Open Access Article
Open Access ArticleMotif-based zwitterionic peptides impact their structure and immunogenicity†
Patrick
McMullen
 a,
Qi
Qiao
b,
Sijin
Luozhong
a,
Lirong
Cai
b,
Liang
Fang
a,
Qing
Shao
a,
Qi
Qiao
b,
Sijin
Luozhong
a,
Lirong
Cai
b,
Liang
Fang
a,
Qing
Shao
 *b and
Shaoyi
Jiang
*b and
Shaoyi
Jiang
 *a
*a
aDepartment of Biomedical Engineering, Cornell University, Ithaca, NY 14853, USA. E-mail: sj19@cornell.edu
bDepartment of Chemical and Materials Engineering, University of Kentucky, Lexington, KY 40506, USA. E-mail: qshao@uky.edu
First published on 7th September 2022
Abstract
The linkage of zwitterionic peptides containing alternating glutamic acid (E) and lysine (K) amino acids exhibits protective effects on protein drugs due to their high hydration capacity. Previously, short EK peptides covering the surface of a protein drug showed significant protective effects and low immunogenicity. However, for high-molecular-weight single-chain (HMWSC) zwitterionic peptides, the incorporation of structure-disrupting amino acids such as proline (P), serine (S), and glycine (G) is necessary to improve their protective ability. Herein, we first probe the immunogenicity of eight EK-containing motif-based peptides, six of which incorporate structure-disrupting amino acids P, S, and G, linked to keyhole limpet hemocyanin (KLH). These studies uncover two sequence motifs, EKS and EKG, which show uniquely higher immunogenicity, while the other motifs, especially those containing P, exhibit lower immunogenicity. Additionally, the structure and dynamics of these sequence motifs are computationally modeled by Rosetta protein predictions and molecular dynamics (MD) simulations to predict properties of higher and lower immunogenicity peptides. These simulations revealed peptides with higher immunogenicity, namely EKS and EKG, exhibit regions of charge imbalance. Then, HMWSC zwitterionic sequences were linked to a typical protein drug, interferon-alpha 2a (IFN), which showed consistent immunogenic behaviors. Finally, epitope mapping and alanine scanning experiments using the serum collected from mice injected with HMWSC sequences also implicated a link between charge imbalance and peptide immunogenicity.
Introduction
The immunogenicity of many therapeutics, including therapeutic proteins, is a key determinant of their safety and efficacy. Immune responses triggered by protein drugs have been shown to induce anti-drug antibodies (ADAs), which can provoke dangerous anaphylactic reactions.1–3 Furthermore, ADAs can facilitate the clearance of therapeutic proteins reducing their circulation time in the body following repeated administration, termed accelerated blood clearance (ABC) effects, thus rendering them ineffective.1,4–6 Zwitterionic molecules containing alternating positive and negative charge domains have a uniquely strong affinity for water and have been shown to inhibit biological interactions due to the formation of a surrounding water barrier functioning to protect the molecule.7–10 Even so, for zwitterionic molecules, charge balance is a critical requirement as a charge imbalance could result in unwanted electrostatic interactions.11,12 Notwithstanding, the zwitterionic approach has proved an invaluable approach. For example, conjugation or encapsulation of protein drugs with zwitterionic poly(carboxybetaine) (PCB) ameliorated the formation of ADAs and did not show ABC effects.13–15Zwitterionic peptides offer another promising approach to protect protein drugs as peptides are biodegradable and can be precisely controlled for their sequences and lengths. Chemical modification of the protein drug asparaginase (Asp) using short zwitterionic peptides containing alternating glutamic acid (E) and lysine residues (K) showed no ABC effects and low immunogenicity.16 As Asp functions via an enzymatic mechanism, its modification with short EK peptides can tolerate complete coating of the Asp surface to produce its protective effects, while still allowing the small-molecule substrate to diffuse to the active site. On the contrary, protein drugs that rely on receptor binding for bioactivity cannot depend upon complete surface coverage because the active site must exhibit significantly greater surface exposure to allow for receptor–ligand interactions.17–19 Therefore, for this class of protein drugs, a single high molecular weight EK peptide is required to provide steric protection, while still allowing access to the active site.
Previously, a high-molecular-weight single-chain (HMWSC) EK peptide was linked to several proteins, which showed stabilizing effects.20 However, further computational and experimental studies uncovered this peptide exhibits alpha-helical and beta-twist structural propensities to a certain degree, which could limit its radius of gyration and surface protection.21,22 While conjugation of short EK peptides evades this concern using multiple conjugation sites, a HMWSC EK peptide could improve its protection by altering its sequence to eliminate these structural tendencies. One such approach is to incorporate structure-disrupting amino acids, such as proline (P), glycine (G), or serine (S), into the primary amino acid sequence,23–25 expanding the possible sequence space. Even so, P, S, and G induce unstructured conformations by unique impacts on the peptide backbone, which could result in rotation of the charged side chains on E and K in unpredicted directions. This unpredictable behavior could lead to charge imbalances and, thus, charge-driven interactions, including immune responses.26–28 These complications warrant a systematic investigation to uncover zwitterionic sequences with favorable behaviors.
Herein, we investigate the immunogenicity of eight motif-based zwitterionic peptides, six of which contain P, S, and G. We conducted computational simulations of zwitterionic peptide motifs to predict properties that of higher and lower immunogenicity motifs. Then, we focus on a few HMWSC EK peptides with P, S, and G linked to a therapeutic protein to assess their applicability.
Results
To examine the immunogenicity of zwitterionic peptides containing the structure-disrupting amino acids, we designed distinct peptide motifs with proline (P), serine (S), and glycine (G). These amino acids disrupt structural tendencies by different mechanisms such as increasing backbone stiffness (P), disrupting hydrogen bonding patterns (S), and increasing backbone flexibility (G).28–32 By systematically examining different sequence motifs with variable amounts of structure-disrupting amino acids, we expected to uncover trends in their immunogenicity. We subsequently modeled these peptides by Rosetta and MD simulations to extract molecular properties correlating with their immunogenicity. Then, we designed three HMWSC zwitterionic sequences linked to IFN, which showed similar immunogenicity behavior as their low molecular weight counterparts. Finally, we examined the serum from mice injected with HMWSC zwitterionic sequences by epitope mapping and alanine scanning to understand the contributions of individual amino acids toward antibody binding.Effects of structure-disrupting amino acids on zwitterionic peptide immunogenicity
To systematically and directly probe the immunogenicity of zwitterionic peptide sequences with structure-disrupting amino acids, eight peptide sequences were designed based on the following sequence motifs: EKX (i, i + 1) and EXKX (i, i + 2) in which X is either P, S, or G (Fig. 1a, Tables S1 and S2†). As well, sequences without structure-disrupting amino acids were designed with the following motifs: EK (i, i + 1) and EEKK (i, i + 2) (Fig. 1a, Tables S1 and S2†). These sequences were designed to test the effects of structure-disrupting amino acids as well as E and K distance in the primary sequence. The distances in oppositely charged E and K residues in the primary amino acid sequence are denoted by i, i + 1, in which E is N-terminally adjacent to K with or without a structure disrupting amino acid C terminally adjacent to K, and i, i + 2, in which E and K are separated by a structure disrupting amino acid on both of their adjacent N-terminal and C-terminal residues. The caveat to this notation is that EEKK, without a structure disrupting amino acid, also contains unavoidable adjacent E and K residues. However, as EEKK contains adjacent E residues and adjacent K residues, we reasoned the i, i + 2 classification to be more appropriate. These peptides were synthesized by solid-phase peptide synthesis to be 12 amino acids long by repeating each motif. Subsequently, zwitterionic peptides were conjugated to Keyhole Limpet Hemacyanogen (KLH) proteins by a free cysteine included on the C terminus of these peptides using maleimide conjugation chemistry. 30 μg of the resulting KLH peptide conjugates were injected into the subcutaneous tissue of C57BL/6 mice (5 per group). Boosters of the same dose and administration route were injected two weeks following the first injection. One week following the booster dose, blood was recovered to determine antibody titers and assess peptide immunogenicity.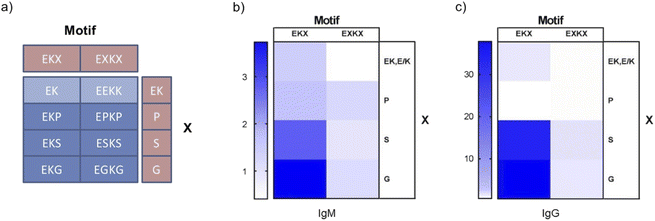 | ||
| Fig. 1 Immunogenicity of i, i + 1 and i, i + 2 EK peptide motifs containing P, S, and G. (a) Peptide motif-based design of 8 zwitterionic peptides of 12 amino acids in length. (b) Heat-map of EK-normalized anti-zwitterionic peptide IgM antibody titers. (c) Heat-map of EK-normalized anti-zwitterionic peptide IgG antibody titers. Antibody titer ELISA data are presented in Fig. S1 and S2.† | ||
To detect peptide immunogenicity, bovine serum albumin (BSA) peptide conjugates were coated onto high binding ELISA plates. Diluted blood serum was incubated with the BSA peptide coated plates to detect antibodies specific to the peptide and not KLH. Titer was determined by the serum dilution required to reduce the antibody binding signal below a defined threshold (OD450 = 0.08) (Fig. S1†). Thus, higher antibody titers reflect increased peptide immunogenicity. Significantly, the EKX (i, i + 1) motif induced X-dependent differences in antibody titers, such that EK and EKP demonstrated lower antibody responses while EKS elicited higher IgG and EKG higher IgM and IgG levels (Fig. 1b, c, S1 and S2†). These results are consistent with those from previous studies in which EK conjugated to asparaginase exhibited low anti-EK antibody titers.16 The EXKX (i, i + 2) motif did not elicit strong antibody responses compared to EKG and EKS, suggesting that these effects were not dependent on direct interactions with the structure-disrupting amino acid (Fig. 1b, c, S1 and S2†).
Structural modeling of zwitterionic peptides with structure-disrupting amino acids
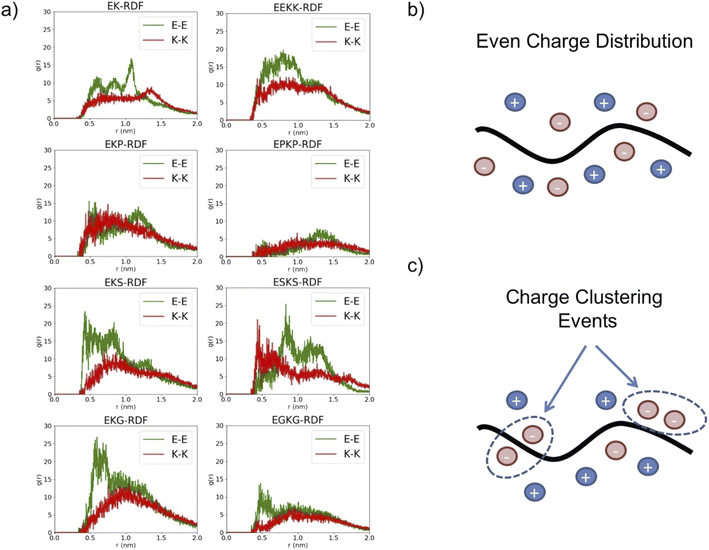
To predict the structural dynamics driving these effects, we investigated the conformational variations of the eight zwitterionic peptide motifs in an explicit solvent using Rosetta protein predictions and molecular dynamics (MD) simulations. Initially, we hypothesized that the immunogenicity of zwitterionic peptides could be related to stable secondary structure formation based on reports of stable structures correlating with higher immune responses.33 However, analysis of the peptide simulations did not show clear a relation connecting prevalence of their secondary structure and their immunogenicity. For instance, the EK and EKS peptides presented a similar percentage of α-helix structure but show different antibody responses (Table S3†). Our second hypothesis was that the immunogenicity of zwitterionic peptides could be related to their hydration. This hypothesis was driven by extensive research that hydrophobicity is correlated with immunogenicity.34 However, the simulations did not show significant differences in peptide–water interactions among the eight peptides (Table S3†).Despite the net charge neutrality of these peptide sequences, we reasoned that the positioning of charged domains from E and K side chains could result in regions of local charge in the peptide structure. These charged regions could induce unwanted electrostatic interactions leading to the observed peptide immunogenicity. To test against this hypothesis, we first analyzed the radial distribution functions (RDFs) between C of the carboxylic group on the E sidechains and N of the amine group on the K sidechains. The E–K RDFs did not show significant differences in the peak location, which was unexpected given E and K in the primary amino acid sequence is variable for i, i + 1 and i, i + 2 sequence motifs (Fig. S3a†). However, the RDF plots for E–E and K–K present distinct variations between sequences. Closer examination of higher immunogenicity EKG and EKS peptides revealed negatively charged E–E RDFs peaked at a shorter distance than positively charged K–K RDFs, while this effect was not observed in other sequences. The increased probability of finding E–E more proximal than K–K suggests EKG and EKS exhibit regions of charge imbalance despite the charge neutrality of the whole peptide (Fig. 2a and S4†). Thus, an ideal, low immunogenicity peptide would exhibit a significant E–K peak with a distance <0.5 nm to maintain the zwitterionic effect and would not exhibit a significant RDF peak within 1.0 nm for either E–E or K–K to avoid charge imbalances.
To further confirm these effects, clustering analysis of the charged atoms was conducted, which cumulatively accounts for the E–E, K–K, and E–K charge distances. Two charged groups were considered in the same cluster if their distance was less than 0.75 nm. This distance criterion was determined based on the RDFs. The percentage of charged clusters as a function of the number of charged groups and net charge for the cluster was plotted for each peptide motif (Fig. S3b†). EKS and EKG exhibited clustering of charged moieties resulting in charge imbalanced regions while lower immunogenicity peptides, namely EK and EKP, were generally charge neutral, which supports the tendencies observed with the RDF analysis (Fig. S3b†).
Immunogenicity of HMWSC zwitterionic peptides linked to IFN
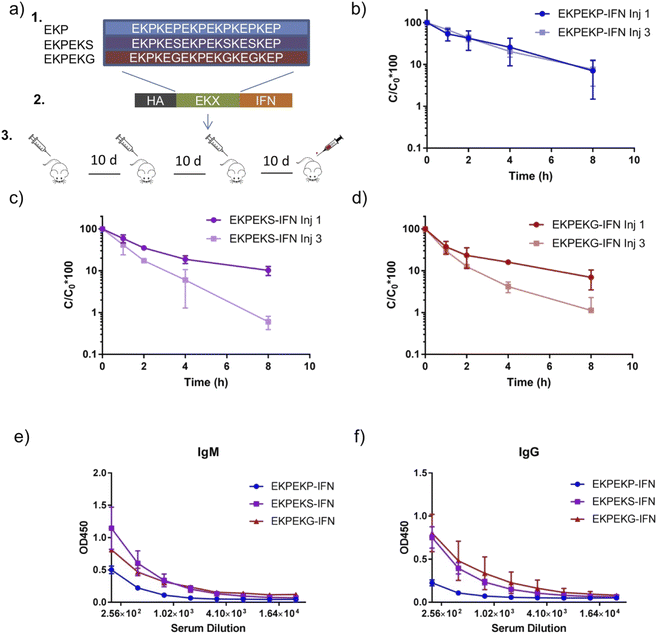
In the initial studies, zwitterionic peptide length was restricted to 12 amino acids for simplicity. However, for the intended applications, these peptides need to be considerably longer. Thus, to validate the previous results, we wondered if the behavior of zwitterionic peptide motifs in initial studies could be extended to HMWSC peptide sequences linked to a protein drug. To do so, we designed zwitterionic peptide sequences containing mixtures of the EKX (i, i + 1) motif with molecular weights near 30 kDa. These sequences contained a mixture of EKP and EKS motifs (termed EKPEKS) and a mixture of EKP and EKG motifs (termed EKPEKG) along with the EKP motif alone (termed EKPEKP) (Fig. 3a). Only half of the EKP motifs were substituted with EKS or EKG to maintain comparable pharmacokinetic properties for a fair comparison of immunogenicity. EKP was selected as the low immunogenic reference as opposed to EPKP to restrain the examination to i, i + 1 motif, which showed the most significant variation in immunogenicity from initial studies.These sequences were genetically appended to the N-terminus of interferon-alpha 2a (IFN), which is an FDA-approved protein drug for the treatment of hepatitis C and hairy cell leukemia. Subsequently, expression constructs were transfected into HEK293F mammalian cells for secreted protein production and purification from the expression media. SDS-PAGE gel analysis of the purified proteins showed a uniform molecular weight purified protein, which was sufficient for in vivo studies (Fig. S5†).
To examine the immunogenicity and ABC effects of EKPEKP-IFN, EKPEKS-IFN, and EKPEKG-IFN, 10 μg of each fusion protein was injected intravenously by the retro-orbital method and its blood clearance post-injection was measured. This process was repeated for two more injections separated by 10 days, and blood was collected 10 days following the third injection to be examined for antibodies reactive against the fusion protein (Fig. 3a). Circulation profiles following intravenous injection of EKPEKP, EKPEKS, and EKPEKG peptide variants revealed significant ABC effects with EKPEKS-IFN and EKPEKG-IFN fusion proteins, while EKPEKP-IFN did not exhibit this effect (Fig. 3b–d and Table S4†). Additionally, EKPEKP-IFN exhibited a longer circulation time following the first injection compared to EKPEKS-IFN and EKPEKG-IFN suggesting these charge effects could also lead to shorter circulation times for the first injection. Notably, EKPEKP-IFN exhibited near 6-fold improvements in circulation half-life, the area under the curve (AUC), and mean residence time (MRT) pharmacokinetic parameters compared to unmodified IFN (Table S4 and Fig. S6†). Subsequent blood collection and antibody analysis correlated well with the observed ABC effects, with EKPEKS-IFN and EKPEKG-IFN both showing higher anti-drug IgG and IgM levels (Fig. 3e and f). These results indicate that the EKG and EKS motifs induce immunogenicity when the peptide length is increased, thereby validating the previous results.
Contribution of individual amino acids to antibody binding in higher immunogenicity HMWSC zwitterionic peptides
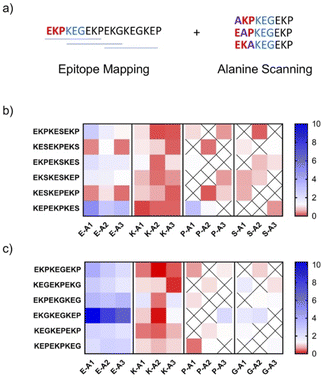
To validate the charge imbalance hypothesis, we sought to determine the contribution of individual amino acid residues towards antibody binding for higher immunogenicity HMWSC zwitterionic peptides. To do so, epitope mapping and alanine scanning experiments of IgG binding to EKPEKS and EKPEKG sequences were conducted. For this study, IgG was selected as its binding patterns tend to be more specific than IgM.35,36 This experiment consisted of immobilizing two categories of peptides to a membrane to measure their binding patterns to IgG from EKPEKS-IFN and EKPEKG-IFN treated mice. The first category of immobilized peptides was designed to reflect unique 9 mer peptides covering the span of the EKPEKS and EKPEKG variants (6 total peptides for each variant), termed epitope mapping. The second category of peptides was designed to reflect single amino acid alanine (A) substitutions from the peptides derived in the first category, termed alanine scanning. Differences in IgG binding signal intensity between the epitope mapping and alanine scanning peptides were used to determine the contribution of specific amino acids (i.e. E, K, P, S, or G) towards IgG binding. This analysis showed that peptides with K to A substitutions in the reduced IgG binding, while E to A changes either did not change or resulted in a slight increase in IgG binding (Fig. 4). Moreover, P to A, S to A, and G to A substitutions resulted in marginal effects on antibody binding. Together, these results indicate the notable contribution of charged residues in antibody binding and validate the role of charge imbalance leading to immunogenicity of EKPEKS and EKPEKG.Discussion
The complex mechanisms driving interactions make a priori predictions of immunogenicity exceedingly challenging given the highly variable environments in biological systems leading to this response. Additionally, technologies with low immunogenicity are increasingly in demand. Zwitterionic peptides are a promising and emerging category of biomolecules containing low immunogenic motifs. As with any emerging technology, its design constraints and dynamics are critical for its development. In this work, we examined motif-based zwitterionic peptides to delineate their design constraints for their application as HMWSC sequences linked to therapeutic proteins. We discovered sequence motifs, namely EKS and EKG, which did not perform well due to their apparent immune response in both short peptide and long genetically engineered formats. Notably, simulations of these two peptide motifs exhibited a conserved charge imbalance property that was not present in the peptides with low immunogenicity (Fig. 2).The critical interplay of charge distance and electrostatic forces are thematic to zwitterionic molecules. For example, studies with other zwitterionic molecules have demonstrated the importance of carbon spacer length between charged domains in preserving stable non-fouling behavior in carboxy betaine.8,12,37 Similar patterns have been also observed with sulfobetaine moieties.38 Although the charge distances of EK containing zwitterionic peptides could differ from carboxy betaine and sulfobetaine due to the location of positive and negative charge moieties being on the same side chain, parallels in these effects may be of scientific significance.
Even though the causal mechanism inducing local charge in EKG or EKS is not fully understood, these effects may be due to different rotational effects of P, S, and G on the peptide backbone. G and S have either no side chain or a very small side chain respectively, while P forms a pyrrolidine loop thus contorting the peptide backbone.27,28,30,31 Therefore, P may be twisting the peptide backbone preventing local charge, while S and G may be forcing or allowing the peptide to twist in a fashion that induces a local charge. Additionally, these twisting effects may be negated for the i, i + 2 motifs as neither ESKS nor EGKG exhibited significant immunogenicity nor regions of charge imbalance.
Furthermore, it is noteworthy that the experimental assignment a specific charge (positive or negative) could be precluded by complex interactions with components in the serum as well as immunological processes such as affinity maturation and somatic hypermutation of antibodies. While alanine scanning experiments could suggest positively charged K residues are directly interacting with antibodies, its additional impact on surrounding ions and water molecules in complex electrostatic and hydrogen bonding networks cannot be ruled out. Nonetheless, the collective experimental and computational evidence points to charge imbalance as a likely rationale for the observed immunogenicity of EKS and EKG as well as to charge balance zwitterionic character for the low immunogenicity of EKP.
Conclusions
In this work, we examined the structure, immunogenicity, and functional behaviour of zwitterionic EK peptide motifs containing P, S, and G, which are structure-disrupting amino acids. When we probed the immunogenicity of eight short peptide motifs in vivo chemically linked to KLH, we uncovered that EKP exhibits lower immunogenicity while EKS and EKG exhibit higher immunogenicity. Results from MD simulations of the eight peptide motifs showed charge imbalance in EKS and EKG, thereby, suggesting a possible causal role. Finally, we synthesized HMWSC zwitterionic peptides and fused them to IFN containing these motifs. The fusion proteins demonstrated similar immunological behaviour to zwitterionic peptides, indicating that these effects are not restricted to small peptides.Methods
Peptide synthesis and KLH conjugation
Peptides EK, EEKK, EKX, or EXKX, in which X is either P, S, or G, were designed to be 12 amino acids in length by repeating the predefined sequence motif. These peptides were conjugated to Keyhole Limpet Hemocyanin (KLH) with a beta-alanine linker and a cysteine-containing a free thiol. Solid-phase peptide synthesis and conjugation to KLH was done courtesy of Kinexus Bioinformatics Inc. In parallel, these peptides were conjugated to bovine serum albumin (BSA) for use in ELISA experiments to detect anti-peptide antibodies following animal injections.Injection schedule of KLH–peptide conjugates
30 μg of KLH–peptide conjugate were administered subcutaneously (SC) near the right dorsal leg of 4 week old C57BL/6 male mice. Two weeks later another thirty-microgram booster dose was administered by the same injection method. One week following the booster dose, 150 μL of blood were harvested by chin bleed. Blood was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g to obtain the serum to be frozen at −20 °C for immunogenicity analysis.
000g to obtain the serum to be frozen at −20 °C for immunogenicity analysis.
Anti-peptide antibody detection by ELISA
150 ng per well of BSA–peptide conjugate were coated on 96 well plates in 50 mM sodium bicarbonate pH 9.6 at a concentration of 3 μg mL−1. Plate coating was performed overnight at 4 °C. Plates were washed twice with 250 μL phosphate buffer saline and 0.03% Tween-20 (PBS-Tween). Then wells were blocked with 200 μL of 3% BSA in PBS-Tween for 2 hours at 37 °C. Following blocking, wells were washed twice with 250 μL PBS-Tween. Then dilutions of serum in 100 μL blocking buffer from mice injected twice with KLH–peptide were added to wells and incubated at 37 °C for 90 minutes. Subsequently, wells were washed three times with 250 μL PBS-Tween, and 100 μL of secondary antibody, either goat anti-mouse IgM HRP conjugate or goat anti-mouse IgG HRP conjugate, diluted 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fold in blocking buffer, were added to wells, which were then incubated at 37 °C for 90 minutes. Again, wells were washed three times with 250 μL of PBS-Tween, and 50 μL of One-Step ELISA TMB Ultra substrate was added to each well and developed at room temperature for 15 minutes. Development was terminated by the addition of 50 μL of sulfuric acid, and absorbance measurements at 450 nm in each well were immediately taken using a Biotek Cytation 5 plate reader.
000 fold in blocking buffer, were added to wells, which were then incubated at 37 °C for 90 minutes. Again, wells were washed three times with 250 μL of PBS-Tween, and 50 μL of One-Step ELISA TMB Ultra substrate was added to each well and developed at room temperature for 15 minutes. Development was terminated by the addition of 50 μL of sulfuric acid, and absorbance measurements at 450 nm in each well were immediately taken using a Biotek Cytation 5 plate reader.
Interferon alpha-2-a zwitterionic peptide fusion protein expression and purification
DNA sequences for recombinant expression of zwitterionic peptides (EKPEKP, EKPEKS, and EKPEKG) fused to interferon-alpha 2a (IFN) were synthesized by Genscript. Zwitterionic sequences were designed to express an approximately 30 kDa peptide fused to the N-terminus of IFN. As well, a nine amino acid hemagglutinin (HA) tag was fused to the N-terminus of the zwitterionic peptide domain for affinity purification. In addition, a transplasminogen activator (TPA) secretion signal was included to induce the secretion of the protein into the expression media. Synthesized DNA constructs were ligated into the plasmid pcDNA3.1(+). Plasmids were amplified in DH5α E. coli and purified by PureLink HiPure Plasmid MaxiPrep (ThermoFisher) kits. Protein expression was carried out in Expi293F cells using F17 expression media (ThermoFisher). Cells were maintained for expansion and expression at 37 °C, 8% CO2, and 85% humidity. For transient transfection, plasmids were sterile filtered and 60 μg plasmid was incubated with 180 μg PEI max in 3 mL expression media. The resulting complex was added to 60 mL of Expi293F cells grown to approximately 2 × 106 cells per mL. Expression was allowed to proceed for 4 days at which point the media containing the fusion protein was isolated by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. The media was concentrated 60-fold in Amicon 30 kDa cut off or 10 kDa cut off (for IFN alone) centrifugal filter units. The resulting concentrate was incubated with 300 μL Pierce anti-HA agarose with rotation at room temperature for 2 h. The concentrate was separated from the protein-bound agarose by centrifugal filters, and it was washed 3× with tris buffered saline with 0.05% Tween-20 (TBS-T). Bound protein was eluted with 0.1 M glycine pH 2.6 and immediately the eluate was neutralized with a 1
000g. The media was concentrated 60-fold in Amicon 30 kDa cut off or 10 kDa cut off (for IFN alone) centrifugal filter units. The resulting concentrate was incubated with 300 μL Pierce anti-HA agarose with rotation at room temperature for 2 h. The concentrate was separated from the protein-bound agarose by centrifugal filters, and it was washed 3× with tris buffered saline with 0.05% Tween-20 (TBS-T). Bound protein was eluted with 0.1 M glycine pH 2.6 and immediately the eluate was neutralized with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratio of 1 M tris pH 9. The resulting protein was buffer exchanged into phosphate-buffered saline (PBS) for subsequent animal studies.
20 ratio of 1 M tris pH 9. The resulting protein was buffer exchanged into phosphate-buffered saline (PBS) for subsequent animal studies.
IFN zwitterionic peptide fusion protein pharmacokinetics and immunogenicity studies
Purified proteins were injected by the retro-orbital method into 4–6 week old C57BL/6 mice. To determine the blood circulation properties of purified proteins, blood was taken by chin bleed (25 μL per bleed) 5 minutes, 1 h, 2 h, 4 h, and 8 h post-injection. Serum was isolated from each blood sample by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. This process was repeated two more times on the same mice for ten days separating injections to detect changes in circulation properties following repeated injection. Interferon alpha-2-a ELISA kits (Invivogen) were used to determine the protein concentration in the serum post-injection. For pharmacokinetic analysis, concentrations were normalized to the protein concentration 5 minutes post-injection to account for slight differences in injection amount. Pharmacokinetic parameters were determined using PKSolver software using one-compartment pharmacokinetic modeling parameters.39 Ten days following the final injection blood was collected by chin bleed and serum was isolated by centrifugation at 13
000g. This process was repeated two more times on the same mice for ten days separating injections to detect changes in circulation properties following repeated injection. Interferon alpha-2-a ELISA kits (Invivogen) were used to determine the protein concentration in the serum post-injection. For pharmacokinetic analysis, concentrations were normalized to the protein concentration 5 minutes post-injection to account for slight differences in injection amount. Pharmacokinetic parameters were determined using PKSolver software using one-compartment pharmacokinetic modeling parameters.39 Ten days following the final injection blood was collected by chin bleed and serum was isolated by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g to detect the presence of anti-EKX-IFN antibodies. Anti-EKX-IFN antibody levels were determined similarly to anti-peptide antibody levels with minor changes. Briefly, 50 μg of protein were coated on high binding ELISA plates in 50 mM sodium bicarbonate pH 9.6 at 4 °C overnight. Blocking was done in 3% BSA PBS Tween for 2 h at 37 °C. 2 fold serial dilutions of serum starting at 1
000g to detect the presence of anti-EKX-IFN antibodies. Anti-EKX-IFN antibody levels were determined similarly to anti-peptide antibody levels with minor changes. Briefly, 50 μg of protein were coated on high binding ELISA plates in 50 mM sodium bicarbonate pH 9.6 at 4 °C overnight. Blocking was done in 3% BSA PBS Tween for 2 h at 37 °C. 2 fold serial dilutions of serum starting at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution were incubated with blocking buffer for 90 minutes. 1
200 dilution were incubated with blocking buffer for 90 minutes. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution of HRP conjugated anti-mouse IgG and anti-mouse IgM (ThermoFisher) were incubated in blocking buffer for 90 minutes. Plates were subsequently developed with TMB Ultra ELISA substrate for 15 minutes and quenched with an equal volume of 2 M sulfuric acid. Absorbance was measured using a Cytation5 plate reader at 450 nm to detect antibody presence. Reduced absorbance values at 450 nm, as well as absorbance at background levels at a lower serum concentration, indicated lower anti-EKX-IFN antibody levels in the serum.
000 dilution of HRP conjugated anti-mouse IgG and anti-mouse IgM (ThermoFisher) were incubated in blocking buffer for 90 minutes. Plates were subsequently developed with TMB Ultra ELISA substrate for 15 minutes and quenched with an equal volume of 2 M sulfuric acid. Absorbance was measured using a Cytation5 plate reader at 450 nm to detect antibody presence. Reduced absorbance values at 450 nm, as well as absorbance at background levels at a lower serum concentration, indicated lower anti-EKX-IFN antibody levels in the serum.
Epitope mapping and alanine scanning EKPEKS and EKPEKG
Peptides were synthesized using TOTD membranes containing a PEG linker via the SPOT synthesis method courtesy of Kinexus Bioinformatics Inc. Membranes were blocked with 3% BSA in PBS/0.03% Tween-20 for two hours at room temperature. Then, membranes were incubated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 dilution of serum in block buffer for 90 minutes at room temperature. Serum was obtained from mice injected 3× with EKPEKS-IFN and EKPEKG-IFN. Serum from animals in each group was mixed to get a representative sample of all animals from each group on one membrane. Anti-mouse IgG–HRP was diluted at 1
200 dilution of serum in block buffer for 90 minutes at room temperature. Serum was obtained from mice injected 3× with EKPEKS-IFN and EKPEKG-IFN. Serum from animals in each group was mixed to get a representative sample of all animals from each group on one membrane. Anti-mouse IgG–HRP was diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 in block buffer and incubated for 90 minutes at room temperature. Membranes were developed using 1-step TMB blotting solution (ThermoFisher). Development was stopped using water and imaged immediately. Signal intensity was determined using ImageJ analysis.
2000 in block buffer and incubated for 90 minutes at room temperature. Membranes were developed using 1-step TMB blotting solution (ThermoFisher). Development was stopped using water and imaged immediately. Signal intensity was determined using ImageJ analysis.
Initial modeling configuration of EKX peptides
All molecules and ions are described using the all-atom model. The initial configurations of all peptides are obtained from the online Rosetta Server using the Rosetta ab initio folding algorithm. The simulation systems are created by placing the peptide in a cubic water box. The initial sizes of the simulation boxes are determined based on the size of the peptide to ensure that the peptide will not interact with their mirrors and the simulation boxes contain enough bulk-phase water molecules. Numbers of Na+ and Cl− are added to make the solution 0.15 M NaCl, equivalent to the physiological condition in the human body.This work utilizes the Amber14 (ref. 40) force field to describe bonded and nonbonded interactions in the systems. The water molecules are described using the TIP3P model41 because the Amber force field is developed based on it. Indeed, various force fields have been developed for molecular simulations. The Amber force field has been widely used for simulating biomolecules. The non-boned interactions are a sum of short-range Lennard-Jones 12–6 potential and long-range coulombic potential, as shown in eqn (1). The bonded interactions are a sum of the bond, angle, and dihedral potentials, as described in the force field.
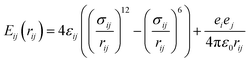 | (1) |
Molecular dynamics simulation of EKX peptides
The simulation procedure includes three steps. First, the energy minimization was conducted to remove any too-close contacts between atoms. Second, a 100 ns isobaric-isothermal (NPT, T = 310 K, P = 1 atm) ensemble MD simulation with a 2 fs integral step was conducted to let the system reach thermodynamic equilibrium. The Berendsen method42 is used to control the temperature and pressure of the system because it allows the system to reach the desired pressure and temperature at a fast pace. The simulation box should reach a proper size so the water molecules present the bulk density when they are away from the protein or the protein–peptide conjugate. Third, a 200 ns isobaric-isothermal (NPT, T = 310 K, P = 1 atm) ensemble MD simulation with a 2 fs integral step was conducted to collect the trajectory at a frequency of 100 ps. The Parrinello–Rahman method43 is used to control the pressure and the velocity-rescaling method44 is used to control the temperature. The short-range van der Waals interactions use a 1.0 nm cut-off and the long-range electrostatic interactions were calculated using the particle mesh Ewald sum.45 All bonds involving H atoms were constrained during the simulations. The energy minimization and MD simulations for all the systems were conducted using Gromacs-2020.46Data availability
Data available on request from the authors.Author contributions
Patrick McMullen, Qing Shao, and Shaoyi Jiang: conceptualition. Patrick McMullen, Sijin Luozhong, and Liang Fang: experimentation. Qi Qiao, Lirong Cai and Qing Shao: molecular simulations and data analysis. Patrick McMullen, Qi Qiao, Qing Shao, and Shaoyi Jiang: manuscript writing.Conflicts of interest
S. J. is a co-founder of ZWI therapeutics, Inc. S. J., P. M., and S. L. are co-inventors of the pending patent US20210324010A1 filed by the University of Washington.Acknowledgements
S. J. acknowledges start-up support from Cornell University, including Robert S. Langer Professorship and Cornell NEXT Nano Initiative. Q. S. acknowledges the support of start-up funds provided by the University of Kentucky. The simulations were conducted on the computational facilities provided by the High-Performance Computing Center of the University of Kentucky.References
- M. Krishna and S. G. Nadler, Immunogenicity to Biotherapeutics - The Role of Anti-drug Immune Complexes, Front. Immunol., 2016, 7, 21 CrossRef PubMed.
- V. Jawa, F. Terry, J. Gokemeijer, S. Mitra-Kaushik, B. J. Roberts, S. Tourdot and A. S. De Groot, T-Cell Dependent Immunogenicity of Protein Therapeutics Pre-clinical Assessment and Mitigation-Updated Consensus and Review 2020, Front. Immunol., 2020, 11, 1301 CrossRef CAS PubMed.
- Z. E. Sauna, S. M. Richards, B. Maillere, E. C. Jury and A. S. Rosenberg, Editorial: Immunogenicity of Proteins Used as Therapeutics, Front. Immunol., 2020, 11, 614856 CrossRef CAS.
- Y. Vugmeyster, X. Xu, F. P. Theil, L. A. Khawli and M. W. Leach, Pharmacokinetics and toxicology of therapeutic proteins: Advances and challenges, World J. Biol. Chem., 2012, 3(4), 73–92 CrossRef.
- A. S. Abu Lila, H. Kiwada and T. Ishida, The accelerated blood clearance (ABC) phenomenon: Clinical challenge and approaches to manage, J. Controlled Release, 2013, 172(1), 38–47 CrossRef CAS PubMed.
- L. Hong, Z. Wang, X. Wei, J. Shi and C. Li, Antibodies against polyethylene glycol in human blood: A literature review, J. Pharmacol. Toxicol. Methods, 2020, 102, 106678 CrossRef CAS PubMed.
- S. Chen, Z. Cao and S. Jiang, Ultra-low fouling peptide surfaces derived from natural amino acids, Biomaterials, 2009, 30(29), 5892–5896 CrossRef CAS PubMed.
- Z. Zhang, H. Vaisocherová, G. Cheng, W. Yang, H. Xue and S. Jiang, Nonfouling Behavior of Polycarboxybetaine-Grafted Surfaces: Structural and Environmental Effects, Biomacromolecules, 2008, 9(10), 2686–2692 CrossRef CAS PubMed.
- H. Vaisocherova, W. Yang, Z. Zhang, Z. Cao, G. Cheng, M. Piliarik, J. Homola and S. Jiang, Ultralow fouling and functionalizable surface chemistry based on a zwitterionic polymer enabling sensitive and specific protein detection in undiluted blood plasma, Anal. Chem., 2008, 80(20), 7894–7901 CrossRef CAS PubMed.
- J. C. Hower, M. T. Bernards, S. Chen, H.-K. Tsao, Y.-J. Sheng and S. Jiang, Hydration of “Nonfouling” Functional Groups, J. Phys. Chem. B, 2008, 113(1), 197–201 CrossRef PubMed.
- Q. Shao and S. Jiang, Molecular Understanding and Design of Zwitterionic Materials, Adv. Mater., 2015, 27(1), 15–26 CrossRef CAS PubMed.
- Q. Shao and S. Jiang, Effect of Carbon Spacer Length on Zwitterionic Carboxybetaines, J. Phys. Chem. B, 2013, 117(5), 1357–1366 CrossRef CAS PubMed.
- P. Zhang, F. Sun, C. Tsao, S. Liu, P. Jain, A. Sinclair, H. C. Hung, T. Bai, K. Wu and S. Jiang, Zwitterionic gel encapsulation promotes protein stability, enhances pharmacokinetics, and reduces immunogenicity, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(39), 12046–12051 CrossRef CAS.
- B. Li, Z. Yuan, H. C. Hung, J. Ma, P. Jain, C. Tsao, J. Xie, P. Zhang, X. Lin, K. Wu and S. Jiang, Revealing the Immunogenic Risk of Polymers, Angew. Chem., Int. Ed. Engl., 2018, 57(42), 13873–13876 CrossRef CAS PubMed.
- P. Zhang, E. J. Liu, C. Tsao, S. A. Kasten, M. V. Boeri, T. L. Dao, S. J. DeBus, C. L. Cadieux, C. A. Baker, T. C. Otto, D. M. Cerasoli, Y. Chen, P. Jain, F. Sun, W. Li, H. C. Hung, Z. Yuan, J. Ma, A. N. Bigley, F. M. Raushel and S. Jiang, Nanoscavenger provides long-term prophylactic protection against nerve agents in rodents, Sci. Transl. Med., 2019, 11(473), eaau7091 CrossRef CAS PubMed.
- Z. Yuan, B. Li, L. Niu, C. Tang, P. McMullen, P. Jain, Y. He and S. Jiang, Zwitterionic Peptide Cloak Mimics Protein Surfaces for Protein Protection, Angew. Chem., Int. Ed. Engl., 2020, 59(50), 22378–22381 CrossRef CAS.
- F. M. Veronese, Peptide and protein PEGylation, Biomaterials, 2001, 22(5), 405–417 CrossRef CAS PubMed.
- S. S. Pai, B. Hammouda, K. Hong, D. C. Pozzo, T. M. Przybycien and R. D. Tilton, The Conformation of the Poly(ethylene glycol) Chain in Mono-PEGylated Lysozyme and Mono-PEGylated Human Growth Hormone, Bioconjugate Chem., 2011, 22(11), 2317–2323 CrossRef CAS PubMed.
- P. B. Lawrence and J. L. Price, How PEGylation influences protein conformational stability, Curr. Opin. Chem. Biol., 2016, 34, 88–94 CrossRef CAS PubMed.
- E. J. Liu, A. Sinclair, A. J. Keefe, B. L. Nannenga, B. L. Coyle, F. Baneyx and S. Jiang, EKylation: Addition of an Alternating-Charge Peptide Stabilizes Proteins, Biomacromolecules, 2015, 16(10), 3357–3361 CrossRef CAS.
- E. J. Liu and S. Jiang, Expressing a Monomeric Organophosphate Hydrolase as an EK Fusion Protein, Bioconjugate Chem., 2018, 29(11), 3686–3690 CrossRef CAS PubMed.
- J. Smith, P. McMullen, Z. Yuan, J. Pfaendtner and S. Jiang, Elucidating Molecular Design Principles for Charge-Alternating Peptides, Biomacromolecules, 2020, 21(2), 435–443 CrossRef CAS PubMed.
- V. Schellenberger, C. W. Wang, N. C. Geething, B. J. Spink, A. Campbell, W. To, M. D. Scholle, Y. Yin, Y. Yao, O. Bogin, J. L. Cleland, J. Silverman and W. P. Stemmer, A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner, Nat. Biotechnol., 2009, 27(12), 1186–1190 CrossRef CAS PubMed.
- M. Schlapschy, U. Binder, C. Borger, I. Theobald, K. Wachinger, S. Kisling, D. Haller and A. Skerra, PASylation: a biological alternative to PEGylation for extending the plasma half-life of pharmaceutically active proteins, Protein Eng., Des. Sel., 2013, 26(8), 489–501 CrossRef CAS.
- J. Breibeck and A. Skerra, The polypeptide biophysics of proline/alanine-rich sequences (PAS): Recombinant biopolymers with PEG-like properties, Biopolymers, 2018, 109(1), e23069 CrossRef PubMed.
- J. S. Richardson and D. C. Richardson, Principles and Patterns of Protein Conformation, in Prediction of Protein Structure and the Principles of Protein Conformation, 1989, pp. 1–98 Search PubMed.
- P. R. Schimmel and P. J. Flory, Conformational energies and configurational statistics of copolypeptides containing L-proline, J. Mol. Biol., 1968, 34(1), 105–120 CrossRef CAS.
- K. Imai and S. Mitaku, Mechanisms of secondary structure breakers in soluble proteins, Biophysics, 2005, 1, 55–65 CrossRef CAS PubMed.
- D. A. Brant and P. J. Flory, The Configuration of Random Polypeptide Chains. II. Theory, J. Am. Chem. Soc., 1965, 87(13), 2791–2800 CrossRef CAS.
- P. R. Schimmel and P. J. Flory, Conformational energy and configurational statistics of poly-L-proline, Proc. Natl. Acad. Sci. U. S. A., 1967, 58(1), 52–59 CrossRef CAS PubMed.
- N. M. Tooney and G. D. Fasman, Conformation of serine polypeptides, J. Mol. Biol., 1968, 36(3), 355–369 CrossRef CAS PubMed.
- S. Blanco, M. E. Sanz, J. C. Lopez and J. L. Alonso, Revealing the multiple structures of serine, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(51), 20183–20188 CrossRef CAS PubMed.
- T. Lengauer, C. J. Camacho, Y. Katsumata and D. P. Ascherman, Structural and Thermodynamic Approach to Peptide Immunogenicity, PLoS Comput. Biol., 2008, 4(11), e1000231 CrossRef PubMed.
- D. Chowell, S. Krishna, P. D. Becker, C. Cocita, J. Shu, X. Tan, P. D. Greenberg, L. S. Klavinskis, J. N. Blattman and K. S. Anderson, TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes, Proc. Natl. Acad. Sci., 2015, 112(14), E1754–E1762 CrossRef CAS PubMed.
- G. J. Wedemayer, P. A. Patten, L. H. Wang, P. G. Schultz and R. C. Stevens, Structural insights into the evolution of an antibody combining site, Science, 1997, 276(5319), 1665–1669 CrossRef CAS PubMed.
- S. A. Frank, in Immunology and Evolution of Infectious Disease, Princeton (NJ), 2002 Search PubMed.
- H. Vaisocherová, Z. Zhang, W. Yang, Z. Cao, G. Cheng, A. D. Taylor, M. Piliarik, J. Homola and S. Jiang, Functionalizable surface platform with reduced nonspecific protein adsorption from full blood plasma—Material selection and protein immobilization optimization, Biosens. Bioelectron., 2009, 24(7), 1924–1930 CrossRef PubMed.
- Y. Higaki, Y. Inutsuka, T. Sakamaki, Y. Terayama, A. Takenaka, K. Higaki, N. L. Yamada, T. Moriwaki, Y. Ikemoto and A. Takahara, Effect of Charged Group Spacer Length on Hydration State in Zwitterionic Poly(sulfobetaine) Brushes, Langmuir, 2017, 33(34), 8404–8412 CrossRef CAS PubMed.
- Y. Zhang, M. Huo, J. Zhou and S. Xie, PKSolver: An add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel, Computer Methods and Programs in Biomedicine, 2010, 99(3), 306–314 CrossRef PubMed.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory Comput., 2015, 11(8), 3696–3713 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79(2), 926–935 CrossRef CAS.
- H. J. C. Berendsen, J. P. M. Postma, W. F. v. Gunsteren, A. DiNola and J. R. Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys., 1984, 81(8), 3684–3690 CrossRef CAS.
- M. Parrinello and A. Rahman, Crystal structure and pair potentials: A molecular-dynamics study, Phys. Rev. Lett., 1980, 45(14), 1196 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126(1), 014101 CrossRef.
- T. Darden, D. York and L. Pedersen, Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems, J. Chem. Phys., 1993, 98(12), 10089–10092 CrossRef CAS.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc03519g |
| This journal is © The Royal Society of Chemistry 2022 |