from the trajectory file in order to perform normal mode analysis on them.
000 steps of conjugate gradient energy minimization and equilibrate the system for 80 ns with temperature controlled at 300 K based on Nanoscale Molecular Dynamics (NAMD).28 The simulation is performed with CHARMM36 all-atom additive protein force field,29 and both electrostatic and van der Waals calculations for non-bonded interactions are truncated smoothly at cutoff distance 12 Å using smooth switching functions. Nosé-Hoover Langevin piston pressure control,30,31 together with Langevin dynamics, are employed to simulate the NPT ensemble. After equilibration, we compute the average root mean square derivations (RMSD) based on the last 20 ns equilibrium period and pick out 10 frames with the nearest RMSD from the trajectory file in order to perform normal mode analysis on them.
| Variant name | WHO label | First reported | Spike mutations | Mutations implemented in this work |
|---|---|---|---|---|
| D614G | — | China | D614G | D614G |
| Cluster 5 | — | Denmark | del69–70, Y453F, D614G, I692V, M1229I | del69 (chain A), Y453F, D614G, I692V |
| Other mink associated mutations | — | Europe | F486L | F486L |
| B.1.1.7 (501Y.V1) | Alpha | UK | del69–70, del Y144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | del69 (chain A), N501Y, A570D, D614G, T716I, S982A, D1118H |
| B.1.351 (501Y.V2) | Beta | South Africa | L18F, D80A, D215G, R246I, K417N, E484K, N501Y, D614G, A701V | D215G, K417N, E484K, N501Y, D614G, A701V |
| P.1 (501Y.V3) | Gamma | Brazil | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I | D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I |
| B.1.617.2 | Delta | India | T19R, G142D, 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N | G142D (chain A/C), L452R, T478K, D614G, D950N |
| Double mutant | — | India | L452R, E484Q, D614G | L452R, E484Q, D614G |
Normal mode analysis
Normal mode analyses (NMA) of coronavirus spike proteins in receptor-accessible state with one upward RBD is conducted to assess the molecular mechanics and vibrational properties from an atom-by-atom perspective. Here, we use the same approach as presented in our previous paper.1 We employ a coarse-grained elastic network model (ENM) available in the Bio3D32–34 package in R to analyze the normal modes of coronavirus spike protein structures and calculate atomic displacements for temperature 300 K to construct the fluctuation profile of each spike protein considered. The coarse-graining model35 is constructed with three to five atoms per residue, where the N, CA, C atoms represent the protein backbone and zero to two selected side chain atoms represent the side chain. The side chain atoms are select based on side chain size and the distance to CA. The energy function is defined by | (1) |
| k(‖r0ij‖) = min {7424·‖r0ij‖–6,1500} | (2) |
We continue to use the similar two important factors that we identify in previous work,1i.e. flexibility level and flexibility ratio, to describe the vibrational behavior, and they are defined as:
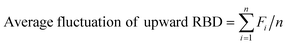 | (3) |
| Fluctuation ratio = mean (F1, F2…Fn)/mean(f1,f2…fm) | (4) |
Epidemiological properties and correlation model
The global confirmed case number and the case fatality rate are updated as of Jul 1, 2022. Korber et al.14 have developed a web tool36 to track mutations of SARS-CoV-2 based on updated GISAID SARS-CoV-2 sequence database. We calculate the portion of D614 on a monthly basis using this tool and evaluate the effective total global confirmed case number for original SARS-CoV-2 with D614 | (5) |
The logarithm of global infection number and the logarithm of case fatality rate in percentage are defined as the infectivity level and lethality level in this paper, ensuring that negative values are also meaningful. When constructing the quantitative correlation model, we investigate the accuracy of polynomial model, mixture model of polynomial and exponential function and exponential model. Given the limited data, only linear dependency is explored, in the form:
| y = a0 + a1f1(x1) + a2f2(x2) | (6) |
Structural analysis
Several important angles and distances are analyzed using VMD. (see Fig. 3(a)) Given the coronavirus spike trimer and the previous finding that the S2 subunit of coronavirus spike protein is quite rigid, we then define the central axis as the perpendicular line that passes the center of the S2 subunit central triangle, which consists of geometrical centers of the S2 subunits in three chains in coronavirus spike protein. The orientation of a certain domain is defined as the orientation of the maximum diameter of the domain, and the angle of a domain is the orientation with respect to the horizontal plane defined by the central axis. The distance of a domain is defined as the distance of the mass center from the central axis, indicating how tightly a domain packs against the central axis. We investigate the influence of various structural factors using a simple linear model:| Y = a0 + a1θRBD-B + a2dRBD-B + a3θRBD-C + a4dRBD-C + a5θNTD-A + a6θNTD-B + a7θNTD-C + a8dNTD | (7) |
Results and discussion
Our study focuses on the spike protein of beta-coronavirus, which is the part that interacts with the human cell receptor and is essential for the infection to take place. In this paper, we mainly study and make predictions for the following SARS-CoV-2 variants: (1) the D614G variant that has quickly become the most prevalent form in the global pandemic since its emergence;14 (2) mink associated mutations including the “Cluster 5” variant identified by Statens Serum Institut (SSI) in Denmark and single spike substitution F486L;19,37 (3) the B.1.1.7 variant (Alpha variant) that was first reported in the UK; (4) the B.1.351 variant (Beta variant) that arose in South Africa, sharing the same mutation N501Y with B.1.1.7 (Alpha variant); (5) the P.1 variant (Gamma variant) that caused widespread infection in Brazil, containing spike substitution E484K and N501Y that present in B.1.351 (Beta variant); (6) the two key mutations that are referred as “double mutant” in B.1.617 variant (parent lineage of Kappa and Delta variant), which emerged in India in late 2020, and also the sub-variant B.1.617.2 (Delta variant).38 (Table 1) Among these variants of interest, the mutation D614G and variants B.1.1.7 (Alpha variant), B.1.351 (Beta variant), P.1 (Gamma variant) and B.1.617.2 (Delta variant) are still circulating globally. In this paper, all the spike mutations are implemented based on the template SARS-CoV-2 spike protein structure in one RBD upward conformation (PDB: 6Z97), so a few mutations are not implemented due to missing regions. Future work may explore these mutations with similar methods as used here.The fluctuation profile of coronavirus spike proteins, obtained through the use of normal mode analysis, reveals the vibrational flexibility of the spike. In previous work, we have shown that while having similar protein structure and sharing the same type of global movement—the swing of upward receptor-binding domain (RBD), MERS-CoV, SARS-CoV and SARS-CoV-2 spike proteins have quite different fluctuation profile.1
Given the small number of spike mutations compared to the total residue number near 3000, will the mutations make a difference to the flexibility profile? The answer is yes. In Fig. 2, we show the flexibility profiles of three notable SARS-CoV-2 variants that are still circulating globally—B.1.1.7, B.1.351 and P.1, identified by WHO as Variants of Concern (VOC) and labelled as Alpha, Beta and Gamma variant respectively. The fluctuation profiles of these three SARS-CoV-2 variants also follow the general characteristics that we discovered for MERS-CoV, SARS-CoV and SARS-CoV-2 spike proteins.
 | ||
| Fig. 2 Flexibility profile and spike mutation spots of the three notable SARS-CoV-2 variants that are still circulating globally—B.1.1.7 (Alpha variant), B.1.351 (Beta variant) and P.1 (Gamma variant). (a) Fluctuation profile of SARS-CoV-2 B.1.1.7 (Alpha variant) spike protein and the flexibility difference between this variant and the original spike protein (PDB ID: 6Z97). The flexibility difference is calculated by subtracting the fluctuation of wild-type virus from that of the mutant. Labels on the top indicate the corresponding subunits S1 and S2 in chain A, B and C. The green labels located in the middle of S1 subunit labels demonstrate the locations of receptor binding domain (RBD). (b) equilibrated structure of SARS-CoV-2 B.1.1.7 (Alpha variant) spike protein, showing only the chain with upward RBD. The upward RBD is colored in green and other parts are colored in cyan. The mutation spots in this chain are colored in red and shown as spheres. The standard deviation (RMSD) that occurs during molecular dynamics (MD) relaxation is about 4.94 Å. (c and d) Fluctuation profile and equilibrated structure of SARS-CoV-2 B.1.351 (Beta variant) spike protein respectively, with RMSD around 4.51 Å during MD relaxation. (e and f) fluctuation profile and equilibrated structure of SARS-CoV-2 P.1 (Gamma variant) spike protein respectively, with RMSD around 3.85 Å during MD relaxation. Although B.1.1.7 (Alpha variant), B.1.351 (Beta variant) and P.1 (Gamma variant), also known as 501Y.V1, 501Y.V2 and 501Y.V3, share the important spike mutation N501Y, their fluctuation profiles behave differently. Compared with B.1.1.7 (Alpha variant) and P.1 (Gamma variant), B.1.351 mutant (Beta variant) has more flexible upward RBD and downward RBD. While spike substitutions generally decrease the mobility of RBDs, several residues in the upward and downward RBDs of B.1.351 (Beta variant) obtain increased flexibility due to mutations. | ||
We find that they all have a much more flexible S1 subunit and a comparatively rigid S2 subunit, with the slowly built-up largest flexibility locating in its upward RBD. Note that the dynamics of stalk hinge (central helix and HR in S2 subunit), which is reported in previous literature,39 is not observed in our analysis because the stalk region is not included in our model. On the other hand, the mutations generally decrease the mobility of N-terminal domains (NTD) and RBDs. No obvious flexibility changes to other regions, which indicates that the rigid regions are pretty stable and retain rigidity. Comparing B.1.1.7 (Alpha variant), B.1.351 (Beta variant) and P.1 (Gamma variant), which also known as 501Y.V1, 501Y.V2 and 501Y.V3 due to the shared N501Y mutation in RBD, there are some differences in their fluctuation profile. In general, the fluctuation difference induced by mutations in B.1.1.7 (Alpha variant) is smaller than the difference due to mutations in B.1.351 (Beta variant) and P.1 (Gamma variant), suggesting B.1.1.7 (Alpha variant) behaves more similar to the original SARS-CoV-2 spike protein. It is also important to note that compared with B.1.1.7 (Alpha variant) and P.1 (Gamma variant), B.1.351 (Beta variant) has larger mobility in both upward and downward RBD. While spike substitutions generally decrease the mobility of RBDs, several residues in the RBD of B.1.351 (Beta variant) obtain increased flexibility due to mutations, showing some specialty of lineage B.1.351 (Beta variant).
We further investigate important angles and distances in the coronavirus spike protein structure to understand the structural parameters that could influence the flexibility level and mobility ratio, the two significant vibrational factors that we identified in previous work.1 According to Fig. 3, among all structural parameters concerned, the factors associated with the upward RBD show a significant positive relationship with the flexibility level. The further upward RBD is from the central axis, the larger its standing angle is, the more “open” the spike conformation is, and thus the higher the vibrational flexibility level is. In terms of the flexibility ratio, the distance of upward RBD and the average distance of NTDs both make significant contributions. While the flexibility ratio increases with the distance of upward RBD, it scales negatively with the average distance of NTDs. Therefore, considering that the flexibility ratio is quantified as the ratio of RBDs in upward and downward conformation, the more tightly the NTDs pack against the central axis, the larger the flexibility ratio is, the more distinct the RBDs in different positions are.
Fig. 4 depicts the quantitative model to predict the infectivity and lethality level of SARS-CoV-2 variants of interest. The nanomechanical vibrational feature map of coronavirus spike proteins and SARS-CoV-2 variants is shown in panel (a). It is demonstrated that generally all variants of concern except B.1.351 (Beta variant) have decreased average fluctuation over upward RBD, which corresponds with the trend we observe with detailed fluctuation profiles in Fig. 2. Compared with the original SARS-CoV-2 spike protein, the variants are moving towards the bottom left side of the diagram, suggesting that their behavior differs even more from the behavior of SARS-CoV and MERS-CoV spike proteins.
 | ||
| Fig. 4 Nanomechanical vibrational feature map of coronavirus spike proteins and SARS-CoV-2 variants, and analyses of correlations with viral infectivity and lethality, including the B.1.617.2 mutant (Delta variant). (a) Nanomechanical vibrational feature map of coronavirus spike proteins and SARS-CoV-2 variants. The largest sphere denotes MERS-CoV S (PDB ID: 5X5F); two middle-size spheres denote SARS-CoV S (PDB ID: 6CRZ and 6CRW); two small spheres denote SARS-CoV-2 S (PDB ID: 6Z97 and 7DDN). Points in diamond shape denote spike proteins of different SARS-CoV-2 variants. (b) Polynomial fitting model linking fluctuation level and ratio to infectivity, where infectivity level is calculated as the logarithm of global confirmed case number. (c) Polynomial fitting model relating fluctuation level and ratio to lethality, where lethality level is calculated as the logarithm of mortality rate in percentage. (d) Nanomechanical vibrational feature map indicating the possibility of more infectious and more lethal variant. Arrow in red indicates the direction where variants are of the same infectivity but more lethal; arrow in magenta indicates the direction where variants are of the same lethality but more infectious. Small area between two arrows is the phase space to generate both more infectious and more lethal variant. (e) Prediction on epidemiological features of SARS-CoV-2 variants. This analysis shows that, generally, most variants of concern are predicted to be more infectious and less lethal than the original SARS-CoV-2. Lineage B.1.351 (Beta variant) and mutant F486L are predicted to be less infectious and more lethal, while lineage B.1.1.7 (Alpha variant) is anticipated to have both higher infectivity and lethality. All containing N501Y substitution in spike, B.1.1.7 (Alpha variant), B.1.351 (Beta variant) and P.1 (Gamma variant) are predicted to have quite different epidemiological features compared to variant with single N501Y mutation. The result suggests that other spike mutations, which have not been discussed in previous literature, also have significant influence on the infectivity and lethality of SARS-CoV-2 variants. The emerging lineage B.1.617.2 (Delta variant), although contains a few key spike mutations other than D614G, behaves quite similar to the single D614G mutation in both vibrational and predicted epidemiological aspects, which might explain its fast circulation given the prevalence of D614G. From panel (d) we can see that the possibility of both more lethal and more infectious variants is quite small for SARS-CoV-2, but it changes with the vibrational features and may become a serious concern when both general flexibility level and fluctuation ratio of the coronavirus spike protein are large. | ||
Aside from the qualitative conclusion drawn from panel (a), we establish our quantitative model to further make predictions about the infectivity and lethality level of SARS-CoV-2 variants of interest based on the relationship observed between vibrational and epidemiological properties.1 As shown in panel (b and c), the polynomial fitting model is able to describe the quantitative relationship appropriately, with correlation coefficient R2 = 0.991 and R2 = 0.998 for infectivity and lethality fitting model respectively. The analysis indicates that most variants of interest are predicted to be more infectious and less lethal than the original SARS-CoV-2. Lineage B.1.351 (Beta variant) and mutant F486L are predicted to be both less infectious and more lethal, while lineage B.1.1.7 (Alpha variant) is anticipated to achieve higher infectivity and higher lethality simultaneously. Our model predictions on lineage B.1.1.7 (Alpha variant) not only correspond with its greater transmissibility reported in previous studies,12 but also agree with the results in literature where analysis on positive SARS-CoV-2 community tests show that B.1.1.7 may lead to increased mortality.40 Within the range of error bars, the qualitative predictions of infectiousness and lethality of most SARS-CoV-2 variants remain the same. The only exception would be mutant F486L, which lies in between the two different performance regions and could result in different qualitative predictions.
Focusing on the mutations associated with mink, we find that single mutation Y453F and cluster 5 variant, which contains Y453F and other two spike substitutions, have similar vibrational behavior and predicted epidemiological properties, and this proves the stability of our model to some extent. However, another mink-associated spike mutation, namely F486L, behaves distinctively from the previous two mutants, which might result from its special location at the receptor binding motif (RBM) of RBD.
Here, we can also further compare the predicted epidemiological characteristics of B.1.1.7 (Alpha variant), B.1.351 (Beta variant), P.1 (Gamma variant) and their shared N501Y spike substitution. It is shown that the four of them behave quite differently, with single mutation N501Y predicted to be the most infectious and the least lethal. The different behavior can be considered as the result of the competition between N501Y mutation and other spike mutations—N501Y pushes the variant to be more infectious and less lethal, and other mutations together drag the variant towards lower infectivity and higher lethality. This analysis result agrees with previous findings revealing that N501Y is associated with increased infectivity and virulence of SARS-CoV-2 in a mouse model.41 Notably, we emphasize here that other spike mutations, which have not been discussed in previous work, could also have significant impact on infectivity and lethality of SARS-CoV-2 variants, providing new aspects for further research on SARS-CoV-2 variants.
Both containing spike substitution in residue 484, B.1.351 (Beta variant) and the “double mutant” variant that has two key mutations in B.1.617 are predicted to have smaller fluctuation ratio than the original SARS-CoV-2 spike protein. Considering the comparatively higher fluctuation ratio of B.1.617.2 (Delta variant), which lacks the E484Q mutation present in the “double mutant”, we suggest that the spike substitution in residue 484 may be associated with lower fluctuation ratio in SARS-CoV-2 spike protein, leading to more similar RBDs in different conformations and larger possibility toward multiple receptor-accessible RBDs.
While all current variants of concern except lineage B.1.1.7 (Alpha variant) show single trend of either higher infectivity and lower lethality or lower infectivity and higher lethality, we demonstrate in panel (d) that that the possibility for the variants to be both more lethal and more infectious could change with different vibrational properties. The possibility is larger for MERS-CoV S compared with that of SARS-CoV S and SARS-CoV-2 S. In the top region of Fig. 4(d), the nonlinearity of epidemiological prediction becomes significant, and thus the likelihood of more lethal and more infectious variants could be quite large. Therefore, dangerous mutations could be a serious concern with future coronavirus whose spike protein has comparatively large flexibility level and fluctuation ratio.
Furthermore, noticing that three of the WHO identified Variants of Concern (VOC), namely B.1.1.7 (Alpha variant), P.1 (Gamma variant) and B.1.617.2 (Delta variant), have similar nanomechanical and epidemiological behavior compared with the single D614G variant, which has quickly become the most prevalent form since it occurred, we suggest that aside from the both more lethal and more infectious variants, there might be some area centering the D614G variant in the nanomechanical vibrational feature map where SARS-CoV-2 variants could be quite dangerous and pose serious threat to global public health.
Conclusions
A predictive model that associates molecular motions and vibrational patterns of the virus spike protein with infectiousness and lethality is constructed and analyzed in this paper. Our findings suggest the possibility to correlate nanomechanical features of spike protein vibration patterns, and how they change due to mutations, with large-scale population-level observation. This is enables us now to explore a variety of mechanistic approaches to understand variants, and the future evolution of variants, based on nanomechanical concepts.As shown in Fig. 4, most variants of concern are predicted to be more infectious and less lethal than the original SARS-CoV-2. Lineage B.1.351 (Beta variant) and mutant F486L are predicted to be less infectious and more lethal, while lineage B.1.1.7 (Alpha variant) is anticipated to have both higher infectivity and lethality. The different behavior of B.1.1.7 (Alpha variant), B.1.351 (Beta variant) and P.1 (Gamma variant) suggests that spike mutations other than N501Y, which hasn’t been discussed in previous literature, also have significant influence on the infectivity and lethality of SARS-CoV-2 variants. Among all variants of interest, the emerging lineage B.1.617.2 (Delta variant), although contains a few key spike mutations other than D614G, behaves quite similar to the single D614G mutation in vibrational feature map and is predicted to have similar infectivity and lethality, which may help explain its fast circulation considering how quickly D614G has become the prevalent form since its occurrence.
For the SARS-CoV-2 virus, its higher infectivity and lethality performance regime appears to be located in the small area above it, as shown in Fig. 4(d). Given the connection between spike mutation at residue 484 and the lower vibrational fluctuation ratio revealed in Fig. 4, we suggest that spike mutations at residue 484 might help drive SARS-CoV-2 variants towards the special performance regime. What's more, the distinct epidemiological predictions found here for B.1.351 (Beta variant) and P.1 (Gamma variant) implies that the combination of spike mutations K417N, E484K and N501Y might be quite dangerous and could possibly lead to higher lethality. Although only lineage B.1.1.7 (Alpha variant), among all variants of interest, is predicted to have higher infectivity and lethality, the possibility of both more lethal and more infectious variants could change with vibrational features and may indeed become a serious concern when both general flexibility level and fluctuation ratio of coronavirus spike protein are large. This can be studied in more detail in future work, perhaps with the combination of advanced protein folding tools, molecular modeling, and experimental assays.
Overall, this work may provide a tool to estimate the epidemiological effects of new variants, offer a pathway to screen mutations against high threat levels and help design possible inhibitors that compensates for the motions induced by mutations. Moreover, the nanomechanical approach, as a novel tool to predict virus-cell interactions, may offer a tool to better understand other viruses. Considering the limitation of implementing mutations based on the current SARS-CoV-2 spike protein templet with several missing regions, future work may analyze certain missing spike mutations with more complete spike protein structures as templets. Other future work may assess such potential mutations from an evolutionary or genetic perspective, to assess the likelihood of their occurrence, or to identify the conditions under which they may emerge.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge support from the MIT-IBM AI Lab, Google Cloud Computing, ONR, and NIH.References
- Y. Hu and M. J. Buehler, Comparative Analysis of Nanomechanical Features of Coronavirus Spike Proteins and Correlation with Lethality and Infection Rate, Matter, 2021, 4(1), 265–275 CrossRef CAS PubMed.
- J. F. W. Chan, S. Yuan and K. H. Kok, et al., A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster, Lancet, 2020, 395(10223), 514–523 CrossRef CAS.
- C. Huang, Y. Wang and X. Li, et al., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet, 2020, 395(10223), 497–506 CrossRef CAS.
- Y. Wan, J. Shang, R. Graham, R. S. Baric and F. Li, Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus, J. Virol., 2020, 94(7), 1–9 CrossRef PubMed.
- COVID-19 Map – Johns Hopkins Coronavirus Resource Center.
- R. Verity, L. C. Okell and I. Dorigatti, et al., Estimates of the severity of coronavirus disease 2019: a model-based analysis, Lancet Infect. Dis., 2020, 20(6), 669–677 CrossRef CAS PubMed.
- D. Wrapp, N. Wang and K. S. Corbett, et al., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science, 2020, 367(6483), 1260–1263 CrossRef CAS PubMed.
- Y. Yuan, D. Cao and Y. Zhang, et al., Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains, Nat. Commun., 2017, 8, 15092 CrossRef CAS PubMed.
- M. Gui, W. Song and H. Zhou, et al., Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding, Cell Res., 2017, 27(1), 119–129 CrossRef CAS PubMed.
- R. N. Kirchdoerfer, C. A. Cottrell and N. Wang, et al., Pre-fusion structure of a human coronavirus spike protein, Nature, 2016, 531(7592), 118–121 CrossRef CAS PubMed.
- R. Yan, Y. Zhang, Y. Li, L. Xia, Y. Guo and Q. Zhou, Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2, Science, 2020, 367(6485), 1444–1448 CrossRef CAS PubMed.
- N. G. Davies, S. Abbott and R. C. Barnard, et al., Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England, Science, 2021, 372(6538), eabg3055 CrossRef CAS PubMed.
- E. Volz, V. Hill and J. T. McCrone, et al., Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity, Cell, 2021, 184(1), 64–75.e11 CrossRef CAS PubMed.
- B. Korber, W. M. Fischer and S. Gnanakaran, et al., Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus, Cell, 2020, 182(4), 812–827.e19 CrossRef CAS PubMed.
- D. J. Benton, A. G. Wrobel and C. Roustan, et al., The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(9), 2–5 CrossRef PubMed.
- J. Zhang, Y. Cai and T. Xiao, et al., Structural impact on SARS-CoV-2 spike protein by D614G substitution, Science, 2021, 372(6541), 525–530 CrossRef CAS PubMed.
- L. Yurkovetskiy, X. Wang and K. E. Pascal, et al., Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant, Cell, 2020, 183(3), 739–751.e8 CrossRef CAS PubMed.
- Y. J. Hou, S. Chiba and P. Halfmann, et al., SARS-CoV-2 D614G variant exhibits enhanced replication ex vivo and earlier transmission in vivo, Science, 2020, 370(6523), 1464–1468 CrossRef CAS PubMed.
- B. B. O. Munnink, R. S. Sikkema and D. F. Nieuwenhuijse, et al., Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans, Science, 2021, 371(6525), 172–177 CrossRef PubMed.
- M. Hoffmann, L. Zhang and N. Krüger, et al., SARS-CoV-2 mutations acquired in mink reduce antibody-mediated neutralization, Cell Rep., 2021, 35(3), 109017 CrossRef CAS PubMed.
- Z. Wang, F. Schmidt and Y. Weisblum, et al., mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants, Nature, 2021, 592(7855), 616–622 CrossRef CAS PubMed.
- J. Chen, R. Wang, M. Wang and G. W. Wei, Mutations Strengthened SARS-CoV-2 Infectivity, J. Mol. Biol., 2020, 432(19), 5212–5226 CrossRef CAS PubMed.
- A. Goyal, D. B. Reeves, E. Fabian Cardozo-Ojeda, J. T. Schiffer and B. T. Mayer, Viral load and contact heterogeneity predict sars-cov-2 transmission and super-spreading events, eLife, 2021, 10, 1–63 CrossRef PubMed.
- E. Pujadas, F. Chaudhry and R. McBride, et al., SARS-CoV-2 viral load predicts COVID-19 mortality, Lancet Respir. Med., 2020, 8(9), 70 CrossRef.
- B. Hie, E. D. Zhong, B. Berger and B. Bryson, Learning the language of viral evolution and escape, Science, 2021, 371(6526), 284–288 CrossRef CAS PubMed.
- H. M. Berman, J. Westbrook and Z. Feng, et al., The Protein Data Bank, Nucleic Acids Res., 2000, 28(1), 235–242 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, VMD: Visual molecular dynamics, J. Mol. Graphics, 1996, 14(1), 33–38 CrossRef CAS PubMed.
- M. T. Nelson, W. Humphrey and A. Gursoy, et al., NAMD: A parallel, object-oriented molecular dynamics program, Int. J. High Perform. Comput. Appl., 1996, 10(4), 251–268 Search PubMed.
- J. Huang and A. D. Mackerell, CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data, J. Comput. Chem., 2013, 34(25), 2135–2145 CrossRef CAS PubMed.
- G. J. Martyna, D. J. Tobias and M. L. Klein, Constant pressure molecular dynamics algorithms, J. Chem. Phys., 1994, 101(5), 4177–4189 CrossRef CAS.
- S. E. Feller, Y. Zhang, R. W. Pastor and B. R. Brooks, Constant pressure molecular dynamics simulation: The Langevin piston method, J. Chem. Phys., 1995, 103(11), 4613–4621 CrossRef CAS.
- K. Hinsen, A. J. Petrescu, S. Dellerue, M. C. Bellissent-Funel and G. R. Kneller, Harmonicity in slow protein dynamics, Chem. Phys., 2000, 261(1-2), 25–37 CrossRef CAS.
- P. Durand, G. Trinquier and Y.-H. Sanejouand, A new approach for determining low-frequency normal modes in macromolecules, Biopolymers, 1994, 34(6), 759–771 CrossRef CAS.
- F. Tama, F. X. Gadea, O. Marques and Y. H. Sanejouand, Building-block approach for determining low-frequency normal modes of macromolecules, Proteins: Struct., Funct., Genet., 2000, 41(1), 1–7 CrossRef CAS.
- X. Q. Yao, L. Skjærven and B. J. Grant, Rapid Characterization of Allosteric Networks with Ensemble Normal Mode Analysis, J. Phys. Chem. B, 2016, 120(33), 8276–8288 CrossRef CAS PubMed.
- COVID-19 Viral Genome Analysis Pipeline.
- L. van Dorp, C. C. S. Tan and S. D. Lam, et al. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2, Nat. Commun., 2020, 11, 5986 Search PubMed.
- P. Mlcochova, S. Kemp and M. S. Dhar, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion, Nature, 2021, 599(7883), 114–119 CrossRef CAS PubMed.
- P. V. Raghuvamsi, N. K. Tulsian and F. Samsudin, et al., SARS-CoV-2 S protein:ACE2 interaction reveals novel allosteric targets, eLife, 2021, 10(M), 1–47 Search PubMed.
- N. G. Davies, C. I. Jarvis and K. van Zandvoort, et al., Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7, Nature, 2021, 593(7858), 270–274 CrossRef CAS PubMed.
- H. Gu, Q. Chen and G. Yang, et al., Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy, Science, 2020, 369(6511), 1603–1607 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d1sm01181b |
| This journal is © The Royal Society of Chemistry 2022 |