Open Access Article
Open Access ArticleStarPEG–heparin biosensors for rapid and portable diagnostics in complex biofluids†
Tim
Thiele
a,
Bergoi
Ibarlucea
 *ab,
Teuku Fawzul
Akbar
c,
Carsten
Werner
*ab,
Teuku Fawzul
Akbar
c,
Carsten
Werner
 c and
Gianaurelio
Cuniberti
*ab
c and
Gianaurelio
Cuniberti
*ab
aInstitute for Materials Science and Max Bergmann Center of Biomaterials, Technische Universität Dresden, Dresden, 01062, Germany. E-mail: bergoi.ibarlucea@tu-dresden.de; gianaurelio.cuniberti@tu-dresden.de
bCenter for advancing electronics Dresden (cfaed), Technische Universität Dresden, Dresden, 01062, Germany
cMax Bergmann Center of Biomaterials, Leibniz Institute of Polymer Research Dresden, Hohe Strasse 6, 01069 Dresden, Germany
First published on 12th April 2022
Abstract
Impedance spectroscopy-based biosensors are typically functionalized following two-dimensional immobilization strategies, with bioreceptors attached through crosslinkers. These methodologies may lead to a decreased receptor activity due to wrong orientation, conformational changes or limited interaction kinetics with the liquid sample. Entrapment of bioreceptors in hydrogels can tackle these issues offering a favourable three-dimensional fluid-like environment, while protecting the electrodes from biofouling in the presence of complex biological fluids at the same time. The star-shaped polyethylene glycol hydrogel doped with heparin (starPEG–heparin) represents a promising candidate, with its excellent hemocompatibility, but its biosensing performance has never been investigated. Here, we show the first demonstration of starPEG–heparin as a biosensor, using antibodies against immunoglobulin G as a model bioreceptor, and we compare it to the performance of other gels with alternative advantages: alginate, which provides easy fabrication and electrode regeneration possibilities, and silicate-based sol–gel, whose porosity can be tuned in a wide range. The starPEG–heparin outperforms the other two, being capable of detecting ultralow antigen concentrations down to the femtomolar levels, implemented in simple photolithography electrodes. We envision its integration in nanomaterial-based sensors which will further improve the sensitivity, and its application in full blood analysis or in implantable devices for in vivo biosensing.
Introduction
The recent outbreak of the pandemic COVID-19 coronavirus disease1 has demonstrated the real need for highly sensitive and fast biosensors to detect diseases at an early stage. In the early phases of the disease, the pathogen concentration is low enough to show no or few symptoms in the host.2 The absence of symptoms could lead to no testing of the individual, followed by a fast spread of the virus. The standard test to confirm infectious diseases such as COVID-19 is the standard polymerase chain reaction (PCR) test.3 Its outcome and communication to the affected individual is rather slow due to the localization in centralized laboratories. Developing new miniaturized and portable diagnostic devices based on alternative techniques would allow taking immediate actions. Such a fast response is ideal to prevent the saturation of medical centers' facilities for critical care in a challenging situation where limited resources and economic constraints are an obstacle.In this context, biosensor development enabled by advances in micro- and nanofabrication4 as well as microfluidics5 has been a growing field for decades. Biosensors relying on transduction mechanisms based on electrical effects (electrochemical, potentiometric, impedimetric, etc.) provide the finest suitability for miniaturization toward portable and point-of-care diagnostics. They require simpler setups where arrays of many sensors can be combined for a label-free and continuous analysis of the analyte of interest.6 Full integration is possible in combination with further circuit packaging for signal processing which require standard electronic microfabrication processes.7
Among the available optics-less sensing formats, impedance spectroscopy8 is endowed with high sensitivity, a label-free nature and excellent miniaturization possibilities. Traditionally, impedance biosensors rely on 2D models. Here bioreceptors are chemically immobilized on electrode surface through a crosslinker molecule. Such an immobilization strategy could lead to a deterioration of receptor activity due to wrong orientation or changes in receptor conformation, as well as increased nonspecific interactions.9 These issues can be overcome by hosting antibodies in hydrogels containing polymers with anti-fouling properties10 by simple entrapment or physisorption. Hydrogels provide a favorable 3D environment, where bioreceptors in large amounts can be immobilized while maintaining their active conformation. The fluid-like environment in a three-dimensional space allows a mobility with much higher interaction probability with the target molecules compared to the limited kinetics of a solid–liquid interface. Beyond simply acting as the host for receptors, the hydrogels can play the additional role of sensor protection. They prevent deterioration by filtering the approaching excess of biological material. This was demonstrated on various challenging applications such as in vivo implantation,11 and full blood12 or sweat analysis.13 Among the existing hydrogels, alginate shows some advantageous characteristics for sensing: (a) localized fabrication via galvanostatic deposition, and (b) facile removal by incubation in calcium chelating buffers, regenerating the transducer. The latter still requires deeper studies. However, PEG finds itself among the most widely used polymers for the fabrication hydrogels with sensing applications.14,15 A particularly interesting PEG-based hydrogel is the star-shaped PEG–heparin (starPEG–heparin). It has shown excellent characteristics for biological and biomedical applications by easily tailoring with additional functional molecules.16,17 More specifically, its functionalization with heparin confers high hemocompatibility to the material,18 making it a promising candidate for full blood analysis or for long-term measurements in vivo. Moreover, the thiol moieties of the used precursors enable its strong attachment on gold electrodes. Its sensing capabilities remain an open topic that needs to be studied. In contrast to most examples, some authors opt for the entrapment within inorganic matrixes such as the sol–gels, where alkoxide precursors are hydrolised, condensated and polymerized.19 The porosity of sol–gels based on silica can be finely tuned20 in such way that a wide variety of species can be entrapped, from large biomolecules such as enzymes21 or antibodies22 to small metal ions, with a certain degree of mobility.23,24 Furthermore, the chemical nature of the polymerization process allows to form covalent bonds with silicon or glass surface for permanent attachment on sensor surfaces. However, research in biosensor performance of sol–gels is scarce.
Here, we propose an evaluation approach in an impedimetric biosensing format (Fig. 1a), aiming at achieving high sensitivity in a miniaturized and portable way. We make use of commercial microelectrodes to enable benchmarking of other surface preparations by the scientific community. For the first time, we provide evidence of the excellent sensing performance of the starPEG–heparin and compare it to the already known alginate and sol–gel matrices. We trapped antibodies against human immunoglobulin G (IgG) as model receptor, which interact with human IgG as target. Such antigen has a similar size to others with interest in the biomedical field such as the spike protein of the SARS-CoV-2. Introducing hydrogels with receptors toward this protein in previously reported rapid SARS-CoV-2 electrical and electrochemical biosensors25–27 that present a more traditional 2D functionalization could help to achieve improved performance.
Results and discussion
Colorimetric confirmation of antibody presence
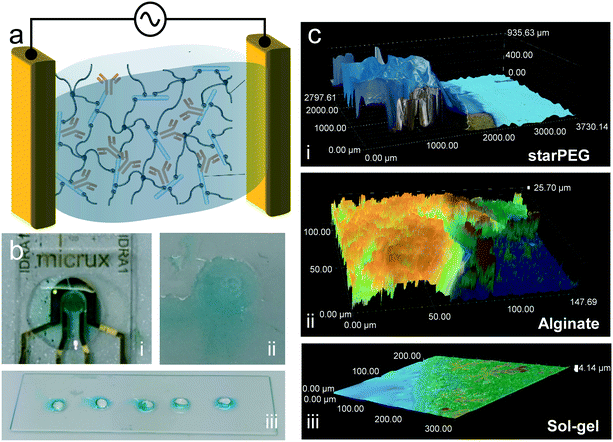
The presence of the antibody in the three tested hydrogels was confirmed by an enzyme-based colorimetric reaction. For this, we trapped antibodies modified with horseradish peroxidase (HRP). This enzyme catalyzes the oxidation of the tetramethylbenzidine substrate solution (TMB), shifting it from colorless to blue. The hydrogels were deposited on glass surfaces without electrodes, with the exception of the alginate, which required the presence of electrodes for its galvanostatic deposition. 5 minutes after drop casting 20 μL of the substrate solution, a clearly visible blue color could be observed in all samples (Fig. 1b), evidencing the presence of the antibody. In the case of starPEG–heparin (Fig. 1b-iii), most of the color develops at the surface of the hydrogel, rather than inside. We attribute this to the faster enzymatic reaction at the surface, where the TMB makes earlier contact with the enyzmes compared to the inside due to the slower diffusion time compared to an aqueous buffer. No color was developed on gels without antibody.Fourier transform infrared spectroscopy (FTIR) confirmation of heparin presence
The presence of heparin in the starPEG gel was confirmed via FTIR analysis. The specific peaks of the sulfate and carboxyl groups are an indicative of its presence. The recorded spectrum is shown in Fig. S1 in the ESI.† Similarly to a previous study of starPEG–heparin,28 we found a prominent absorbance peak at 1247 cm−1, which can be ascribed to the O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S
S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations. The COO– band is shown at 1624 cm−1.
O stretching vibrations. The COO– band is shown at 1624 cm−1.
Hydrogel microscopy characterization
Optical microscopy imaging at consecutive height layers were taken to obtain a three-dimensional profile of the hydrogels (Fig. 1c and S2†). StarPEG–heparin deposition resulted in a thick receptor layer, in the range of 1000 μm, with a well-defined shape due to the use of the cover glass slide during the polymerization process. This thickness could be appropriate for point-of-care diagnostics but may need further optimization for in vivo applications where friction or space limitations may be an issue. In the case of alginate, the chosen electrodeposition conditions resulted in a thinner layer in the range of few tens of micrometers (25–50 μm). The thickness range is comparable to that obtained in our previous work.13 The variability can be attributed to the difficult manipulation due to the viscosity of the precursor mix. However, it is known that varying electrodeposition current density and time can vary its density and thickness,29 and the shape is given by the geometry of the electrodes. The sol–gel formed the thinnest layer (4–15 μm) of the three compared receptor layers due to the fast spreading of the precursor on the hydrophilic glass used as substrate. Branched crystal-like structures were observed, indicating excessive drying of the film during the image acquisition. This could result in increased risk of antibody activity loss if storage conditions are not appropriate until use. The undefined shape of the sol–gel can be avoided by implementing a cuvette to contain the precursor mix, although we expect that the passivation layer of the sensor chip prevents contribution to the signal by the excess gel area.Biosensing response
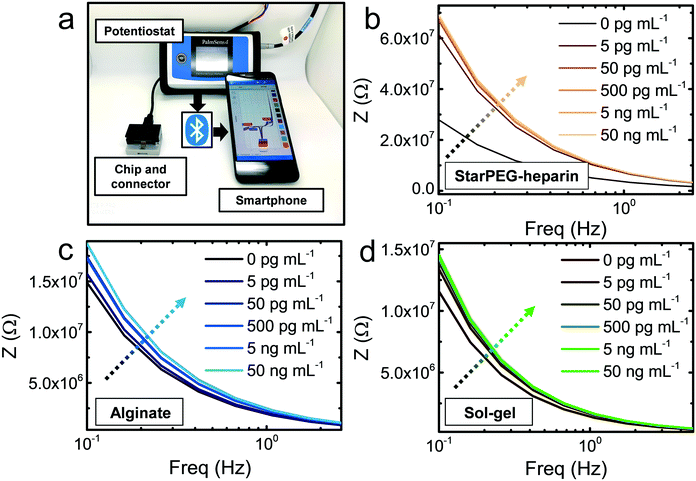
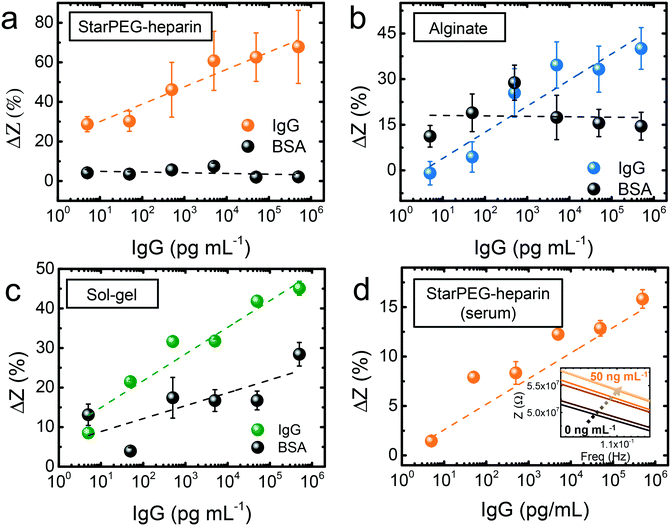
The biosensing approach was fully compatible with portable diagnostics (Fig. 2a). The commercial gold thin film electrodes can be placed in the designated socket of a drop-cell connector with mini-USB to banana connectors for the connection to the potentiostat. The Bluetooth connectivity of the potentiostat allows the wireless operation via smartphone.The hydrogels were deposited on the electrodes, which were subsequently placed on the connector. 5 μL drops of IgG solutions with concentration ranging from 5 pg mL−1 to 50 ng mL−1 were drop casted and incubated for 15 min. After thorough rinsing to remove loosely bound antigen, 5 μL buffer (see Experimental section) were once more drop casted to measure the resulting frequency versus impedance signal. In the three cases (Fig. 2b and c), a growing impedance signal with increasing IgG concentration could be observed in the smallest frequencies (0.1–1 Hz), and the differences decreased at higher frequencies, becoming negligible above 10 Hz. The calibration at the frequency where the strongest signal variations were observed (0.1 Hz) showed that in all cases (Fig. 3a–c) the impedance varied upon incubation of the smallest concentration of 5 pg mL−1. However, control incubations with the nonspecific bovine serum albumin (BSA) evidenced that the starPEG–heparin was the only one capable of preventing nonspecific adsorption through the same whole concentration range. Alginate could only discriminate the IgG from the nonspecific target at concentrations higher than 5 ng mL−1, which can be convenient for other medical applications such as the monitoring of thrombin, where the target is found at higher concentrations.30 The discrimination with the sol–gel was possible at 50 pg mL−1, however the impedance after BSA incubation was 10-fold higher more than with the starPEG–heparin, making the starPEG–heparin the most reliable of the three hydrogels when it comes to prevent nonspecific adsorption.
In view of the results, we proceeded to test the performance of the starPEG–heparin-based sensors in real samples, fetal bovine serum spiked with human IgG. The impedance signal increased once more through the whole tested concentration range (Fig. 3d), being possible to observe the signal change from the smallest tested concentration (5 pg mL−1). The impedance change was smaller compared to the measurements in buffer, which may be due to the decreased probability of interaction in such a complex medium where many other molecules can interact with both antigen and antibody. Considering an estimated size of 150 kDa for the IgG, the biosensor could detect IgG in an equivalent concentration of 33 fM in both buffer and serum samples.
The results are at the level of recently reported ultrasensitive and label-free electrical biosensors.31,32 Some examples outperform our results.8 However, the key element providing the high sensitivity is typically a nanomaterial as part of the transducer. Our approach demonstrates an excellent detection capability without the need of nanomaterials or signal amplification techniques. Rather than compete with other nanoscopic transducers such as metallic33 or semiconductor nanowires,34 their synergy with the hydrogel technology could boost the results to subfemtomolar detection limits.
While we expect that antibodies are entrapped or physisorbed in the starPEG–heparin, the possibility of covalent bonds still exists in case of free cysteine residues in the antibody sequence. Ionic interactions can also occur with the negative charges of the heparin if there are repeated positively charged amino acids (lysine and arginine). Hydrogen bonds are possible as well with the presence of asparagine, glutamine, and histidine.35 The knowledge on the sequence of antibodies and bioreceptors in general is therefore important to choose the most appropriate hydrogel fabrication approach, or otherwise to find the most appropriate receptors. This would help minimizing possible effects that could limit the activity of the bioreceptors.
Electrode regeneration
Although the starPEG–heparin outperformed the alginate and the sol–gel in sensitivity terms, alginate can be advantageous in certain scenarios. The sensing response of the alginate-modified biosensor was reliable at concentrations higher than 5 ng mL−1. This is still a sensitive approach for applications such as thrombin monitoring. Alginate is a suitable material for sweat analysis,13 given the slightly acidic nature of such biofluid. The low pH ensures that the calcium ions will remain crosslinking the alginic acid. Certain important analytes such as glucose or lactate can be found at high concentrations in sweat or other biofluids, appropriate for the sensitivity of alginate-based biosensors.12 One of the most interesting features of this material is the possibility of electrode regeneration in an easy way. In order to achieve this goal, the calcium ions that crosslink the alginic acid monomers can be removed, hence dissolving the gel. Previous works claimed the success of the regeneration by simply incubating the sensors in a buffer like phosphate buffered saline (PBS).13,29 In spite of that, PBS is not an intrinsic calcium chelating agent. It is known that calcium could precipitate in phosphate containing buffers,36 leading to an imperfect regeneration that could prevent from multiple regeneration steps. A simple optical microscopy observation can lead to the false assumption that the alginate film is removed, while a submicrometer layer of hydrogel or calcium precipitates on the electrode surface may still be present. Such layer may affect the future use of electrodes. Thus, we proceeded to carry out the confirmation by observing the regeneration of the impedance signal itself as a more reliable indicator.The alginate deposition resulted in an overall decreased impedance of the system. Incubation for 30 min in PBS in an orbital shaker helped to remove partially the alginate, and the signal was not fully recovered (Fig. 4a). When PBS contained 0.5 m EDTA, the same incubation period resulted in a full recovery of the impedance signal (Fig. 4b), evidencing full regeneration of the electrodes as well. We suggest that the EDTA helped to chelate the calcium ions that crosslink the alginic acid, and will serve for improved reusability of the electrodes in the future. This preliminary findings indicate that alginate deserves a further attention concerning its reusability possibilities, with further reformation, sensing, and regeneration steps to study the limitations of the protocols.
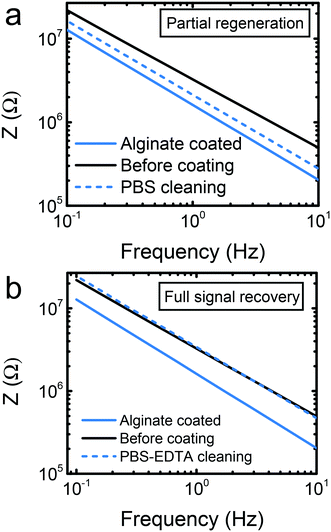 | ||
| Fig. 4 Electrode recovery test. (a) Partial recovery of the signal by incubation in PBS. (b) Full recovery of the signal by incubation in PBS with EDTA. | ||
On the contrary, starPEG–heparin and silicate sol–gel form covalent bonds with the gold electrodes and with the glass support. This makes electrode regeneration more difficult, requiring treatments such as plasma cleaning or immersion in piranha solutions. The first one is not compatible with the idea of a miniaturized and portable point-of-care device, while the second one could harm the passivation layer. However, the antigen may be separated from the antibody by immersing in acidic buffers complemented with glycerol.37 This would restore the activity of the antibody without requiring the repetition of the coating.
Conclusions
We present the first demonstration of the starPEG–heparin hydrogel as sensitive layer of biosensors. We compare it with two materials, alginate and sol–gel, for the noncovalent entrapment of antibodies for IgG detection by impedance spectroscopy. Commercial electrodes were used, enabling future benchmarking of hydrogels developed by any work group without introducing variables related to the transducers. The starPEG–heparin outperforms the others by detecting smaller antigen concentrations down to femtomolar levels (5 pg mL−1) as well as in terms of prevention of the nonspecific adsorption. The hydrogel maintains its sensing capabilities with real samples, as tested with serum. The size of the used antigen as model (human IgG in this work) is relatively similar to that of the spike protein of SARS-CoV-2 and, given the compatibility of the setup with fully portable smartphone diagnostics, we predict that the proposed biosensing approach can play an important role as ubiquitous rapid testing approach in pandemic situations. Currently commercial rapid antigen tests are based on the well-known paper-based fluidics.38,39 Although as downside electrochemical sensors require a specialized reader, quantification is possible in a less complex setup easier to miniaturize, as shown recently.25The hemocompatible properties of the starPEG–heparin gel can be fully exploited in the future in full blood analysis or in vivo measurements. The application of the coating to nanomaterial-containing transducers may further boost the detection capabilities. While alginate and the sol–gel can still be valid for applications where the target molecules are found at higher concentrations, at the smallest ones the signal change may be screened by variations of nonspecific species. However, the alginate can be benefited from the easy removal for electrode regeneration by incubation in a buffer complemented with a calcium chelating agent. The tendency to dry shown by the sol–gel material suggests that further optimization of its fabrication is needed. The use of protocols which result in dry but more stable coatings can host non-biological material.24 This can serve in the future for scenarios such as non-enzymatic sensing.
Experimental
Hydrogel preparation
All reagents were from Sigma Aldrich, Germany, unless otherwise stated.StarPEG–heparin was fabricated by employing Michael-type addition reaction adapted from the previously reported protocol.40 Briefly, thiol terminated 4-arm star polyethylene glycol (starPEG-SH, MW ≈ 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Polymer Source, Inc., Canada) and maleimide conjugated heparin (MW ≈ 15
000, Polymer Source, Inc., Canada) and maleimide conjugated heparin (MW ≈ 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, synthesized in-house following a previous method)40 were dissolved in PBS pH 7.4. Both solutions were put in an ice bath and quickly mixed together with anti-IgG to achieve anti-IgG concentration of 0.1 mg mL−1. The final concentration of both starPEG and heparin were 1.5 μmol mL−1 after mixing. 5 μL of the overall mix was drop casted on the sensor chip and left to polymerize for 1 hour under a glass coverslip to obtain a homogeneous surface coating.
000, synthesized in-house following a previous method)40 were dissolved in PBS pH 7.4. Both solutions were put in an ice bath and quickly mixed together with anti-IgG to achieve anti-IgG concentration of 0.1 mg mL−1. The final concentration of both starPEG and heparin were 1.5 μmol mL−1 after mixing. 5 μL of the overall mix was drop casted on the sensor chip and left to polymerize for 1 hour under a glass coverslip to obtain a homogeneous surface coating.
For the alginate preparation, we followed a galvanostatic deposition process already detailed in a previous work.13 Briefly, a precursor mix with alginic acid (1%), calcium carbonate (0.5%) and antibodies (0.1 mg mL−1) was prepared. 5 μL of the mix were drop casted on the sensor chip. An applied current density of 1.25 A m−2 initiated a water splitting process releasing protons which acidified the vicinity of the working electrode, dissolving the calcium carbonate and producing calcium ions. These ions crosslinked the alginic acid forming the hydrogel with trapped antibodies. After 2 min of the electrodeposition process, the hydrogel was rinsed with NaCl (145 mm) and imidazole buffer (10 mM, pH 7.4) supplemented with KCl (145 mm) and CaCl2 (10 mm) to avoid loss of Ca2+ ion and the subsequent dissolution of the gel.
Sol–gel was fabricated following a previously reported methodology.20 Tetramethoxysilane (TMOS) was used as silicate precursor and it was mixed with HCl (2.5 mm) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 ratio and in presence of 10% PEG (molecular weight 8 kDa). The mixture was sonicated for 30 min and mixed afterwards with HEPES buffer (pH 7.6) in a 1
8 ratio and in presence of 10% PEG (molecular weight 8 kDa). The mixture was sonicated for 30 min and mixed afterwards with HEPES buffer (pH 7.6) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio containing antibodies. 5 μL of the final mixture, where the total antibody concentration was 0.1 mg mL−1, were deposited on the sensor chip and left to gelate for 30 min under humid atmosphere to prevent excessive drying. The gel was finally rinsed with HEPES buffer.
1 ratio containing antibodies. 5 μL of the final mixture, where the total antibody concentration was 0.1 mg mL−1, were deposited on the sensor chip and left to gelate for 30 min under humid atmosphere to prevent excessive drying. The gel was finally rinsed with HEPES buffer.
Hydrogel characterization
The three materials tested in this work were thoroughly characterized in the past. The mesh size of the starPEG–heparin is calculated to be ca. 30 nm.28 Alginate and sol–gel with PEG have also been well studied in the past and they are known to form networks where protein-based bioreceptors can be trapped for biosensing, as also demonstrated in various works.12,20,29 Here, we proceeded to confirm the entrapment of the antibodies as well as the presence of the crosslinked heparin in the starPEG–heparin gel. Finally, we estimated the thickness of the formed coatings. For the colorimetric confirmation of antibody presence, the same procedure above was followed for hydrogel fabrication on glass slides, avoiding unnecessary waste of electrodes, with the exception of the alginate whose fabrication requires them for the galvanostatic deposition. HRP-modified antibodies were used in the mixture instead of the regular antibodies. After rinsing the hydrogels, 5 μL of the TMB substrate solution were drop casted and the color change was observed during 5 min. The FTIR analysis was done in the attenuated total reflection (ATR) mode using an IFS Equinox 55 from Bruker Optik GmbH. For the 3D profiling via optical microscopy, a Keyence VHX-7000 digital microscope was used.Impedance measurements
Commercial electrodes with thin films of gold were used as basic sensor chips (ED-SE1-Au, MicruX Technologies, Spain). The chips modified with hydrogels were placed in the drop cell connector (MicruX Technologies, Spain) and plugged to the potentiostat (Palmsens 4, Palmsens BV, The Netherlands). The wireless connectivity was possible through the PStouch Android app (Palmsens BV, The Netherlands). Impedance was measured at an applied AC signal of 10 mV, sweeping the frequency from 0.1 Hz to 100 kHz. Calibrations in buffer were done using PBS except for alginate, which was done in the previously described imidazole buffer to prevent hydrogel dissolution. 5 μL of each IgG concentration was incubated for 15 min under humid atmosphere to prevent excessive drying, followed by rinsing with buffer and drop casting of new buffer for the measurement obtaining the Bode plots.Author contributions
T. T. and B. I. contributed equally to the work. T. T. modified the sensor surface with hydrogels and performed biosensing experiments under the supervision of B. I. B. I. analysed the resulting data and prepared the figures. T. F. A. assisted T. T. with sensor modification and characterization. C. W. and G. C. coordinated the work and carried out the project management work. B. I. wrote the manuscript with input from all coauthors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the Sächsische AufbauBank project 100525920.References
- World Health Organization, Situation Report-29 SITUATION IN NUMBERS total and new cases in last 24 hours, https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_8, (accessed 12 March 2020).
- J. M. Sharfstein, S. J. Becker and M. M. Mello, JAMA, 2020, 323(15), 1437–1438 CrossRef CAS PubMed.
- R. Service, Science, 2020, 1–10 Search PubMed.
- P. M. Kosaka, V. Pini, J. J. Ruz, R. A. Da Silva, M. U. González, D. Ramos, M. Calleja and J. Tamayo, Nat. Nanotechnol., 2014, 9, 1047–1053 CrossRef CAS PubMed.
- B. Ibarlucea, X. Munoz-Berbel, P. Ortiz, S. Büttgenbach, C. Fernández-Sánchez and A. Llobera, Sens. Actuators, B, 2016, 237, 16–23 CrossRef CAS.
- J. M. Rothberg, W. Hinz, T. M. Rearick, J. Schultz, W. Mileski, M. Davey, J. H. Leamon, K. Johnson, M. J. Milgrew, M. Edwards, J. Hoon, J. F. Simons, D. Marran, J. W. Myers, J. F. Davidson, A. Branting, J. R. Nobile, B. P. Puc, D. Light, T. A. Clark, M. Huber, J. T. Branciforte, I. B. Stoner, S. E. Cawley, M. Lyons, Y. Fu, N. Homer, M. Sedova, X. Miao, B. Reed, J. Sabina, E. Feierstein, M. Schorn, M. Alanjary, E. Dimalanta, D. Dressman, R. Kasinskas, T. Sokolsky, J. A. Fidanza, E. Namsaraev, K. J. McKernan, A. Williams, G. T. Roth and J. Bustillo, Nature, 2011, 475, 348–352 CrossRef CAS PubMed.
- X. Luo and J. J. Davis, Chem. Soc. Rev., 2013, 42, 5944–5962 RSC.
- M. Medina-Sanchez, B. Ibarlucea, N. Perez, D. D. Karnaushenko, S. M. Weiz, L. Baraban, G. Cuniberti and O. G. Schmidt, Nano Lett., 2016, 16, 4288–4296 CrossRef CAS PubMed.
- R. Randriantsilefisoa, J. L. Cuellar-Camacho, M. S. Chowdhury, P. Dey, U. Schedler and R. Haag, J. Mater. Chem. B, 2019, 7, 3220–3231 RSC.
- A. Herrmann, R. Haag and U. Schedler, Adv. Healthcare Mater., 2021, 10, 2100062 CrossRef CAS PubMed.
- Y. J. Heo, H. Shibata, T. Okitsu, T. Kawanishi and S. Takeuchi, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 13399–13403 CrossRef CAS PubMed.
- A. Márquez, J. Aymerich, M. Dei, R. Rodríguez-Rodríguez, M. Vázquez-Carrera, J. Pizarro-Delgado, P. Giménez-Gómez, Á. Merlos, L. Terés, F. Serra-Graells, C. Jiménez-Jorquera, C. Domínguez and X. Muñoz-Berbel, Biosens. Bioelectron., 2019, 136, 38–46 CrossRef PubMed.
- B. Ibarlucea, A. Pérez Roig, D. Belyaev, L. Baraban and G. Cuniberti, Microchim. Acta, 2020, 187, 520 CrossRef CAS PubMed.
- X. Dai, R. Vo, H. H. Hsu, P. Deng, Y. Zhang and X. Jiang, Nano Lett., 2019, 19, 6658–6664 CrossRef CAS PubMed.
- H. H. Bay, R. Vo, X. Dai, H. H. Hsu, Z. Mo, S. Cao, W. Li, F. G. Omenetto and X. Jiang, Nano Lett., 2019, 19, 2620–2626 CrossRef CAS PubMed.
- U. Freudenberg, A. Hermann, P. B. Welzel, K. Stirl, S. C. Schwarz, M. Grimmer, A. Zieris, W. Panyanuwat, S. Zschoche, D. Meinhold, A. Storch and C. Werner, Biomaterials, 2009, 30, 5049–5060 CrossRef CAS PubMed.
- A. Zieris, S. Prokoph, K. R. Levental, P. B. Welzel, M. Grimmer, U. Freudenberg and C. Werner, Biomaterials, 2010, 31, 7985–7994 CrossRef CAS PubMed.
- C. Johnbosco, S. Zschoche, M. Nitschke, D. Hahn, C. Werner and M. F. Maitz, Mater. Sci. Eng., C, 2021, 128, 112268 CrossRef CAS PubMed.
- T. R. Besanger and J. D. Brennan, Anal. Chem., 2003, 75, 1094–1101 CrossRef CAS PubMed.
- M. Shalev and A. Miriam, Materials, 2011, 4, 469–486 CrossRef CAS PubMed.
- S. Braun, S. Rappoport, R. Zusman, D. Avnir and M. Ottolenghi, Mater. Lett., 1990, 10, 1–5 CrossRef CAS.
- M. Altstein and A. Bronshtein, Immunoass. Other Bioanal. Tech., 2006, pp. 357–384 Search PubMed.
- K. Nych, E. Baek, C. K. Baek, B. Ibarlucea, L. Baraban and G. Cuniberti, in 2019 26th IEEE International Conference on Electronics, Circuits and Systems, ICECS, 2019, vol. 2019, pp. 795–798 Search PubMed.
- E. Baek, N. R. Das, C. V. Cannistraci, T. Rim, G. S. C. Bermúdez, K. Nych, H. Cho, K. Kim, C. K. Baek, D. Makarov, R. Tetzlaff, L. Chua, L. Baraban and G. Cuniberti, Nat. Electron., 2020, 3, 398–408 CrossRef CAS.
- P. Lasserre, B. Balansethupathy, V. J. Vezza, A. Butterworth, A. Macdonald, E. O. Blair, L. McAteer, S. Hannah, A. C. Ward, P. A. Hoskisson, A. Longmuir, S. Setford, E. C. W. Farmer, M. E. Murphy, H. Flynn and D. K. Corrigan, Anal. Chem., 2022, 94, 2126–2133 CrossRef CAS PubMed.
- G. Seo, G. Lee, M. J. Kim, S. H. Baek, M. Choi, K. B. Ku, C. S. Lee, S. Jun, D. Park, H. G. Kim, S. I. S. J. Kim, J. O. Lee, B. T. Kim, E. C. Park and S. I. S. J. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- R. M. Torrente-Rodríguez, H. Lukas, J. Tu, C. Xu, H. B. Rossiter and W. Gao, Matter, 2020, 3(6), 1981–1998 CrossRef PubMed.
- A. Zieris, R. Dockhorn, A. Röhrich, R. Zimmermann, M. Müller, P. B. Welzel, M. V. Tsurkan, J. U. Sommer, U. Freudenberg and C. Werner, Biomacromolecules, 2014, 15, 4439–4446 CrossRef CAS PubMed.
- A. Márquez, C. Jiménez-Jorquera, C. Domínguez and X. Muñoz-Berbel, Biosens. Bioelectron., 2017, 97, 136–142 CrossRef PubMed.
- B. Ibarlucea, L. Römhildt, F. Zörgiebel, S. Pregl, M. Vahdatzadeh, W. M. Weber, T. Mikolajick, J. Opitz, L. Baraban, G. Cuniberti, B. Ibarlucea, L. Römhildt, F. Zörgiebel, S. Pregl, M. Vahdatzadeh, W. M. Weber, T. Mikolajick, J. Opitz, L. Baraban and G. Cuniberti, Appl. Sci., 2018, 8, 950 CrossRef.
- B. Ibarlucea, T. F. Akbar, K. Kim, T. Rim, C.-K. C.-K. C.-K. K. Baek, A. Ascoli, R. Tetzlaff, L. Baraban, G. Cuniberti, T. Fawzul Akbar, K. Kim, T. Rim, C.-K. C.-K. C.-K. K. Baek, A. Ascoli, R. Tetzlaff, L. Baraban and G. Cuniberti, Nano Res., 2018, 11, 1057–1068 CrossRef CAS.
- R. Akter, B. Jeong, Y. M. Lee, J. S. Choi and M. A. Rahman, Biosens. Bioelectron., 2017, 91, 637–643 CrossRef CAS PubMed.
- J. Schuett, D. I. Sandoval Bojorquez, E. Avitabile, E. Sergio, O. Mata, G. Milyukov, J. Colditz, L. G. Delogu, M. Rauner, A. Feldmann, S. Koristka, J. M. Middeke, K. Sockel, J. Fassbender, M. Bachmann, M. Bornhauser, G. Cuniberti and L. Baraban, Nano Lett., 2020, 20(9), 6572–6581 CrossRef CAS PubMed.
- D. Karnaushenko, B. Ibarlucea, S. Lee, G. Lin, L. Baraban, S. Pregl, M. Melzer, D. Makarov, W. M. Weber, T. Mikolajick, O. G. O. G. Schmidt and G. Cuniberti, Adv. Healthcare Mater., 2015, 4, 1517–1525 CrossRef CAS PubMed.
- D. Xu and J. D. Esko, Annu. Rev. Biochem., 2014, 83, 129–157 CrossRef CAS PubMed.
- M. C. Larson, M. R. Luthi, N. Hogg and C. A. Hillery, Cytometry, Part A, 2013, 83, 242–250 CrossRef PubMed.
- P. Zhang, S. Yang, R. Pineda-Gómez, B. Ibarlucea, J. Ma, M. R. Lohe, T. F. Akbar, L. Baraban, G. Cuniberti and X. Feng, Small, 2019, 15, 1901265 CrossRef PubMed.
- W. W. W. Hsiao, T. N. Le, D. M. Pham, H. H. Ko, H. C. Chang, C. C. Lee, N. Sharma, C. K. Lee and W. H. Chiang, Biosensors, 2021, 11, 295 CrossRef CAS PubMed.
- B. Jin, Z. Li, G. Zhao, J. Ji, J. Chen, Y. Yang and R. Xu, Anal. Chim. Acta, 2022, 1192, 339388 CrossRef CAS PubMed.
- M. V. Tsurkan, K. Chwalek, S. Prokoph, A. Zieris, K. R. Levental, U. Freudenberg and C. Werner, Adv. Mater., 2013, 25, 2606–2610 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: FTIR spectrum of the starPEG–heparin. See DOI: https://doi.org/10.1039/d2sd00036a |
| This journal is © The Royal Society of Chemistry 2022 |