Open Access Article
Open Access ArticleProgrammable mismatch-fueled high-efficiency DNA signal amplifier†
Xiao-Long
Zhang‡
,
Sha-Sha
Li‡
,
Wei-Wei
Liu
,
Ling-Qi
Kong
,
Ya-Qin
Chai
* and
Ruo
Yuan
 *
*
Key Laboratory of Luminescence Analysis and Molecular Sensing (Southwest University), Ministry of Education, College of Chemistry and Chemical Engineering, Southwest University, Chongqing 400715, PR China. E-mail: yuanruo@swu.edu.cn; yqchai@swu.edu.cn
First published on 26th September 2022
Abstract
Herein, by introducing mismatches, a high-efficiency mismatch-fueled catalytic multiple-arm DNA junction assembly (M-CMDJA) with high-reactivity and a high-threshold is developed as a programmable DNA signal amplifier for rapid detection and ultrasensitive intracellular imaging of miRNA. Compared with traditional nucleic acid signal amplification (NASA) with a perfect complement, the M-CMDJA possesses larger kinetic and thermodynamic favorability owing to the more negative reaction standard free energy (ΔG) as driving force, resulting in much higher efficiency and rates. Once traces of the input initiator react with the mismatched substrate DNA, it could be converted into amounts of output multiple-arm DNA junctions via the M-CMDJA as the functional DNA conversion nanodevice. Impressively, the mismatch-fueled catalytic four-arm DNA junction assembly (M-CFDJA) exhibits high conversion efficiency up to 1.05 × 108 in 30 min, which is almost ten times more than those of conventional methods. Therefore, the M-CMDJA could easily address the challenges of traditional methods: slow rates and low efficiency. In application, the M-CFDJA as a DNA signal amplifier was successfully used to develop a biosensing platform for rapid miRNA detection with a LOD of 6.11 aM and the ultrasensitive intracellular imaging of miRNA, providing a basis for the next-generation of versatile DNA signal amplification methods for ultimate applications in DNA nanobiotechnology, biosensing assay, and clinical diagnoses.
Introduction
Benefiting from perfectly predictable base pairings, outstanding biocompatibility, and low cost, a considerable number of DNA-based rigid motifs1,2 have been developed to prepare different dimensional DNA nanostructures as biomolecular scaffolds or nanoparticle templates3,4 like DNA origami5,6 and DNA dynamic nanodevices.7,8 Remarkably, in the development of dynamic DNA nanostructural chemistry, the catalytic multiple-arm DNA junction assembly (CMDJA)9,10 as a flexible and ideal basic building block for DNA structural nanotechnology11–15 has attracted increasing attention. Compared to the branched DNA obtained by conventional annealing approaches with tedious temperature control,16,17 the isothermal CMDJA was obtained simply by relying on the base-pairing principle, in which the self-assembly can be well controlled by catalysts. Nevertheless, the CMDJA has also suffered from multiple-step reactions and weak driving forces, resulting in inherent time-consuming and suboptimal kinetic and thermodynamic properties, which further limits its deeper exploitation and wider application.18–20 On account of these disadvantages, it is of critical significance to improve the reaction speed and efficiency of CMDJAs to advance their superiority and applicability in the areas of nucleic acid signal amplification, biomarker assay, biomolecular nanomachines, and computing.2,21Since the performance of CMDJAs that relies on the toehold strand displacement reaction (TSDR) is immensely limited by the branch migration step in the displacement domain,22,23 the producing rate and yield of a multiple-arm DNA junction would be influenced by the reaction equilibrium of the step of branch migration in the TSDR. Therefore, the introduction of suitable mismatches in the displacement domain of the subsequent hairpin DNA can make the corresponding ΔG more negative than the one in a traditional CMDJA consisting of completely matched reactant DNA,24–27 realizing a stronger driving force accompanied with higher reactivity for improving the reaction rate and efficiency. In this study, we utilized NUPACK28 to find some possibly useful mismatches in the displacement domain of the substrate hairpin DNAs and compared the thermodynamic and kinetic parameters as essential indices to pick out the optimal mismatches for a mismatch-fueled catalytic multiple-arm DNA junction assembly (M-CMDJA). As a result, we carved out a mismatch-fueled catalytic four-arm DNA junction assembly (M-CFDJA) with optimal properties and used it as a high-efficiency DNA signal amplifier for rapid biomarker detection and ultrasensitive intracellular miRNA imaging, which helps in the development of the next generation of nucleic acid signal amplification techniques for biosensing assay and early disease diagnosis.
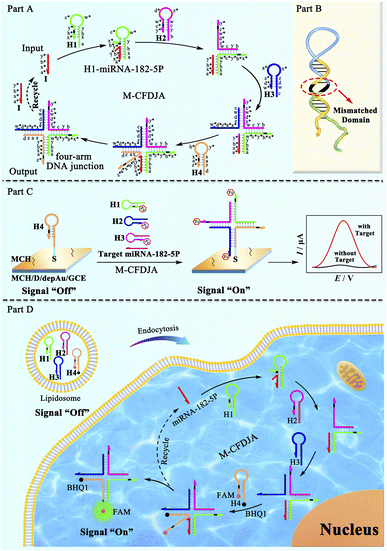
The reaction mechanism and the practical application of the M-CFDJA are shown in Scheme 1. First, the mechanism illustration of the catalytic formation pathway of the M-CFDJA is depicted in part A. The complementarity relationships of the segments of hairpin DNAs were specified so that it is only with the initiator target miRNA-182-5P that the hairpin DNAs are kinetically prevented from generating the four-arm DNA junctions that can be predicted to dominate at equilibrium. The first hairpin DNA (H1) is opened by the initiator miRNA-182-5P from the toehold to form the duplex H1–miRNA-182-5P and the pre-locked stem region is exposed, then the second hairpin DNA (H2) will be opened by H1's new sticky-end. Similarly, the rest of the hairpin DNAs (H3 and H4) will be opened to form the corresponding multiple-arm DNA junction and the initiator will be released to participate in the next pathway. Most importantly, the optimal mismatches in the neck (displacement domain) of the hairpin DNAs (part B) were selected to engineer the M-CFDJA with a fast reaction rate and high conversion efficiency. With the help of the M-CFDJA, traces of initiator can be rapidly converted into amounts of four-arm DNA junction for constructing a DNA conversion nanodevice. As a proof of concept, the evolved M-CFDJA was applied to develop a highly sensitive electrochemical biosensing platform (part C) for rapid miRNA-182-5P detection with a LOD (6.11 aM) that is nearly two orders of magnitude that of the wild CFDJA. Impressively, the M-CFDJA was further applied in rapid intracellular miRNA imaging to monitor miRNA-182-5P expression in cancer cells (part D), and it was observed to be much more sensitive than the typical FISH approach. The evolved M-CFDJA therefore showed great potential for ultimate application in biological nanobiotechnology and biomarker assay in the nascent stages of diseases.
Results and discussion
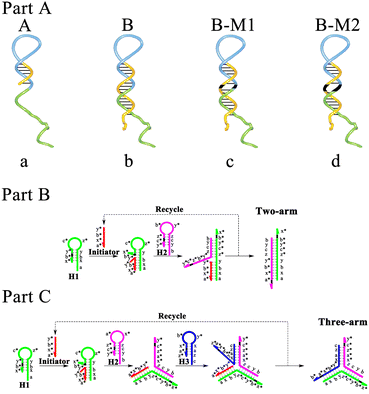
First, through polyacrylamide gel electrophoresis (PAGE), it was certified that the reactions in the CMDJA could successfully occur (Fig. S1†). Moreover, the three-arm and four-arm DNA junctions obtained via the CMDJA were respectively characterized using atomic force microscopy (AFM), and the results (Fig. S2†) were as expected. Next, based on the ΔG obtained from NUPACK as an evaluation index of hairpin DNA stability, we selected a suitable number of positions of mismatches following the principle that these hairpin DNAs as reactants with enough stability must possess less negative free energy to lead to a more negative reaction standard free energy as the driving force of the M-CMDJA, thus resulting in a higher amplification efficiency with low background leakage. In conclusion, one mismatch or double mismatches could be introduced into the neck of hairpin DNA (3′-terminal) for realizing enough stability and a strong driving force, and the suitable mismatches are listed in Tables S1 and S2.† Subsequently, to verify the high efficiency of the mismatch-fueled catalytic multiple-arm DNA junction assembly (M-CMDJA), as displayed in Scheme 2 (part A), we produced a series of CMDJAs consisting of substrate hairpin DNAs with a shorter neck (A), wild hairpin DNAs (B), hairpin DNAs with one mismatch (B-M1), and hairpin DNAs with two mismatches (B-M2), respectively. And the schematic illustrations of a further two typical CMDJAs (two-arm and three-arm) are also shown in part B and part C, separately.Efficiency comparison of annealing and self-assembly in the different M-CMDJA and CMDJAs
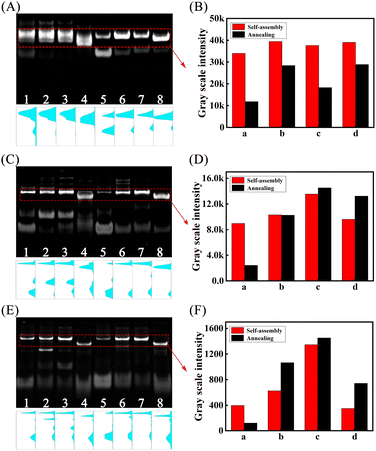
We first compared the yield of different multiple-arm DNA junctions by annealing and self-assembly respectively through PAGE experiments. As shown by the results in Fig. 1, with the introduction of mismatches into the hairpin DNA substrates, the corresponding yields of the multiple-arm DNA junction formed by either self-assembly (lanes 1–4) or annealing (lanes 5 and 6) were all dramatically improved (two-arm, Fig. 1A and B; three-arm, Fig. 1C and D; four-arm, Fig. 1E and F), demonstrating that the introduction of mismatches into hairpin DNA substrates can not only enhance the thermodynamic tendency of the self-assembly pathway, but also solve the problem of the free-energy landscapes with kinetic traps29 that cannot be addressed via conventional annealing methods.Kinetic performance comparison of the M-CMDJAs and CMDJAs under different conditions
To study the kinetic performance of the developed M-CMDJA accurately, we adopted real-time fluorescence analysis to characterize the corresponding reaction progress of the M-CMDJA and the CMDJA (Fig. 2). Through the fluorophore (FAM) and quencher (BHQ1) labeled on the ends of the hairpin DNA substrate (Table S1†), the corresponding fluorescence response increased as the multiple-arm DNA junction was formed. As shown by the results in Fig. 2 (Fig. 2A, two-arm; Fig. 2C, three-arm; Fig. 2E, four-arm), in the presence of miRNA-182-5P, the fluorescence response of the M-CMDJA (Scheme 2, part A, B-M1 and B-M2) increased much faster to a higher value beyond those of the wild CMDJA (Scheme 2, part A, A and B), manifesting the obviously improved kinetic and thermodynamical performance of the M-CMDJA. In order to obtain a comparison of the accurate kinetic parameters of these control groups, the reaction rate was obtained based on the value of fluorescence response intensity versus the time. The results in Fig. 2B (two-arm), Fig. 2D (three-arm), and Fig. 2F (four-arm) show that not only were the initial reaction rates of the M-CMDJA obviously faster than those of the wild CMDJA (Table 1), but the instantaneous reaction rates of the M-CMDJA in the reaction process were also improved to some extent. Subsequently, through the relationship of fluorescence response and fluorophore concentration depicted in Fig. S7,† the specific average reaction rates (the ratio between the resultant concentration and the reaction time) of the M-CMDJA were all faster than those of the wild CMDJA (Table 1) (the concentration of multiple-arm DNA junction assembled after 5200 s was computed based on Fig. 2). Impressively, compared with other comparison groups (two-arm DNA junction and three-arm DNA junction), the mismatch-fueled catalytic four-arm DNA junction assembly (M-CFDJA) with two mismatched bases possesses the best kinetic performance (Table 1, initial reaction rate: 274.31 a.u. s−1, average reaction rate: 1.97 × 10−10 M s−1). To explain this phenomenon, based on the conclusion that a stronger thermodynamic driving force from a more negative ΔG allows acceleration of the TSDR verified by previous works,30–32 we speculate two reasons as follows: (1) though the four-arm system requires more reaction steps that could dramatically decrease the reaction rate, its much stronger thermodynamic driving force beyond those of the two-arm system and three-arm system (Table S3†) offsets the kinetic decay, resulting in a just slightly faster reaction of the four-arm system (Table 1); (2) the target miRNA-182-5p as catalyst in the four-arm system could be recycled more easily to further accelerate the reaction of the four-arm system.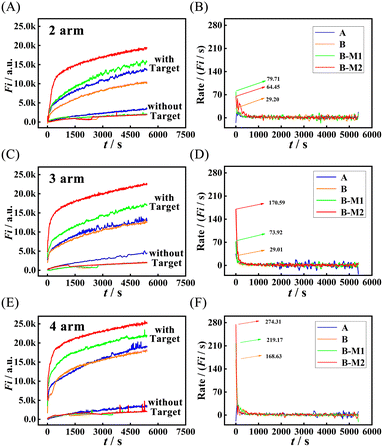 | ||
| Fig. 2 Real-time fluorescence responses of different M-CMDJAs and CMDJAs ((A) two-arm, (C) three-arm, and (E) four-arm) with and without the target miRNA-182-5P (1 nM) and the reaction rate of different M-CMDJA and CMDJAs ((B) two-arm, (D) three-arm, and (F) four-arm) in response to 1 nM target miRNA-182-5P (the meanings of A, B, B-M1, and B-M2 are shown in Scheme 2; EX WL: 492 nm, EM WL: 525 nm, gain: 75, time: 5200 s, interval: 15 s). | ||
| M-CMDJAs and CMDJAs | Comparison groups | Initial rate in fluorescence/(a.u. s−1) | Average rate/(M s−1) |
|---|---|---|---|
| Two-arm | Wild | 29.20 | 7.96 × 10−11 |
| Mismatched (B-M1) | 79.71 | 1.24 × 10−10 | |
| Mismatched (B-M2) | 64.45 | 1.51 × 10−10 | |
| Three-arm | Wild | 29.01 | 9.83 × 10−11 |
| Mismatched (B-M1) | 73.92 | 1.33 × 10−10 | |
| Mismatched (B-M2) | 170.59 | 1.77 × 10−10 | |
| Four-arm | Wild | 168.63 | 1.40 × 10−10 |
| Mismatched (B-M1) | 219.17 | 1.71 × 10−10 | |
| Mismatched (B-M2) | 274.31 | 1.97 × 10−10 |
Moreover, it is worth mentioning that the CMDJA not only hardly exhibited improved kinetic performance but also suffered from an extremely obvious background leakage when reducing the matched bases in the neck of the substrate hairpin DNAs (Scheme 2, part A, A) to make the ΔG more negative for obtaining a stronger driving force, which was also verified by the PAGE experiments shown in Fig. S6,† further demonstrating the superiority of introducing suitable mismatches in the M-CMDJA that can successfully improve the performance and decrease the background leakage simultaneously.
Conversion efficiency comparison of the M-CMDJAs and CMDJAs using fluorescence
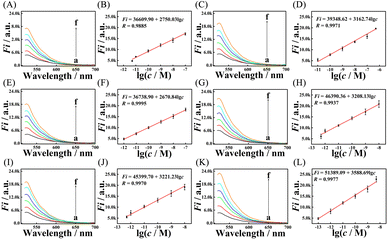
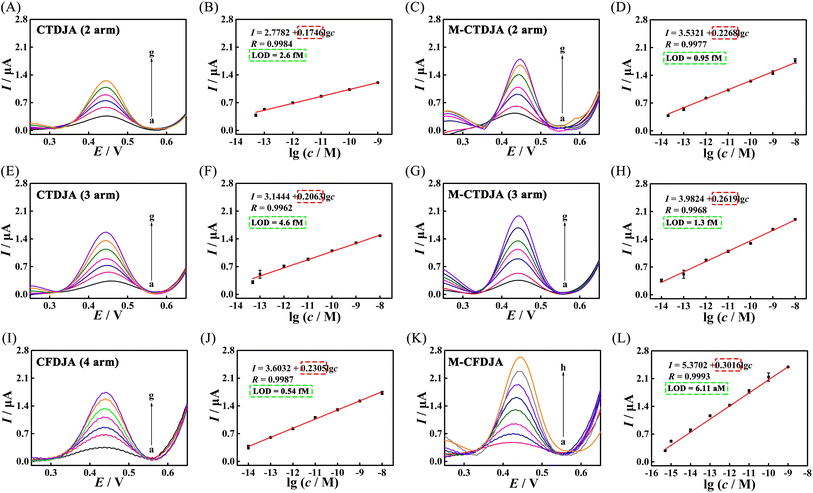
To further explore the conversion efficiency of the M-CMDJA and wild CMDJA, the relationships between fluorescence response value and concentration of the initiator miRNA-182-5P were studied respectively, and the results are shown in Fig. 3 (Fig. 3A–D, two-arm; Fig. 3E–H, three-arm; Fig. 3I–L, four-arm). When the concentration of miRNA-182-5P increased, the fluorescence intensity of these comparison groups all increased obviously and showed good linear relationships with the logarithm concentration. Among these results, the M-CFDJA showed the most sensitive response to miRNA-182-5P with the regression equation for the value of fluorescence response vs. c of Fi = 51389.09 + 3588.69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c (from 100 fM to 10 nM, R = −0.9977) (Fi: the fluorescence intensity, c: the miRNA concentration). Then, the accurate conversion efficiency (N) at different concentrations of initiator miRNA-182-5P in fluorescence was calculated (Table S4†) based on Fig. S7.† Impressively, the M-CFDJA obtained the highest conversion efficiency (NM-CFDJA-F: 1.96 × 106), almost five times that of the wild CFDJA (NCFDJA-F: 4.15 × 105) and at least four times that of the other comparison groups (two-arm and three-arm, Table 2), suggesting the outstanding thermodynamic performance of the M-CFDJA and that mismatch introduction could improve the thermodynamic performance of the CMDJA. These results reported above demonstrate that the developed M-CMDJA indeed excelled either with a faster reaction rate or a higher conversion efficiency compared to the wild CMDJA, precisely due to the introduction of mismatches in the substrate hairpin DNA that could obviously improve the driving force, promoting the whole hybridization reaction.
c (from 100 fM to 10 nM, R = −0.9977) (Fi: the fluorescence intensity, c: the miRNA concentration). Then, the accurate conversion efficiency (N) at different concentrations of initiator miRNA-182-5P in fluorescence was calculated (Table S4†) based on Fig. S7.† Impressively, the M-CFDJA obtained the highest conversion efficiency (NM-CFDJA-F: 1.96 × 106), almost five times that of the wild CFDJA (NCFDJA-F: 4.15 × 105) and at least four times that of the other comparison groups (two-arm and three-arm, Table 2), suggesting the outstanding thermodynamic performance of the M-CFDJA and that mismatch introduction could improve the thermodynamic performance of the CMDJA. These results reported above demonstrate that the developed M-CMDJA indeed excelled either with a faster reaction rate or a higher conversion efficiency compared to the wild CMDJA, precisely due to the introduction of mismatches in the substrate hairpin DNA that could obviously improve the driving force, promoting the whole hybridization reaction.
| Method | Wild CMDJA | M-CMDJA |
|---|---|---|
| Two-arm | 1.44–1.95 × 104 | 8.26–4.37 × 104 |
| Three-arm | 7.69–1.96 × 105 | 8.79 × 101 to 5.00 × 105 |
| Four-arm | 8.08 × 101 to 4.15 × 105 | 1.92 × 102 to 1.96 × 106 |
Conversion efficiency comparison of the M-CMDJAs and CMDJAs in electrochemistry
In order to apply the M-CMDJA to develop an ultrasensitive electrochemical biosensing platform, we explored the response of a biosensing platform based on different CMDJAs and M-CMDJAs to miRNA-182-5P at different concentrations, respectively (Fig. 4). Next, the corresponding conversion efficiency using the electrochemistry method (Table S5†) was also computed based on the relationship of the electrochemical signal response with the concentration of multiple-arm DNA junction (Fig. S8†). Significantly, the M-CFDJA (four-arm) obtained the highest conversion efficiency (NM-CFDJA-E: 1.05 × 108), almost eight times that of the wild CFDJA (NCFDJA-E: 1.38 × 107, Table 3).| Method | Wild | Mismatched |
|---|---|---|
| Two-arm | 6.14 × 102 to 1.88 × 106 | 9.09 × 101 to 4.75 × 106 |
| Three-arm | 6.65 × 101 to 1.39 × 106 | 9.04 × 101 to 5.93 × 106 |
| Four-arm | 5.78 × 102 to 1.38 × 107 | 8.38 × 102 to 1.05 × 108 |
Moreover, the conversion efficiency of the M-CFDJA obtained using the electrochemistry method was nearly two orders of magnitude that obtained using the fluorescence method (NM-CFDJA-F: 1.96 × 106, Table 2), which was because of the high sensitivity of the electrochemistry method and the probably fewer interferences of the biphasic electrode operation.33 These results reported above confirm that the M-CMDJA has obviously surpassed the wild CMDJA in both kinetic and thermodynamic performance, enabling it to be further used in electrochemical biosensing platform construction for ultrasensitive miRNA detection.
Additionally, we chose some conventional isothermal amplification methods as control groups and compared their corresponding performance. As the results show in Table 4, the M-CFDJA possesses the lowest reaction time, and widest range and highest conversion efficiency even at a moderate temperature, breaking the bottleneck of long reaction times and low conversion efficiencies in conventional NASA.
Application evaluation of the M-CFDJA in cancer biomarker miRNA detection
As a practical application, the M-CFDJA was used as an efficient DNA signal amplifier to develop an ultrasensitive electrochemical biosensing platform for rapid miRNA-182-5P detection. Firstly, the successful construction of the biosensing platform was verified through CV (Fig. S3A and S4†), EIS (Fig. S3B†), and CC experiments (Fig. S5†). Then, after the reaction time was optimized (Fig. S9†), the prepared electrochemical biosensing platform displayed a sensitive response to miRNA-182-5P, as shown in Fig. 4K and L, and the corresponding regression equation is I = 0.3016![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c + 5.3702 (cmiRNA-182-5P: 0.5 fM–1 nM, R = 0.9993). The corresponding detection limit was as found to be low as 6.11 aM for miRNA-182-5P, which is 88.4-fold lower than that of the electrochemical biosensor constructed with the wild CFDJA (0.54 fM) (Table S6†), showing the excellent sensitivity of the biosensor constructed based on the M-CFDJA. Moreover, as shown in Table 5, the proposed sensing platform showed quite a wide linear range and a much lower detection limit compared to those of other biosensors applied for nucleic acid detection, which was ascribed to the versatile nature of the M-CFDJA with hyper conversion efficiency.
c + 5.3702 (cmiRNA-182-5P: 0.5 fM–1 nM, R = 0.9993). The corresponding detection limit was as found to be low as 6.11 aM for miRNA-182-5P, which is 88.4-fold lower than that of the electrochemical biosensor constructed with the wild CFDJA (0.54 fM) (Table S6†), showing the excellent sensitivity of the biosensor constructed based on the M-CFDJA. Moreover, as shown in Table 5, the proposed sensing platform showed quite a wide linear range and a much lower detection limit compared to those of other biosensors applied for nucleic acid detection, which was ascribed to the versatile nature of the M-CFDJA with hyper conversion efficiency.
| Analytical methods | Linear range | Detection limit | Ref. |
|---|---|---|---|
| Fluorescence | 1 pM to 10 nM | 0.35 pM | 44 |
| Electrochemiluminescence | 10 fM to 0.1 nM | 6.6 fM | 45 |
| Chronocoulometry | 2.0 fM to 1.0 nM | 2.0 fM | 46 |
| Electrochemical | 0.14 nM to 10 nM | 40 pM | 47 |
| Electrochemical | 1 pM to 1 nM | 10 pM | 48 |
| Electrochemical | 0.5 fM to 1 nM | 6.11 aM | This work |
Moreover, the biosensing platform constructed using the evolved M-CFDJA also showed outstanding reproducibility (Fig. S10A†), selectivity (Fig. S10B†), and stability (Fig. S10C†). Importantly, as shown in Fig. S10D,† the feasibility of the prepared sensing platform for the detection of miRNA-182-5P in practical samples was studied using total RNA extracted from the human cancer cells 22RV1, A549, and HeLa. The obtained results (Fig. S10D†) were in line with the previous reports,49,50 suggesting the superiority of the M-CFDJA in ultrasensitive biosensor development for rapid miRNA detection from cancer cells.
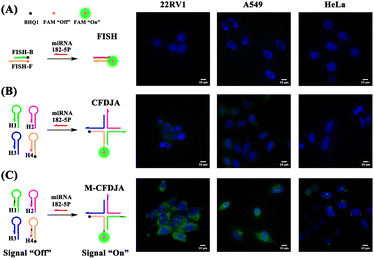
Total internal reflection fluorescence microscope (TIRFM) imaging of miRNAs in living cancer cells
In order to verify the outstanding performance of the M-CFDJA for imaging of cancer biomarker miRNA in living cells, we then applied the system to image and monitor miRNA-182-5P in living cells using the TIRFM. 22RV1 cells (high expression of miRNA-182-5P), A549 cells (high expression of miRNA-182-5P), and HeLa cells (low expression of miRNA-182-5P) were chosen to investigate the capability of the M-CFDJA-mediated intracellular miRNA-182-5P imaging strategy compared with the wild CFDJA and conventional fluorescence in situ hybridization (FISH) method. As shown by the results in Fig. 5, the M-CFDJA displayed quite a strong fluorescence response in 22RV1 cells and A549 cells, whereas nearly no fluorescence response (FAM, green) of the FISH system and a weak fluorescence response of the CMDJA in 22RV1 cells and A549 cells could be observed, respectively. Considering these results, it can be concluded that the M-CFDJA system could convert miRNA in living cells into reaction products with higher concentration in comparison with the traditional FISH method and wild CMDJA method, suggesting that the M-CFDJA can still achieve excellent performance for ultrasensitive miRNA detection in living cells.Conclusions
In conclusion, unlike the conventional NASA, the mismatch-fueled catalytic four-arm DNA junction assembly (M-CFDJA) we proposed realizes an improved driving force by introducing suitable mismatches in reactant DNA, which lead to stronger kinetic and thermodynamic trends realizing fast reaction rates and outstanding conversion efficiency. Based on these advantages, the M-CFDJA is successfully used as an efficient DNA signal amplifier in an electrochemical biosensor for the rapid and highly sensitive detection of miRNA-21 and the highly sensitive intracellular imaging of miRNA, providing an ingenious approach to explore potential strategies for ultimate application in biosensing detection and clinical diagnoses. Moreover, the M-CFDJA provides novel insight into the investigation of high-efficiency nucleic acid signal amplification methods for biological research and DNA nanobiotechnology.In comparison with other catalytic DNA circuit systems, like HCR,51 CHA,52 and EFTRA,53 the M-CFDJA we proposed could facilely generate much more stable and programmable DNA nanostructure units with faster reaction rates and higher amplification efficiency, from which not only a higher sensitivity of a biosensor based on the M-CFDJA can be realized, but also the obtained DNA nanostructure unit possesses significant potential for application in other areas of DNA nanodevices54 and biochemical research.2 Nevertheless, the M-CFDJA system still suffers from an inherently low driving force, which largely depends on the random collisions in Brownian motion and the difference between the reaction standard free energy before and after reaction32 (in comparison with the enzyme-driven strategies NASBA,55 HRCA,56 and LAMP57), limiting further improvement of its amplification performance. Additionally, when used in live cell analysis, the M-CFDJA system can only be delivered into cells via complicated liposome-based transfection, from which the limited quantity of transfected probes may restrict the sensitivity.58–60
In order to address these limitations, some possible methods as follows may be adopted in the future: (1) introducing some energy sources as an additional driving force, such as chemical fuels,61,62 magnetic fields,63 acoustic fields,64 electric fields,65 and light,66 which may endow the M-CFDJA system with strong thermodynamical and dynamical favorability; (2) choosing alternative measures to avoid the complicated liposome-based transfection step in the delivery of nucleic acid probes into live cells, like constructing functional DNA nanostructures67,68 and combining DNA with biocompatible materials,69,70 which have much better cell internalization efficiencies via macropinocytosis and caveolae-mediated endocytosis pathways. In this way, it may be possible to further improve the comprehensive performance of the M-CFDJA for ultimate application in biochemical and medical fields.
Data availability
Research data are not shared.Author contributions
Xiao-Long Zhang and Sha-Sha Li conceived the project. Xiao-Long Zhang and Sha-Sha Li designed the sequences, performed the experiments and analyzed the data. Ruo Yuan supervised the project. Xiao-Long Zhang wrote the paper. Xiao-Long Zhang, Ruo Yuan, and Ya-Qin Chai commented and revised the paper. Sha-Sha Li, Wei-Wei Liu, and Ling-Qi Kong assisted in revising the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This paper was financially supported by the National Natural Science Foundation of China (22174113, 22176153, and 21974108), and the Fundamental Research Funds for the Central Universities (XDJK2020TY002), China.References
- W. H. Fu, L. L. Tang, G. H. Wei, L. Fang, J. Zeng, R. J. Zhan, X. M. Liu, H. Zuo, C. Z. Huang and C. D. Mao, Rational design of pH-responsive DNA motifs with general sequence compatibility, Angew. Chem., Int. Ed., 2019, 58, 16405–16410 CrossRef CAS.
- Y. H. Dong, C. Yao, Y. Zhu, L. Yang, D. Luo and D. Y. Yang, DNA functional materials assembled from branched DNA: design, synthesis, and applications, Chem. Rev., 2020, 120, 9420–9481 CrossRef CAS PubMed.
- F. A. Aldaye, A. L. Palmer and H. F. Sleiman, Assembling materials with DNA as the guide, Science, 2008, 321, 1795–1799 CrossRef CAS.
- Y. Hu and C. M. Niemeyer, From DNA nanotechnology to material systems engineering, Adv. Mater., 2019, 31, 1806294 CrossRef.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- S. Loescher, S. Groeer and A. Walther, 3D DNA origami nanoparticles: from basic design principles to emerging applications in soft matter and (bio-) nanosciences, Angew. Chem., Int. Ed., 2018, 57, 10436–10448 CrossRef CAS.
- E. Kopperger, J. List, S. Madhira, F. Rothfischer, D. C. Lamb and F. C. Simmel, A self-assembled nanoscale robotic arm controlled by electric fields, Science, 2018, 359, 296–300 CrossRef CAS.
- F. N. Gür, S. Kempter, F. Schueder, C. Sikeler, M. J. Urban, R. Jungmann, P. C. Nickels and T. Liedl, Double- to single-strand transition induces forces and motion in DNA origami nanostructures, Adv. Mater., 2021, 33, 2101986 CrossRef.
- P. Yin, H. M. Choi, C. R. Calvert and N. A. Pierce, Programming biomolecular self-assembly pathways, Nature, 2008, 451, 318–322 CrossRef CAS.
- R. Liu, S. B. Zhang, T. T. Zheng, Y. R. Chen, J. T. Wu and Z. S. Wu, Intracellular nonenzymatic in situ growth of three-dimensional DNA nanostructures for imaging specific biomolecules in living cells, ACS Nano, 2020, 14, 9572–9584 CrossRef CAS PubMed.
- U. Feldkamp and C. M. Niemeyer, Rational design of DNA nanoarchitectures, Angew. Chem., Int. Ed., 2006, 45, 1856–1876 CrossRef CAS PubMed.
- J. Zheng, J. J. Birktoft, Y. Chen, T. Wang, R. Sha, P. E. Constantinou, S. L. Ginell, C. Mao and N. C. Seeman, From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal, Nature, 2009, 461, 74–77 CrossRef CAS.
- Q. Q. Hu, H. Li, L. H. Wang, H. Z. Gu and C. H. Fan, DNA nanotechnology-enabled drug delivery systems, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed.
- P. S. Kwon, S. Ren, S. J. Kwon, M. E. Kizer, L. Kuo, M. Xie, D. Zhu, F. Zhou, F. Zhang, D. Kim, K. Fraser, L. D. Kramer, N. C. Seeman, J. S. Dordick, R. J. Linhardt, J. Chao and X. Wang, Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition, Nat. Chem., 2020, 12, 26–35 CrossRef CAS PubMed.
- S. K. Ren, K. Fraser, L. L. Kuo, N. Chauhan, A. T. Adrian, F. M. Zhang, R. J. Linhardt and P. S. Kwon, Designer DNA nanostructures for viral inhibition, Nat. Protoc., 2022, 17, 282–326 CrossRef CAS.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS.
- H. Kang, T. Lin, X. J. Xu, Q. S. Jia, R. Lakerveld and B. Wei, DNA dynamics and computation based on toehold-free strand displacement, Nat. Commun., 2021, 12, 4994 CrossRef CAS.
- B. Yurke, A. J. Turberfield, A. P. Mills, F. C. Simmel and J. L. Neumann, A DNA-fueled molecular machine made of DNA, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- A. L. Prinzen, D. Saliba, C. Hennecker, T. Trinh, A. Mittermaier and H. F. Sleiman, Amplified self-immolative release of small molecules by spatial isolation of reactive groups on DNA-minimal architectures, Angew. Chem., Int. Ed., 2020, 59, 2–11 CrossRef.
- F. Li, Z. Y. Lv, X. Zhang, Y. H. Dong, X. H. Ding, Z. M. Li, S. Li, C. Yao and D. Y. Yang, Supramolecular self-assembled DNA nanosystem for synergistic chemical and gene regulations on cancer cells, Angew. Chem., Int. Ed., 2021, 60, 25557–25566 CrossRef CAS.
- J. Deng and A. Walther, Fuel-driven transient DNA strand displacement circuitry with self-resetting function, J. Am. Chem. Soc., 2020, 142, 21102–21109 CrossRef CAS PubMed.
- A. J. Genot, D. Y. Zhang, J. Bath and A. J. Turbereld, Remote toehold: a mechanism for flexible control of DNA hybridization kinetics, J. Am. Chem. Soc., 2011, 133, 2177–2182 CrossRef CAS.
- D. Y. Zhang and E. Winfree, Control of DNA strand displacement kinetics using toehold exchange, J. Am. Chem. Soc., 2009, 131, 17303–17314 CrossRef CAS PubMed.
- J. S. Wang and D. Y. Zhang, Simulation-guided DNA probe design for consistently ultraspecific hybridization, Nat. Chem., 2015, 7, 545–553 CrossRef CAS.
- C. Wu, S. Cansiz, L. Zhang, I. Teng, L. Qiu, J. Li, Y. Liu, C. Zhou, R. Hu, T. Zhang, C. Cui, L. Cui and W. Tan, A nonenzymatic hairpin DNA cascade reaction provides high signal gain of mRNA imaging inside live cells, J. Am. Chem. Soc., 2015, 137, 4900–4903 CrossRef CAS.
- Z. K. Wu, H. H. Fan, N. S. R. Satyavolu, W. J. Wang, R. Lake, J. H. Jiang and Y. Lu, Imaging Endogenous Metal Ions in Living Cells Using a DNAzyme-Catalytic Hairpin Assembly Probe, Angew. Chem., Int. Ed., 2017, 56, 8721–8725 CrossRef CAS.
- W. H. Dai, H. F. Dong, K. K. Guo and X. J. Zhang, Near-infrared triggered strand displacement amplification for MicroRNA quantitative detection in single living cells, Chem. Sci., 2018, 9, 1753–1759 RSC.
- Y. Li, G. A. Wang, S. D. Mason, X. Yang, Z. Yu, Y. Tang and F. Li, Simulation-guided engineering of an enzyme-powered three dimensional DNA nanomachine for discriminating single nucleotide variants, Chem. Sci., 2018, 9, 6434–6439 RSC.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Programming biomolecular self-assembly pathways, Nature, 2008, 451, 318–323 CrossRef CAS PubMed.
- N. Srinivas, T. E. Ouldridge, P. Šulc, J. M. Schaeffer, B. Yurke, A. A. Louis, J. P. K. Doye and E. Winfree, On the biophysics and kinetics of toehold-mediated DNA strand displacement, Nucleic Acids Res., 2013, 41, 10641–10658 CrossRef CAS PubMed.
- N. E. C. Haley, T. E. Ouldridge, I. M. Ruiz, A. Geraldini, A. A. Louis, J. Bath and A. J. Turberfield, Design of hidden thermodynamic driving for non-equilibrium systems via mismatch elimination during DNA strand displacement, Nat. Commun., 2020, 11, 2562 CrossRef CAS PubMed.
- P. Irmisch, T. E. Ouldridge and R. Seidel, Modeling DNA-strand displacement reactions in the presence of base-pair mismatches, J. Am. Chem. Soc., 2020, 142, 11451–11463 CrossRef CAS PubMed.
- S. Liu, L. Fang, Y. Wang and L. Wang, Universal dynamic DNA assembly-programmed surface hybridization effect for single-step, reusable, and amplified electrochemical nucleic acid biosensing, Anal. Chem., 2017, 89, 3108–3115 CrossRef CAS PubMed.
- G. T. Walker, M. S. Fraiser, J. L. Schram, M. C. Little, J. G. Nadeau and D. P. Malinowski, Strand displacement amplification—an isothermal, in vitro DNA amplification technique, Nucleic Acids Res., 1992, 20, 1691–1696 CrossRef CAS PubMed.
- C. Shi, Q. Liu, C. Ma and W. Zhong, Exponential strand-displacement amplification for detection of microRNAs, Anal. Chem., 2014, 86, 336–339 CrossRef CAS.
- T. Murakami and J. Sumaoka, Sensitive isothermal detection of nucleic-acid sequence by primer generation–rolling circle amplification, Nucleic Acids Res., 2009, 37, e19 CrossRef.
- T. Murakami, J. Sumaoka and M. Komiyama, Sensitive RNA detection by combining three-way junction formation and primer generation-rolling circle amplification, Nucleic Acids Res., 2012, 40, e22 CrossRef CAS.
- M. Vincent, Y. Xu and H. Kong, Helicase-dependent isothermal DNA amplification, EMBO Rep., 2004, 5, 795–800 CrossRef CAS.
- A. Cao and C. Y. Zhang, Real-Time detection of transcription factors using target-converted helicase-dependent amplification assay with zero-background signal, Anal. Chem., 2013, 85, 2543–2547 CrossRef CAS PubMed.
- F. B. Dean, S. Hosono, L. Fang, X. Wu, A. F. Faruqi, P. BrayWard, Z. Sun, Q. Zong, Y. Du, J. Du, M. Driscoll, W. Song, S. F. Kingsmore, M. Egholm and R. S. Lasken, Comprehensive human genome amplification using multiple displacement amplification, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5261–5266 CrossRef CAS PubMed.
- W. Henke, K. Herdel, K. Schnorr, D. Jung and S. A. Loening, Betaine improves the PCR amplification of GC-rich DNA sequences, Nucleic Acids Res., 1997, 25, 3957–3958 CrossRef CAS.
- Y. Jiang, B. Li, J. N. Milligan, S. Bhadra and A. D. Ellington, Real-Time detection of isothermal amplification reactions with thermostable catalytic hairpin assembly, J. Am. Chem. Soc., 2013, 135, 7430–7433 CrossRef CAS.
- Z. Wu, H. Fan, N. R. Satyavolu, W. Wang, R. Lake, J. Jiang and Y. Lu, Imaging endogenous metal ions in living cells using a DNAzyme–catalytic hairpin assembly probe, Angew. Chem., 2017, 129, 8847–8851 CrossRef.
- Z. Z. Yang, X. Peng, P. Yang, Y. Zhuo, Y. Q. Chai, W. B. Liang and R. Yuan, A Janus 3D DNA nanomachine for simultaneous and sensitive fluorescence detection and imaging of dual microRNAs in cancer cells, Chem. Sci., 2020, 11, 8482–8488 RSC.
- P. Zhang, J. Jiang, R. Yuan, Y. Zhuo and Y. Q. Chai, Highly ordered and field-free 3D DNA nanostructure: the next generation of DNA nanomachine for rapid single-step sensing, J. Am. Chem. Soc., 2018, 140, 9361–9364 CrossRef CAS PubMed.
- C. Fang, K. Kim, B. Yu, S. Jon, M. Kim and H. Yang, Ultrasensitive electrochemical detection of miRNA-21 using a zinc finger protein specific to DNA–RNA hybrids, Anal. Chem., 2017, 89, 2024–2031 CrossRef CAS PubMed.
- A. D. Castañeda, N. J. Brenes, A. Kondajji and R. M. Crooks, Detection of microRNA by electrocatalytic amplification: a general approach for single-particle biosensing, J. Am. Chem. Soc., 2017, 139, 7657–7664 CrossRef PubMed.
- R. Bruch, J. Baaske, C. Chatelle, M. Meirich, S. Madlener, W. Weber, C. Dincer and G. A. Urban, CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics, Adv. Mater., 2019, 31, 1905311 CrossRef CAS.
- L. Yang, Y. Dou, Z. X. Sui, H. Cheng, X. Liu, Q. L. Wang, P. F. Gao, Y. Qu and M. Xu, Upregulated miRNA-182-5p expression in tumor tissue and peripheral blood samples from patients with non-small cell lung cancer is associated with downregulated Caspase 2 expression, Exp. Ther. Med., 2020, 19, 603–610 CAS.
- C. H. Gu, K. Y. Zhao, N. C. Zhou, F. Liu, F. Xie, S. L. Yu, Y. J. Feng, L. Chen, J. J. Yang, F. Y. Tian and G. S. Jiang, UBAC2 promotes bladder cancer proliferation through BCRC-3/miRNA-182-5p/p27 axis, Cell Death Dis., 2020, 11, 733 CrossRef CAS PubMed.
- H. M. Wang, C. X. Li, X. Q. Liu, X. Zhou and F. A. Wang, Construction of an enzyme-free concatenated DNA circuit for signal amplification and intracellular imaging, Chem. Sci., 2018, 9, 5842–5849 RSC.
- H. Wang, H. M. Wang, Q. Wu, M. J. Liang, X. Q. Liu and F. A. Wang, A DNAzyme-amplified DNA circuit for highly accurate microRNA detection and intracellular imaging, Chem. Sci., 2019, 10, 9597–9604 RSC.
- X. L. Zhang, Z. H. Yang, Y. Y. Chang, D. Liu, Y. R. Li, Y. Q. Chai, Y. Zhuo and R. Yuan, Programmable mismatch-fueled high-efficiency DNA signal converter, Chem. Sci., 2020, 11, 148–153 RSC.
- S. L. Liu, Q. Jiang, X. Zhao, R. F. Zhao, Y. N. Wang, Y. M. Wang, J. B Liu, Y. X. Shang, S. Zhao, T. T. Wu, Y. L. Zhang, G. J. Nie and B. Q. Ding, A DNA nanodevice-based vaccine for cancer immunotherapy, Nat. Mater., 2021, 20, 421–430 CrossRef CAS PubMed.
- J. C. Guatelli, K. M. Whitfield, D. Y. Kwoh, K. J. Barringer, D. D. Richman and T. R. Gingeras, Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 1874–1878 CrossRef CAS PubMed.
- P. M. Lizardi, X. Huang, Z. Zhu, P. Bray-Ward, D. C. Thomas and D. C. Ward, Mutation detection and single-molecule counting using isothermal rolling-circle amplification, Nat. Genet., 1998, 19, 225–232 CrossRef CAS PubMed.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products, Nat. Protoc., 2008, 3, 877–882 CrossRef CAS PubMed.
- Z. Cheglakov, T. M. Cronin, C. He and Y. Weizmann, Live cell microRNA imaging using cascade hybridization reaction, J. Am. Chem. Soc., 2015, 137, 6116–6119 CrossRef CAS.
- C. Xue, S. X. Zhang, C. H. Ouyang, D. Chang, B. J. Salena, Y. F. Li and Z. S. Wu, Target-induced catalytic assembly of Y-shaped DNA and its application for in situ imaging of microRNAs, Angew. Chem., 2018, 130, 9887–9891 CrossRef.
- X. D. Meng, W. H. Dai, K. Zhang, H. F. Dong and X. J. Zhang, Imaging multiple microRNAs in living cells using ATP self-powered strand-displacement cascade amplification, Chem. Sci., 2018, 9, 1184–1190 RSC.
- S. Sengupta, K. K. Dey, H. S. Muddana, T. Tabouillot, M. E. Ibele, P. J. Butler and A. Sen, Enzyme molecules as nanomotors, J. Am. Chem. Soc., 2013, 135, 1406–1414 CrossRef CAS PubMed.
- Y. Yuan, C. Du, C. J. Sun, J. Zhu, S. Wu, Y. L. Zhang, T. J. Ji, J. L. Lei, Y. M Yang, N. Gao and G. J. Nie, Chaperonin-GroEL as a smart hydrophobic drug delivery and tumor targeting molecular machine for tumor therapy, Nano Lett., 2018, 18, 921–928 CrossRef CAS PubMed.
- W. Gao, X. Feng, A. Pei, C. R. Kane, R. Tam, C. Hennessy and J. Wang, Bioinspired helical microswimmers based on vascular plants, Nano Lett., 2014, 14, 305–310 CrossRef CAS PubMed.
- W. Wang, S. Li, L. Mair, S. Ahmed, T. J. Huang and T. E. Mallouk, Acoustic propulsion of nanorod motors inside living cells, Angew. Chem., 2014, 126, 3265–3268 CrossRef.
- G. Loget and A. Kuhn, Electric field-induced chemical locomotion of conducting objects, Nat. Commun., 2011, 2, 535 CrossRef.
- H. Q. Chu, J. Zhao, Y. S. Mi, Y. L. Zhao and L. L. Li, Near-infrared light-initiated hybridization chain reaction for spatially and temporally resolved signal amplification, Angew. Chem., Int. Ed., 2019, 58, 14877–14881 CrossRef CAS PubMed.
- Y. F. Lv, R. Hu, G. Z. Zhu, X. B. Zhang, L. Mei, Q. L. Liu, L. P. Qiu, C. C. Wu and W. H. Tan, Preparation and biomedical applications of programmable and multifunctional DNA nanoflowers, Nat. Protoc., 2015, 10, 1508–1524 CrossRef CAS.
- Q. L. Huang, B. Chen, J. L. Shen, L. Liu, J. J. Li, J. Y. Shi, Q. Li, X. L. Zuo, L. H. Wang, C. H. Fan and J. Li, Encoding fluorescence anisotropic barcodes with DNA frameworks, J. Am. Chem. Soc., 2021, 143, 10735–10742 CrossRef CAS PubMed.
- J. Zhao, H. Q. Chu, Y. Zhao, Y. Lu and L. L. Li, A NIR light gated DNA nanodevice for spatiotemporally controlled imaging of microRNA in cells and animals, J. Am. Chem. Soc., 2019, 141, 7056–7062 CrossRef CAS.
- N. Yan, X. J. Wang, L. Lin, T. J. Song, P. J. Sun, H. Y. Tian, H. J. Liang and X. S. Chen, Gold nanorods electrostatically binding nucleic acid probe for in vivo microRNA amplified detection and photoacoustic imaging-guided photothermal therapy, Adv. Funct. Mater., 2018, 28, 1800490 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section and related experimental data. See https://doi.org/10.1039/d2sc04814k |
| ‡ X. L. Zhang and S. S. Li contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |