Open Access Article
Open Access ArticlePlasmonic nanometal surface energy transfer-based dual excitation biosensing of pathogens†
Milad
Torabfam
 a,
Hasan
Kurt
bcd,
Mustafa Kemal
Bayazıt
a,
Hasan
Kurt
bcd,
Mustafa Kemal
Bayazıt
 e and
Meral
Yüce
e and
Meral
Yüce
 *e
*e
aSabanci University, Faculty of Engineering and Natural Sciences, 34956, Istanbul, Turkey
bIstanbul Medipol University, School of Engineering and Natural Sciences, 34810, Beykoz, Istanbul, Turkey
cIstanbul Medipol University, Research Institute for Health Sciences and Technologies (SABITA), 34810, Beykoz, Istanbul, Turkey
dNanosolar Plasmonics Ltd., 41400, Gebze, Kocaeli, Turkey
eSabanci University, SUNUM Nanotechnology Research and Application Centre, 34956, Istanbul, Turkey. E-mail: meralyuce@sabanciuniv.edu
First published on 9th May 2022
Abstract
In this assay, the simultaneous screening of foodborne bacterial pathogens, namely Escherichia coli and Salmonella typhimurium, was investigated by developing a highly specific dual-excitation biosensor which works based on plasmonic nanoparticle surface energy transfer (PSET) between aptamer capped plasmonic gold nanostructures (AuNSs) as capture probes and luminescent nanoparticles (LNPs) as signal probes labeled with complementary single-strand DNA of the utilized aptamers. For the characterization of the provided sensing probes, techniques such as UV-visible spectroscopy, dynamic light scattering, scanning electron microscopy, and circular dichroism spectroscopy were used. While CdSe/ZnS core/shell quantum dots (QDs) became excited with ultraviolet (UV) radiation at 350 nm, the light source utilized for excitation of NaYF4:Yb,Er upconverting nanoparticles (UCNPs) was near-infrared (NIR) at 980 nm, and also the signal cross-talk possibility between QDs and UCNPs was removed using the dual-excitation technique. The limit of detection (LOD) was calculated to be as low as 7.38 and 9.31 CFU mL−1 for simultaneous monitoring of E. coli and S. typhimurium in one experimental batch. The biosensor was also evaluated for detecting bacteria simultaneously in actual lake samples. The results proposed the viability of the technique for real-time sample analysis. Using numerous AuNSs and their corresponding UCNPs and QDs benefiting from distinct luminescence emission profiles, the suggested NSET-based biosensor may be utilized to simultaneously detect a wide range of analytes, posing good application prospects in various fields ranging from food safety analysis to biomedical applications.
Introduction
Biosensors that use fluorescence resonance energy transfer (FRET) for monitoring targets stand out among detection assays owing to their features, such as high sensitivity and selectivity, and being labor-saving.1–4 Although FRET, which is affected by many factors including the orientation of pairs and the distance between acceptor and donor species, is a highly suitable technique for sensing, the nature of dipole–dipole interaction restricts FRET to a maximum separation distance of 80 Å.5–8 Since most of biological interactions happen in distances beyond 100 Å, nano surface energy transfer (NSET) as a longer-distance compatible alternative energy transfer (ET) method is imperative for biosensing.9–11 NSET includes non-radiative ET benefiting from dipole–dipole interaction.10,12 Acceptor metallic nanoparticles (MNPs) with suitable Fermi levels and favorable surfaces can facilitate collecting such dipoles and provide additional energy oscillations such as surface plasmon polaritons with high absorption cross-sections resulting in a higher ET efficiency.9,13 NSET is solely based on the interaction between the electromagnetic field of the donor dipole and free conduction electrons of the acceptor MNPs.8,14 To date, NSET has been utilized in numerous applications such as bioimaging15 and spectroscopic analysis.16QDs are highly stable against photo-bleaching and their small size leads to the confinement of their energy levels.17 They also benefit from improved brightness and high signal-to-noise ratio.17–19 Furthermore, size-dependent and relatively narrow emission and broad absorption spectra make QDs outstanding donors in sensing.10 Also, producing core/shell QDs solved the toxicity caused by core QDs such as cadmium and improved optical features including an enhancement of quantum yield and improvement in resistance against photodegradation.20 On the other hand, lanthanide-doped UCNPs possess an anti-Stokes shift.21 Thanks to this feature, they can emit light from the NIR region to the UV-visible region.22 Since autofluorescence of biomaterials is low in the NIR region, the design of a detection system for monitoring biological materials using UCNPs will be more desirable due to the enhancement of the signal-to-background noise ratio.23,24 Besides exhibiting a longer luminescence lifetime, the optical features of UCNPs are adjustable by changing the lanthanide dopants. These features bring LNPs to the center of attention for multiple target detection.
In the case of NSET acceptors, properties such as high resistance against photobleaching, sufficient binding to biological molecules, broad absorption spectrum, and tunable optical features motivate the growing appearance of AuNSs in ET-based detections.25,26 The usage of sphere NPs in fluorescence sensors suffers from a lack of effective absorption in the electromagnetic spectrum's red region. However, the usage of gold nanorods (AuNRs) and gold nanourchins (AuNUs) overcomes this challenge since diverse forms of AuNSs lead to different absorption spectra.27 Regarding AuNRs, modifying the aspect ratio leads to tuning the longitudinal SPR from the visible region to NIR which contributes to the improvement of the quenching efficiency.28 In the case of the SPR band, the absorption cross-section is higher for AuNRs compared with that for AuNPs.29 Regarding AuNUs, the spiky and rough surface leads to a redshift in the SPR absorption peak compared with sphere AuNPs, which opens the way for effective quenching of LNPs.30 In parallel, a redshift can be noticed in AuNRs with a comparatively higher surface area and adequate mass transfer features, making them excellent candidates for ET systems.31
Over the past few years, AuNPs have been widely utilized in developing ET-based systems. For instance, the AuNR–oligonucleotide hybrid nanocomplexes with dye established by Fu et al.32 exhibited a considerably higher fluorescence emission than pure oligonucleotide-modified dye, which signifies the high effectiveness of the NSET method. In another experiment, Darbha et al.33 designed an NSET-based detection platform for sensing mercury in water and soil through fluorescence quenching of rhodamine B (RhB) with modified gold NPs. The binding of RhB as a donor to the surface of gold NPs led to a strong fluorescence quenching efficiency. In another work, an NSET-based sensor between AuNRs and fluorescent tetrakis(4-sulfophenyl)porphyrin (TPPS4) as an NSET pair was structured for detection of heparin.34 Table S1† also summarizes NSET-based detection platforms for monitoring targets in the past few years. Moreover, an extensive explanation of the NSET assay can be found in the recent review articles by Chen and Hildebrandt et al.12
Considering the advantages of LNPs over fluorophores along with the significance of donor/acceptor spectral overlap in ET-based conjugations, UCNPs and QDs were chosen as a donor to form donor/acceptor pairs for NSET with AuNUs and AuNRs, respectively. Another dominant aspect of our constructed detection system is the usage of aptamers due to their high capability in binding to their complementary oligonucleotides in the absence of a target and their high affinity to target bacteria in the presence of targets, which improves the sensitivity and selectivity of biosensors.35 To the best of our knowledge, a dual excitation NSET-based aptasensor has been developed for the first time for sensitive, rapid, and simultaneous sensing of two targets in a single test tube, and bacteria such as S. typhimurium and E. coli were chosen as targets to investigate the efficiency of the provided platform. Also, the improved detection technique was tested on artificially spiked samples to evaluate its effectiveness for real sample measurements. Considering the robustness, selectivity, and cost-effectiveness of the provided multicolor sensing probes, they can be ideal substitutes for previous conventional techniques.
Materials and methods
Reagents
Carboxylic acid-functionalized erbium-doped UCNPs (NaYF4:Yb,Er; main λems = 545 nm) and CdSe/ZnS core/shell QDs (λems = 620 nm) were obtained from Ocean NanoTech (San Diego, CA, USA). N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide sodium salt (sulpho-NHS) were purchased from Sigma-Aldrich Chemie GmbH (Germany). Carboxylic acid-functionalized AuNRs and AuNUs were obtained from Cytodiagnostics (Canada).Emission and absorption spectra of both QDs and UCNPs are shown in ESI† Fig. S1. Similarly, emission/absorption spectra of Er-doped UCNPs/AuNUs and QDs/AuNRs as NSET pairs are provided in ESI† Fig. S2.
Escherichia coli O157:H7 (ATCC® 25922™) and Staphylococcus aureus (ATCC® 14028™) were obtained from the American Type Culture Collection (ATCC, USA). The single-stranded DNA aptamers specific to E. coli O157:H7 and S. typhimurium and their partially complementary single-stranded DNA oligomers (cDNAs) 5′-NH2-C6 modification were obtained from Integrated DNA Technologies (IDT DNA, USA), and based on research performed by Joshi et al.36 and Wu et al.,37 the sequences of DNA presented in Table S2† are chosen for Salmonella typhimurium and Escherichia coli, respectively. A DNase footprint assay was performed by Joshi et al.36 to evaluate the sensitivity and binding affinity of the aptamer to S. typhimurium which indicates a high affinity of the aptamer. Compared with primer regions, the binding of the aptamer to bacteria in the randomized region is significantly higher. Also, the aptamer selected by Wu et al.37 (mentioned in Table S2†) was developed based on the ATACGGGAGCCAACACCATTCTATCGTTCCGGACGCTTATGCCTTGCCATCTACAGAGCAGGTGTGACGGAT sequence which has the ability to bind to lipopolysaccharides of E. coli. Regarding the preparation of complementary DNA for the abovementioned aptamers, under conditions where the bases match with all of the bases in the aptamer, the detachment of the aptamer from cDNA and binding to the target will be almost impossible. As a result, the bases of cDNA pair with ones related to the aptamer partially. The complementary sequences in the aptamer are also highlighted in Table S2.† Considering the fact that the most important rule for the formation of hydrogen bonds between two oligonucleotides is their complementarity to each other, each aptamer modified AuNS will only form a conjugation with a LNP which has a complementary oligonucleotide.
Preparation of NSET based target-specific nanoprobes
The functionalization of each carboxylic acid-functionalized NP with a corresponding amine-functionalized aptamer is realized using EDC-NHS based covalent linkage.38 A 2-(N-morpholino)ethanesulfonic acid buffer (MES, 25 mM, and pH 6.0) was used to prepare a solution of each NP species at a concentration of 0.25 mg mL−1. For EDC-NHS coupling, sulpho-NHS (2 mg mL−1) and EDC (10 mg mL−1) solutions were prepared in MES buffer (at 4 °C) separately. For activation of the carboxylic acid groups on NPs, 32 μL EDC solution was added to the solutions and left for 30 s vigorous mixing which was followed by addition of 80 μl sulpho-NHS solution and mixing for an additional 10 minutes. Before adding 50 μl amine-functionalized aptamer solutions (at a concentration of 100 mM) to the related surface activated NP solutions, the solutions of aptamers were heated to 95 °C for 4 minutes and cooled in a block of ice for an additional 4 minutes and left at room temperature for minimum energy secondary structure formation of aptamers. The E. coli specific and S. typhimurium specific aptamers were covalently coupled with AuNRs and AuNUs, respectively. The coupling mixtures were left at room temperature for a few hours. Two successive steps of centrifugation were applied to eliminate the excess aptamers from the solution. The remaining aptamer-modified AuNR and AuNU pellets were redispersed in 1× phosphate-buffered saline (PBS). ESI† Fig. S3 presents the graphical illustration of the procedure.To modify LNPs with complementary DNA oligomers, 1 mg mL−1 of each LNP was provided in 25 mM MES buffer (pH 6.0). As described above, the EDC/sulfo-NHS coupling method was applied to activate the carboxylic acid groups on the surface of the NPs. The surface-activated LNPs were functionalized with 50 μl of the respective cDNA solution (100 μM) at room temperature for a couple of hours. The final solution was centrifuged two times to remove excess cDNAs and redispersed in 1× PBS. A schematic of LNP modification with the respective cDNA is also shown in ESI† Fig. S3.
Furthermore, both aptamer–AuNS solutions were mixed (at 250 rpm) and incubated for 2 hours (at 37 °C) with their corresponding cDNA–LNP solution. For NSET quenching of LNPs, AuNU–aptamer and UCNP–cDNAs and AuNR–aptamer and QD–cDNAs were combined at a ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v), respectively. The experimental optimization of NSET quenching is described in ESI† Fig. S4. For the separation of AuNS–LNP conjugations, the time and speed of centrifugation required for sedimentation of each AuNSs–aptamer and LNPs–cDNA were studied separately, followed by optimization of the centrifugation step of the AuNS–LNP conjugates in terms of time and speed of centrifugation. Eventually, two consecutive washing steps were applied to the final solution for separating the conjugated AuNS–aptamer–cDNA–LNPs from non-conjugated NPs which were redispersed in 1× PBS for further analysis.
1 (v/v), respectively. The experimental optimization of NSET quenching is described in ESI† Fig. S4. For the separation of AuNS–LNP conjugations, the time and speed of centrifugation required for sedimentation of each AuNSs–aptamer and LNPs–cDNA were studied separately, followed by optimization of the centrifugation step of the AuNS–LNP conjugates in terms of time and speed of centrifugation. Eventually, two consecutive washing steps were applied to the final solution for separating the conjugated AuNS–aptamer–cDNA–LNPs from non-conjugated NPs which were redispersed in 1× PBS for further analysis.
Characterization of NSET based target-specific nanoprobes
The UV-vis absorption spectra of the samples were measured with quartz cuvettes (Quartz-Suprasil, Hellma GmbH & Co. K.G., Mulheim, Germany), using a UV-vis-NIR spectrophotometer (Cary 5000, Varian, USA). It is worth noting that 100-fold dilution was applied on the samples before UV-vis measurements to eliminate contributions from multiple scattering events. A circular dichroism (CD) spectrophotometer, J-815 (Jasco International Co., Tokyo, Japan), was utilized to investigate the chirality of the samples using a quartz cuvette with a path length of 1.0 mm. The scanning speed, band width, and number of scans of CD measurements were chosen as 50 nm min−1, 1 nm, and 10, respectively. The hydrodynamic size distribution profiles of AuNSs, LNPs, aptamer–AuNSs, and cDNA–LNPs in glass DLS (dynamic light scattering) cuvettes were measured using a Malvern Zetasizer Nano ZS equipped with a 10 mW 632.8 nm red laser (Malvern Instruments, Worcestershire, U.K.) with a detector measurement angle of 173°. 1000-times dilution using sterilized Milli-Q water was applied for all samples to prevent multiple scattering events, followed by filtration with 0.2 mm PTFE filters. The number of measurements for each sample was set to three, and ten individuals runs were performed for each measurement. The intensity-based results were used to demonstrate the size distribution of the three samples. For the measurement of zeta potential, all samples were diluted 10 times using 1× PBS to ensure precise measurement and transferred to a disposable capillary cell. Analysis of the aptamer functionalized AuNSs, cDNA modified UCNPs, AuNR–UCNP conjugates, and E. coli–AuNRs and S. typhimurium–AuNUs was performed using a scanning electron microscope (SEM) with a focused electron beam (Zeiss Leo Supra 35 VP, Germany). The luminescence spectra of the naive QDs, naive UCNPs, and corresponding conjugates were recorded using a Cary Eclipse fluorescence spectrophotometer (Agilent, Santa Clara, US). While an internal laser of the spectrophotometer mentioned above was utilized for UV excitation of QDs at a wavelength of 350 nm, both excitation and emission slits were set to 5 nm, and all spectra were recorded between 500 and 700 nm. However, for UCNPs, an external 500 mW continuous-wave diode laser (Dragon Lasers, China) was installed orthogonal to the measurement plane for NIR excitation at 980 nm. The bio/chemiluminescence mode was chosen with an emission slit of 2.5 nm for collecting all spectra between the wavelengths of 400 and 800 nm.Preparation of bacterial cultures
Firstly, S. typhimurium and E.coli cultures were cultivated on nutrient agar media for 24 hours using an incubator at 37 °C. After adding a single colony to the nutrient broth medium, it was incubated overnight at 37 °C. A Bio-Rad Smart Spec Plus spectrophotometer (Bio-Rad Laboratories Inc., USA) was used to measure the colony-forming units (CFU) per mL of the culture. The pathogens were centrifuged and washed 2 times using 1× PBS and redispersed in the same buffer before use. After the preparation of bacterial cultures, NSET pairs were incubated with related target bacteria at 37 °C for melting of the partially complementary DNA duplex.Results and discussion
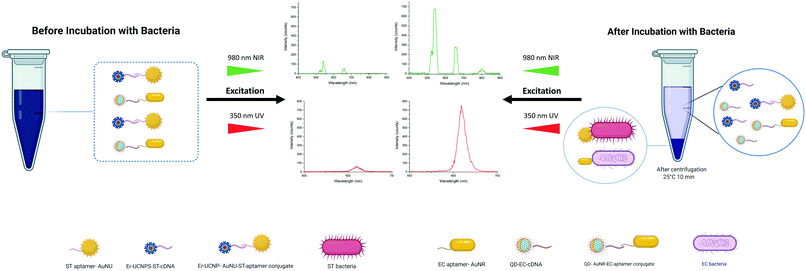
In the developed NSET-based detection method, aptamer functionalized AuNSs, including EC-aptamer–AuNRs and ST-aptamer–AuNUs, served as capture probes, and cDNA-functionalized LNPs, including EC-cDNA–QDs and ST-cDNA–UCNPs, act as signal transduction elements for multiplex sensing of pathogens, E. coli and S. typhimurium. The carboxylic acid-functionalized AuNRs and AuNUs were further modified covalently with the respective 5′-amino-terminated aptamers to prepare aptamer–AuNS conjugates. In parallel, 5′-amino-terminated cDNAs of the aptamers mentioned previously were used to modify the carboxylic acid labeled NaYF4:Yb,Er UCNPs and carboxyl Cd-based core/shell QDs for the formation of cDNA–LNP assemblies. Then, the as-prepared aptamer–AuNSs were incubated with cDNA–LNPs to form a detection probe via extensive interaction between the aptamer and its complementary oligonucleotide. As a result, UCNP/AuNU and QD/AuNR conjugated biosensors were designed for quantifying S. typhimurium and E. coli, respectively. The principle of this NSET based bioassay is shown in Fig. 1.In the literature, the NSET based sensing method was applied for the detection of glutathione through exploiting 5-aminofluorescein and AuNPs39 and sensing C-reactive protein using fluorescein and AuNPs.40 An NSET-based biosensing platform between SiO2 coated CdTe (CdTe/SiO2) core/shell NPs and AuNPs was also reported for the same aim, facilitating facile and sensitive detection of DNA.41 Furthermore, an effective fluorescence quenching of rhodamine 6G was achieved by Patel et al.42 using rhodamine 6G and AuNPs as NSET pairs. In this assay, DNA-modified AuNPs act as an acceptor of fluorescence emitted by the rhodamine 6G compound. In another assay, Li et al.43 successfully constructed an NSET-based sensor to detect mercury(II) by bringing the DNA-conjugated QDs and AuNPs into a suitable proximity, which enables effectual quenching of the fluorescence emission of the QDs. In another study, a highly selective and sensitive AuNP-based NSET probe was improved for screening hepatitis C virus (HCV) RNA through exploiting ET between the Cy3 dye and AuNPs.13 In nearly all the recently designed NSET detection platforms, fluorophores and AuNPs were used as NSET pairs. Based on the advantages mentioned above of LNPs and the advantages of AuNSs with different shapes, they can be exploited to design more effective and sensitive multiplex NSET biosensing platforms. Limitations were faced while assembling the explained ET-based biosensors for multiple screening of targets, including a limited number of colors in UCNPs because of the low number of luminescent activators and broad emission spectra of QDs. However, the distinct excitation spectra of used LNPs enabled their usage as signal reagents in multiple detection bioassays, which resulted in effective utilization of the visible-NIR region of the electromagnetic spectrum. Regarding QDs, they can be excited by photons with energies higher than their bandgap and convert them to photons with lower levels of energies at 620 nm. UCNPs effectively absorb photons at a wavelength of 980 nm and pose two weak and robust emission peaks at 540 and 655 nm. In other words, a substantial 340-nm anti-Stokes shift of UCNPs along with a 10 nm Stokes shift of QDs offers a convenient solution for the challenge of spectral overlap in developing multiple sensing stages. One particularly challenging aspect of exploiting these LNPs in biosensors is the absorption of UCNP-emitted photons by QDs. However, FRET only happens when the gap separating them is less than 10 nm. Also, the employment of single-strand oligonucleotides can address this issue as a labeling agent for LNPs, which have a high affinity for binding to the aptamer. Their complementary structure is used as a capping agent for AuNSs. Moreover, the highly opposing surface of these oligonucleotides leads to strong repulsive Coulomb force. In addition, in order to suppress the issue in which QDs and UCNPs have multiple binding sites for AuNSs, the conjugations were performed at dilute concentrations. Also, the QDs are significantly smaller than the AuNRs and the aggregations would be limited to multiple QDs attaching on the more accessible shorter edge of the AuNRs. The QD size is well below the spatial confinement of the EM field. In the UCNP case, the conjugation ratio is maintained using low concentrations in the conjugation step. As a result, the system works in longitudinal mode.
Characterization of AuNSs–apt probes and LNPs–cDNA complexes
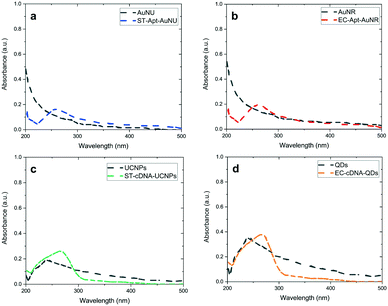
UV-vis analysis was performed to demonstrate the covalent attachment of aptamers and the corresponding cDNA to –COOH functionalized AuNSs and LNPs, respectively. Comparing the spectra obtained for unmodified and modified AuNSs and LNPs indicates the appearance of a specific absorption peak related to ssDNA at a wavelength of 260 nm. Regarding AuNS–aptamer conjugations, since the absorption intensity for each of the AuNSs was low at approximately 260 nm, the aptamer absorption peak was observable (Fig. 2a and b). As shown in Fig. 2c, the well-defined peak related to ST-cDNA was noticeable in the UCNP–ST-cDNA solution since the comparatively high bandgap of the core material (NaYF4) does not overlap with the peak of ST-cDNA. On the other hand, Fig. 2d reveals an absorption shoulder for EC-cDNA despite the shadowing effect of a strong absorption peak associated with QDs in the QD–EC-cDNA complex. Furthermore, UV-vis spectra of S. typhimurium and E. coli with and without the related AuNS–aptamer as another control experiment are also provided in ESI† Fig. S5a and b, respectively, which indicate the successful attachment of aptamer modified AuNSs to related bacteria. The applied robust centrifugation using Amicon® Ultra centrifugal filters promoted the separation of single-strand DNA modified NPs from unbound DNAs based on the difference in their mass (kDa) and led to the recovery of NPs. Considering this step, the obtained results demonstrate the successful modification of nanomaterials with DNA oligomers, which is in line with previous findings.21,44Even though the existence of chosen aptamers and their complementary oligonucleotides on capture probes and signal transducers and validation of their effective attachment were verified using UV-vis spectrophotometry (this will also be supported by DLS and SEM analysis which is provided in the following parts), confirming the maintenance of the DNA oligomers' secondary structure and their stability under experimental conditions is of great significance. Within this framework, the chiral features of the exploited oligonucleotide after being introduced to the LNPs and AuNSs can be investigated via CD spectroscopy, a technique working based on the difference between the absorption of left- and right-handed circularly polarized light.2 Given that there is almost no difference between left- and right-circularly polarized light absorption by AuNSs and LNPs, they do not substantially impact the CD spectra of the aptamers and cDNAs utilized in the capture and signal probes, respectively.45,46
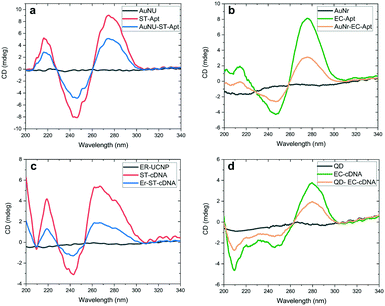
Fig. 3a and b provide the CD spectra related to uncapped AuNSs, free aptamers, and aptamer–AuNS complexes. Since AuNUs lack chirality features, polarization-dependent absorption was not detected in the CD spectra of this AuNS between 200 and 340 nm. In the case of AuNRs, the chirality was insignificant in comparison with that of aptamer-modified AuNRs. An alteration was only detected in the secondary structure of aptamers caused by their attachment on AuNRs, which leads to the loss of the amine group at the end of these DNA oligomers. The same CD analyses were conducted for evaluation of the cDNA conjugation with the carboxyl functionalized LNPs. As shown in Fig. 3c and d, restricted chirality was observed in the CD spectra of LNPs between 200 and 300 nm. The result was the same for the spectral range of 300–340 nm. One exciting fact highlighted by this figure is the chirality alteration for cDNAs attached to LNPs in the range of 200–240 nm. The leading cause of this minor alteration is that these oligonucleotides are less stable than the aptamers in terms of the secondary structure due to fewer nucleotides. Based on the literature, the obtained results signify the successful modification of nanomaterials with DNA oligomers.38,45
Next, DLS analysis was applied to confirm further the functionalization of AuNSs and LNPs with the aptamer and cDNA based on the change in the NP hydrodynamic size by modification (Table S3,†Fig. 4, and ESI† Fig. S6). Attachment of aptamers and their complementary oligonucleotides to NPs increases the number of DNA oligomers around them and leads to an increase in the hydration sphere of NPs. An increase took place in the uncapped AuNU hydrodynamic radius after functionalization with ST-aptamer from 91.3 nm to 105.8 nm. Likewise, labeling AuNRs with EC-aptamer led to a rise in the hydrodynamic radius of AuNRs, changing from 55.7 nm to 66.2 nm. For LNPs, increasing the hydrodynamic radius of unlabeled QDs from 25.93 nm to 61.76 nm occurred by modifying them with EC-cDNA while capping UCNPs with ST-cDNA altered the size from 41.8 nm to 87.7 nm.
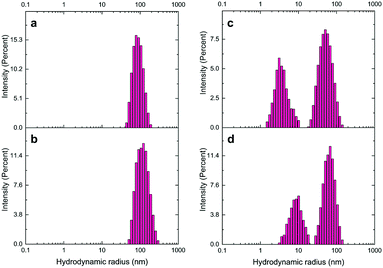 | ||
| Fig. 4 Hydrodynamic size distribution of the a) unmodified AuNUs, b) ST aptamer-labeled AuNUs, c) naive AuNRs, and d) EC aptamer-modified AuNRs. | ||
Another technique for demonstrating the successful binding of DNA oligomers to nanomaterials is the measurement of zeta potential. This value can be defined as the difference between the potential at the particle's slipping plane and the solution potential. As a fundamental index of the charge of the particle surface, this scientific term shows the stability of particles in solution.47 A glance at Table S4† reveals the surface potential (ζ) of AuNSs and LNPs before and after surface functionalization with aptamers and cDNAs, respectively. Before aptamer modification, a zeta potential of −22.2 mV and −20.8 mV obtained from the table indicates the negative charge on the surface of AuNUs and AuNRs, respectively, because of being modified with carboxyl groups. However, after binding amine-terminated aptamers to their surface, a positive shift of the surface potential of AuNS–aptamer was noticed, which reached −9.73 mV and −8.27 mV for ST-aptamer–AuNUs and EC-aptamer–AuNRs, respectively. The same positive shift was detected in the modification of LNPs with cDNAs. The electrostatic potential of QDs and UCNPs was −32.5 mV and −30.7 mV, respectively, because of negatively charged carboxyl groups. Nonetheless, after modification with the respective amine-functionalized cDNA, QD–EC-cDNA's zeta potential moved to −12.9 mV, while it became −14.3 mV for UCNP–ST-cDNA. The results provided by zeta potential measurements, which are additional confirmation to the findings through UV-vis, DLS, and CD analysis, revealed the adequate preparation of aptamer–AuNS and cDNA-LNP conjugates, which is in agreement with the literature.38,44,48 As presented in Fig. S7.† SEM micrographs are provided for AuNU–ST-Apt, AuNR–EC-Apt, and UCNP–ST-cDNA to supplement the above-mentioned results. SEM micrographs depict the hydrodynamic sizes of NPs. The ionic halo around particles due to the attachment of ST-Apt, EC-Apt, and ST-cDNA as a single strand to AuNUs, AuNRs, and UCNPs, respectively, causes an increase in sizes obtained by DLS measurement of each NS. As can be noticed in the SEM micrographs provided in ESI† Fig. S7, there is no aggregation in ST-Apt coated AuNUs, EC-Apt coated AuNRs, and ST-cDNA coated UCNPs. Following this, SEM images of the ST-AuNU nanoprobe with S. typhimurium (ESI† Fig. S8a), EC-AuNR nanoprobe with E. coli (ESI† Fig. S8b), and UCNP–AuNR conjugate (ESI† Fig. S9) are presented to confirm that the utilized binding method and sensing platform are working properly.
Aptasensor based on dual-excitation for foodborne bacterial pathogen detection
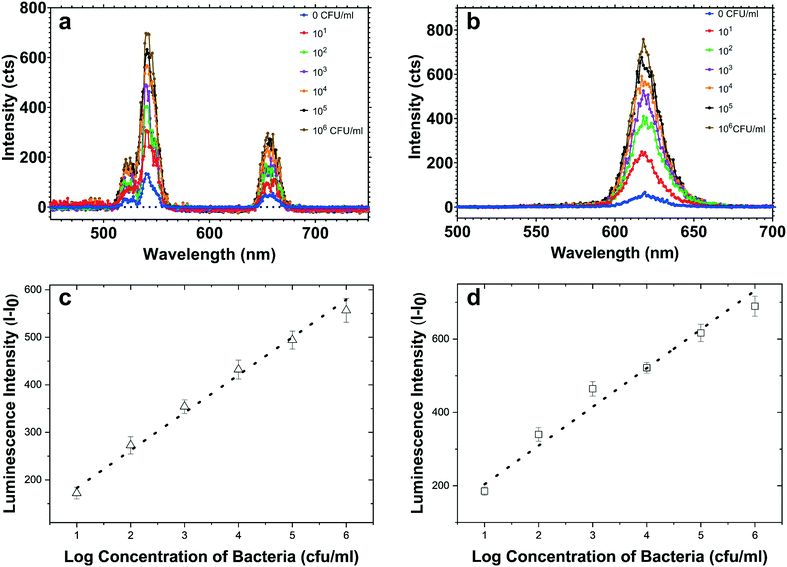
At first, both AuNU–UCNP and AuNR–QD aptasensors were provided in separate batches. Each of the designed complexes was then assessed individually in samples containing its corresponding target bacterial pathogens. For this aim, incubation of each aptasensor with a related bacterial pathogen was performed at 37 °C for 45 minutes in distinct experimental batches. This step was followed by removing the bacteria-bound AuNS–aptamer system from the solution through a robust and optimized centrifugation process. The AuNSs–aptamer sensing probes break their bonds with the respective complementary oligonucleotides and attach to the target bacteria due to the aptamers' high affinity for pathogens. The steps mentioned above were applied for each bacterium at logarithmically increasing concentrations (10–106 CFU mL−1). Next, fluorescence spectroscopy was utilized to measure the intensity of luminescence related to the preliminary complex before bacteria addition and the luminescence intensity of the LNPs–cDNA remaining in the supernatant after centrifugation in the step of bacteria addition.By exposure of the UCNP- and QD-labeled samples to light sources at wavelengths of 980 and 350 nm, the luminescence of each complex was recorded as a spectrum. As shown in Fig. 5, a simultaneous increase in the luminescence intensity of EC-aptamer–AuNR–QD and ST-aptamer–AuNU–UCNP was detected after adding increasing the concentrations of E. coli and S. typhimurium, which is an indicator of the turn-on (signal-on) detection method. For quantifying E. coli, the linear range of the aptasensor was 102–106 CFU mL−1, and a LOD of 4.94 CFU mL−1 was achieved for this foodborne bacterium. Regarding S. typhimurium, the linear range of the employed aptamer-based biosensing platform was 102–106 CFU mL−1 with 7.55 CFU mL−1 as the LOD. The values related to the detection limit were calculated as an approximation to the nearest higher integral number. The calculations were performed based on the standard method provided by the Clinical and Laboratory Standards Institute.49 According to this method, in order to define the lowest concentration which can be measured by the proposed sensing platform, terms such as the limit of blank (LOB), and LOD were utilized. Regarding LOB, it can be defined as the highest amount of apparent analyte which is expected to be distinguished when an analyte-free blank sample is analyzed in many replications. The formula for LOB calculation is also provided as LOB = mean blank + 1.645 (SD blank). Besides, LOD is the lowest concentration of analyte which is defined using LOB. Both LOB and test replicates of a sample with a low concentration of analyte are required for LOD calculation which can be written as LOD = LOB + 1.645 (SD low concentration sample). The calibration graphs for luminescence emission of LNPs in terms of the corresponding bacteria are plotted in Fig. 5, and their parameters are tabulated in Table S5.† Based on the results obtained for single detection of bacteria using the respective developed AuNS–LNP conjugate, the LOD calculated for each bacterium is significantly lower than those of previously employed ET-based biosystems.38
Multiplex biosensor for simultaneous detection of bacteria
As previously mentioned, signal crosstalk/overlap is still a challenge in developing multiplex NSET-based screening platforms. So, this issue must be investigated besides major sensing experiments to observe the impact of the emission spectrum of each LNP on the other one. Based on our investigations regarding simultaneous utilization of QDs and UCNPs, sample exposure to NIR radiation at 980 nm only leads to excitation of UCPs because this light source's energy is less than the bandgap of the utilized core/shell QDs. Similarly, being exposed to UV radiation at 350 nm, QDs are the only NPs that become excited, and UCNPs will not be affected. As shown in ESI† Fig. S1, the UCNPs used in this experiment as a donor exhibit a strong emission peak at 545 nm, which may have a considerable role in the excitation of CdSe/ZnS core/shell QDs. The experiments demonstrate that signal overlap is not probable in our assay, thanks to the low quantum yields of UCNPs. Furthermore, as described in the Introduction part, due to factors such as distance between acceptor and donor species, the luminescence resonance energy transfer (LRET) possibility is almost zero due to modification of LRET pairs in the provided sensing biosystems.To evaluate the detection efficiency of the multiplex aptamer-based sensor, the designed UCNP–AuNU and QD–AuNR biosystems were utilized at the same time in one experimental batch. Similar to the single aptasensor developed in the previous section, logarithmically increasing concentrations of both bacteria ranging from 10 to 105 CFU mL−1 were provided for multiplex sensing. ESI† Fig. S10 reveals a noticeable increase in the luminescence intensity of each conjugate by increasing the concentration of the respective target bacteria due to the unbounding of an increasing number of AuNS–aptamer probes from LNPs and binding to a particular target pathogen. The calibration plot between the luminescence intensity of UCNPs and S. typhimurium concentration was linear in the range of 102–105 CFU mL−1 with a LOD of 9.31 CFU mL−1. In the calibration curve for detection of E. coli through the QD-based aptasensor, the linear range was calculated as 103–105 while the LOD was as low as 7.38 CFU mL−1. Also, the calibration curve parameters, along with the respective LOD values, are tabulated in Table S6.† One point that can be noted from our simultaneous NSET detection biosystem is the lower LOD values than those of previously developed ET-based multiplex ones.38,50,51
Real sample detection
The applicability and reliability of the suggested NSET-based aptasensors were assessed and confirmed using lake water as a real sample. As a pretreatment step, the sample was kept in standard centrifuge tubes in a refrigerator for 24 hours. Then, a particular volume was obtained from the supernatant part and filtered. Following this, the pretreated solution was spiked with specified concentrations of bacteria ranging from 102–104 CFU mL−1. As shown in Table S7,† which provides the spiked concentrations and concentrations measured by the proposed biosensor for lake water, the lowest error of detection is shown by ST-aptamer–AuNU–UCNP. The applicability evaluation results confirmed that the developed multiplex aptamer-based detection platform possesses outstanding potential in the effective detection and quantification of bacterial pathogens in real samples.Specificity of the NSET aptasensor
Each conjugate NSET probe was evaluated against a set of non-specific target bacteria introduced to the system individually to validate the specificity features of the employed biosensors. The concentration of all bacteria was fixed at 104 CFU mL−1, and the luminescence signal readouts of UCNP–ST-cDNA and QD–EC-cDNA after incubation with non-specific pathogens were measured and normalized based on the target-specific signal and are shown in Fig. 6. The increase in luminescence intensity related to incubation of the conjugate with S. typhimurium is considerably higher than those of non-specific ones. A similar outcome was observed from the QD–AuNR conjugate in E. coli detection, and a slight change was detected when incubated with non-specific pathogens. It is worth mentioning that the luminescence intensity increases related to incubation of the provided UCNP and QD-labeled conjugations with non-specific bacteria were similar to 113.81 and 128.96 count intercepts of the linear calibration curves which confirmed the high selectivity of the proposed multiplex sensing platform.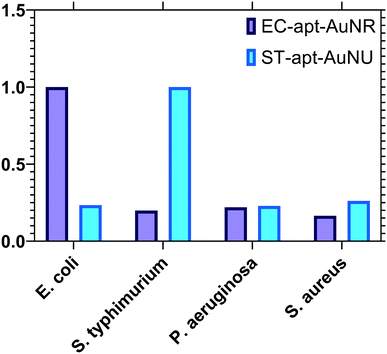 | ||
| Fig. 6 Target selectivity of the developed NSET-based aptasensors against non-specific bacterial targets with a concentration of 104 CFU mL−1. | ||
Conclusion
By way of conclusion, a multiplex NSET-based biosensor was reported based on the conjugation of AuNSs with the respective LNPs as NSET pairs and validated for sensitive, facile, one-pot, and simultaneous screening of foodborne bacterial pathogens, namely S. typhimurium and E. coli. Using AuNSs as acceptors of NSET pairs paved the way for effectual fluorescence quenching of LNPs and consequently enhanced ET in the developed sensing batch. More importantly, though, exploiting the anti-Stokes shift in NaYF4:Yb,Er UCNPs and Stokes shift in CdSe/ZnS core/shell QDs significantly contributes to effectual light utilization at the NIR and visible regions and also removed the possibility of signal cross-talk in the dual-excitation ET-based biosensor. In addition to this, efficacious functionalization of AuNSs and LNPs with highly stable target-specific aptamers and their complementary DNA oligomers respectively was confirmed by methods such as DLS, UV-vis, SEM, and CD analysis. Using fluorescence spectroscopy, the increase in the luminescence intensity of QDs and UCNPs was utilized to screen E. coli and S. typhimurium at the same time with a linear range of 103–105 and 102–105 and a low LOD of 7.38 and 9.31 CFU mL−1 respectively in one experimental tube. Moreover, the multiplex detection bioassay was also efficaciously exploited for sensing E. coli and S. typhimurium in lake water. Furthermore, specificity assays confirmed that the provided sensing technique has a negligible response against non-specific bacterial targets and consequently possesses high sensitivity. Additional enhancement of AuNS features will ensure improvement in the sensing potential of the provided detection method. Considering the suggested enhancements, screening other targets such as cancer biomarkers, etc. in their original place and bioimaging via a developed method will be applicable. Based on our anticipation, thrilling and highly specific and sensitive detection strategies would be provided for environmental and biochemical applications by making simultaneous use of AuNSs of various shapes and other potential LNPs. Thus, the number of targets for simultaneous screening of real samples can be increased. Additionally, the effect of parameters such as the size of exploited NPs on the efficiency of detecting platforms can be a potential topic for further experimental studies.Abbreviations
| AuNPs | Gold nanoparticles |
| AuNRs | Gold nanorods |
| AuNSs | Gold nanostructures |
| AuNUs | Gold nanourchins |
| CD | Circular dichroism spectroscopy |
| cDNA | Complementary DNA |
| CFU | Colony-forming units |
| DLS | Dynamic light scattering |
| EDC | N-(3-Dimethylaminopropyl)-N′-ethyl carbodiimide hydrochloride |
| ET | Energy transfer |
| LNPs | Luminescent nanoparticles |
| MNPs | Metallic nanoparticles |
| NIR | Near-infrared |
| NSET | Nano surface energy transfer |
| NPs | Nanoparticles |
| PSET | Plasmonic nanoparticle surface energy transfer |
| QDs | Quantum dots |
| Sulpho-NHS | N-Hydroxysulfosuccinimide sodium salt |
| SPR | Surface plasmon resonance |
| UCNPs | Upconverting nanoparticles |
| UV-vis | Ultraviolet-visible |
Author contributions
MT conducted the experiments, and wrote and edited the original draft. HK, MKB, and MY participated in the study design and reviewed the manuscript. All the authors contributed to and approved the final version of the manuscript.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
The authors acknowledge Individual Research Funding (IRF) from the Sabanci University Nanotechnology Research and Application Centre (SUNUM).References
- Z. Jiang, B. Feng, J. Xu, T. Qing, P. Zhang and Z. Qing, Biosens. Bioelectron., 2020, 166, 112471 CrossRef CAS PubMed.
- M. Yüce and H. Kurt, RSC Adv., 2017, 7, 49386–49403 RSC.
- R. Zhang, T. Belwal, L. Li, X. Lin, Y. Xu and Z. Luo, Compr. Rev. Food Sci. Food Saf., 2020, 19, 1465–1487 CrossRef PubMed.
- D. Bahari, B. Babamiri, A. Salimi and H. Salimizand, Talanta, 2021, 221, 121619 CrossRef CAS PubMed.
- H. Kurt, E. Alpaslan, B. Yildiz, A. Taralp and C. W. Ow-Yang, J. Colloid Interface Sci., 2017, 488, 348–355 CrossRef CAS PubMed.
- M. M. Elsutohy, A. Selo, V. M. Chauhan, S. J. B. Tendler and J. W. Aylott, RSC Adv., 2018, 8, 35840–35848 RSC.
- C. J. Breshike, R. A. Riskowski and G. F. Strouse, J. Phys. Chem. C, 2013, 117, 23942–23949 CrossRef CAS.
- T. Sen, S. Sadhu and A. Patra, Appl. Phys. Lett., 2007, 91, 1–4 Search PubMed.
- A. Samanta, Y. Zhou, S. Zou, H. Yan and Y. Liu, Nano Lett., 2014, 14, 5052–5057 CrossRef CAS PubMed.
- Q. Lai, Y. Liu, L. Ge, Y. Yang, X. Ji and Z. He, Anal. Methods, 2021, 13, 2092 RSC.
- C. Chen, C. Midelet, S. Bhuckory, N. Hildebrandt and M. H. V. Werts, J. Phys. Chem. C, 2018, 122, 17566–17574 CrossRef CAS.
- C. Chen and N. Hildebrandt, TrAC, Trends Anal. Chem., 2020, 123, 115748 CrossRef CAS.
- J. Griffin, A. K. Singh, D. Senapati, P. Rhodes, K. Mitchell, B. Robinson, E. Yu and P. C. Ray, Chem. – Eur. J., 2009, 15, 342–351 CrossRef CAS PubMed.
- C. S. Yun, A. Javier, T. Jennings, M. Fisher, S. Hira, S. Peterson, B. Hopkins, N. O. Reich and G. F. Strouse, J. Am. Chem. Soc., 2005, 127, 3115–3119 CrossRef CAS PubMed.
- M. Ku, Y. Hong, D. Heo, E. Lee, S. Hwang, J. S. Suh and J. Yang, Biosens. Bioelectron., 2016, 77, 471–477 CrossRef CAS PubMed.
- W. Yang, Y. Shen, D. Zhang, C. Li, R. Yuan and W. Xu, Anal. Chem., 2019, 91, 7782–7789 CrossRef CAS PubMed.
- R. M. Renuka, M. Nikhil, J. Achuth, K. Ponmalai, P. Kolandaivel and K. Kadirvelu, Spectrochim. Acta, Part A, 2020, 243, 118662 CrossRef PubMed.
- J. Ren, G. Liang, Y. Man, A. Li, X. Jin, Q. Liu and L. Pan, PLoS One, 2019, 14, 1–13 Search PubMed.
- L. Yang and Y. Li, Analyst, 2006, 131, 394–401 RSC.
- J. A. Stride and F. Mirnajafizadeh, SDRP Journal of Nanotechnology & Material Science, 2020, 3, 121–126 Search PubMed.
- B. Jin, S. Wang, M. Lin, Y. Jin, S. Zhang, X. Cui, Y. Gong, A. Li, F. Xu and T. J. Lu, Biosens. Bioelectron., 2017, 90, 525–533 CrossRef CAS PubMed.
- B. Zhou, B. Shi, D. Jin and X. Liu, Nat. Nanotechnol., 2015, 10, 924–936 CrossRef CAS PubMed.
- T. Mairal, V. Cengiz Özalp, P. Lozano Sánchez, M. Mir, I. Katakis and C. K. O'Sullivan, Anal. Bioanal. Chem., 2008, 390, 989–1007 CrossRef CAS PubMed.
- B. Zhang, H. Li, W. Pan, Q. Chen, Q. Ouyang and J. Zhao, Food Anal. Methods, 2017, 10, 2036–2045 CrossRef.
- M. Yu, H. Wang, F. Fu, L. Li, J. Li, G. Li, Y. Song, M. T. Swihart and E. Song, Anal. Chem., 2017, 89, 4085–4090 CrossRef CAS PubMed.
- A. B. Chinen, C. M. Guan, J. R. Ferrer, S. N. Barnaby, T. J. Merkel and C. A. Mirkin, Chem. Rev., 2015, 115, 10530–10574 CrossRef CAS PubMed.
- H. Xing, T. Wei, X. Lin and Z. Dai, Anal. Chim. Acta, 2018, 1042, 71–78 CrossRef CAS PubMed.
- K. Park, M. S. Hsiao, Y. J. Yi, S. Izor, H. Koerner, A. Jawaid and R. A. Vaia, ACS Appl. Mater. Interfaces, 2017, 9, 26363–26371 CrossRef CAS PubMed.
- Y. Xia, L. Song and C. Zhu, Anal. Chem., 2011, 83, 1401–1407 CrossRef CAS PubMed.
- M. KGaA, https://www.sigmaaldrich.com, 2021.
- J. Wang, M. You, G. Zhu, M. I. Shukoor, Z. Chen, Z. Zhao, M. B. Altman, Q. Yuan, Z. Zhu, Y. Chen, C. Z. Huang and W. Tan, Small, 2013, 9, 3678–3684 CrossRef CAS PubMed.
- Y. Fu, J. Zhang and J. R. Lakowicz, J. Am. Chem. Soc., 2010, 132, 5540–5541 CrossRef CAS PubMed.
- G. K. Darbha, A. Ray and P. C. Ray, ACS Nano, 2007, 1, 208–214 CrossRef CAS PubMed.
- J. J. Liu, D. Yuan, H. Z. Zhang, Y. D. Lu, N. Wang, H. Y. Zou and J. Wang, Sens. Actuators, B, 2018, 274, 318–323 CrossRef CAS.
- V. Pavlov, Y. Xiao, B. Shlyahovsky and I. Willner, J. Am. Chem. Soc., 2004, 126, 11768–11769 CrossRef CAS PubMed.
- R. Joshi, H. Janagama, H. P. Dwivedi, T. M. A. Senthil Kumar, L. A. Jaykus, J. Schefers and S. Sreevatsan, Mol. Cell. Probes, 2009, 23, 20–28 CrossRef CAS PubMed.
- W. Wu, J. Zhang, M. Zheng, Y. Zhong, J. Yang, Y. Zhao, W. Wu, W. Ye, J. Wen, Q. Wang and J. Lu, PLoS One, 2012, 7, e48999 CrossRef CAS PubMed.
- H. Kurt, M. Yüce, B. Hussain and H. Budak, Biosens. Bioelectron., 2016, 81, 280–286 CrossRef CAS PubMed.
- Q. Li, A. Sun, Y. Si, M. Chen and L. Wu, Chem. Mater., 2017, 29, 6758–6765 CrossRef CAS.
- C. Bravin and V. Amendola, Biosens. Bioelectron., 2020, 169, 112591 CrossRef CAS PubMed.
- W. Guo, Y. Wei, Z. Dai, G. Chen, Y. Chu and Y. Zhao, Materials, 2018, 11, 272 CrossRef PubMed.
- A. S. Patel, H. Sahoo and T. Mohanty, J. Fluoresc., 2016, 26, 1849–1855 CrossRef CAS PubMed.
- M. Li, Q. Wang, X. Shi, L. A. Hornak and N. Wu, Anal. Chem., 2011, 83, 7061–7065 CrossRef CAS PubMed.
- H. Li, W. Ahmad, Y. Rong, Q. Chen, M. Zuo, Q. Ouyang and Z. Guo, Food Control, 2020, 107, 106761 CrossRef CAS.
- M. Yüce, H. Kurt, B. Hussain, C. W. Ow-Yang and H. Budak, ChemistrySelect, 2018, 3, 5814–5823 CrossRef.
- J. H. Choi, K. H. Chen and M. S. Strano, J. Am. Chem. Soc., 2006, 128, 15584–15585 CrossRef CAS PubMed.
- R. Vogel, A. K. Pal, S. Jambhrunkar, P. Patel, S. S. Thakur, E. Reátegui, H. S. Parekh, P. Saá, A. Stassinopoulos and M. F. Broom, Sci. Rep., 2017, 7, 1–13 CrossRef CAS PubMed.
- P. Wang, A. Wang, M. M. Hassan, Q. Ouyang, H. Li and Q. Chen, Sens. Actuators, B, 2020, 320, 128434 CrossRef CAS.
- D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29(Suppl 1), S49–S52 Search PubMed.
- N. Duan, S. Wu, Y. Yu, X. Ma, Y. Xia, X. Chen, Y. Huang and Z. Wang, Anal. Chim. Acta, 2013, 804, 151–158 CrossRef CAS PubMed.
- L. Xu, Z. T. Callaway, R. Wang, H. Wang, M. F. Slavik, A. Wang and Y. Li, Trans. ASABE, 2015, 58, 891–906 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d1sd00073j |
| This journal is © The Royal Society of Chemistry 2022 |