Open Access Article
Open Access ArticleLipid coated protein condensates as stable protocells with selective uptake abilities for biomolecules†
Juyoung
Son
and
Yongwon
Jung
 *
*
Department of Chemistry, KAIST, 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea. E-mail: ywjung@kaist.ac.kr; Fax: +82-42-350-2810; Tel: +82-42-350-2817
First published on 27th September 2022
Abstract
To create cell-like synthetic systems, spatial confinement that is stable against environmental changes and selective uptake of diverse biomolecules into these compartments are key initial conditions. However, fabrication of protocells with these two features has been extremely difficult. Here, we used fully protein-based liquid condensates and a lipid coating on these condensates to construct highly stable protocells with an uptake ability for outside biomolecules. Condensates with an extremely high density of 6His-tagged proteins were coated with Ni(II)-NTA(nitrilotriacetic acid)-modified lipids. High condensate rigidity and specific 6His-Ni-NTA interactions enabled the formation of lipid–protein protocells, which are stable even after centrifugations. In addition, immobile lipid coatings on condensates were permeable to outside biomolecules. When binding modules were fused into condensate-forming proteins, the resulting functionalized condensate-protocells could strongly and selectively uptake various outside proteins through specific protein interactions.
Introduction
Artificial cell-like entities have been actively built with the aim of gaining knowledge on existing cellular systems and to develop new synthetic machineries with abilities to perform complex processes of living cells.1 An ultimate goal is to produce a minimal synthetic object that can execute key life activities such as energy conversion, growth, reproduction, and even evolution.2–5 Compartmentalization is an essential initial condition for synthetic as well as natural cells. Synthetic protocells have been assembled and compartmentalized with various materials including lipid vesicles (liposomes),6 amphiphilic polymers (polymersomes),7 and colloidal particles (colloidosomes).8 Liposomes have been the most widely used component to construct protocell liquid compartments since lipid bilayers are also natural boundary membranes for cells and organelles. However, liposome compartments are relatively unstable compared to natural membranes, and only limited structural variations have been reported.9 Compartment boundary properties such as rigidity, stability, and thickness are more widely varied by applying unnatural polymers10 and particles.8 For example, colloidosomes (also called pickering emulsions) could offer more permeable barriers than liposomes, which might be more suitable for bioreactors.11Recently, coacervate-based protocells have gained strong interest as they offer condensed cell-size liquid droplets.12 In general, coacervates are formed via liquid–liquid phase separation (LLPS) of two opposite charged polymers. Coacervates offer extremely crowded cytosol-like environments,13 and multiple studies reported highly efficient (sometimes enhanced) biochemical reactions inside these condensed droplets.14,15 Free coacervates, however, can be easily merged with neighboring coacervates and mostly are unstable against environmental changes. Recently, multiple coacervate-based protocells were thereby developed by protecting coacervates with amphiphiles such as copolymers, fatty acids, and even cell lipid membrane fragments.11,16–18 Diverse cell processes such as enzyme reactions,16 vasodilation,17 phagocytosis,18 and predatory action19 were demonstrated with these protocells. Currently, most coacervate-based protocells are constructed with coacervates of charged polycarbohydrates (e.g., dextran, amylose, hyaluronate). Expanding coacervate materials will greatly increase the opportunity to add new features to these protocells. For example, by modifying polycarbohydrates with Ni2+-nitrilotriacetic acid (Ni-NTA), various His-tagged proteins were effectively contained inside polymer-coated coacervates.20
An ability to exchange matter with the surrounding environment is another essential condition for protocells.1 However, for most protocells with compartmentalization barriers, selective incorporation of outside materials (particularly large biomolecules) is still extremely difficult. In addition, an increase of protocell permeability might lead to decreased stability of protocells against external stimuli such as physical stresses and environmental changes. Ideally, protocell boundaries must be designed to allow selective uptake of outside materials including large biomolecules, while withstanding environmental changes. In cells, biomolecular coacervates (often called condensates) act as membrane-less organelles, which are enriched with a selective set of biomolecules, likely by selective uptake of surrounding biomolecules.21 These condensates are formed by LLPS of diverse phase separable proteins, and the resulting protein condensates show great diversities in physical and chemical properties.22,23 We believe that protein condensates will be attractive materials to build protocells with their abilities to selectively confine biomolecules and to have diverse structures and functions.
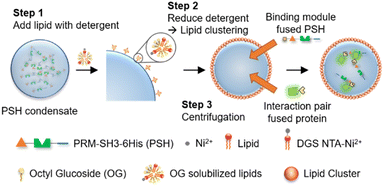
Here, we developed highly stable protocells with an ability to uptake diverse biomolecules by implementing fully protein-based condensates and a surface lipid coating. Ni-NTA-modified lipids were used for the specific lipid coating on protein condensates, which consist of highly dense 6His-tagged phase separable proteins (Scheme 1). The constructed protocells could be centrifuged without structural deformation, and were also stable against changes of environmental conditions such as osmolarity and temperature. Interestingly, these lipid-coated protein condensates were permeable to diverse molecules including small dyes, DNAs, and proteins, unlike liposomes. In addition, condensate-forming scaffold proteins could be functionalized with binding moieties, which allowed strong recruitment of specific outside proteins into these protein condensate-protocells (Scheme 1).
Results and discussion
Fabrication of lipid-coated protein condensates as stable protocells
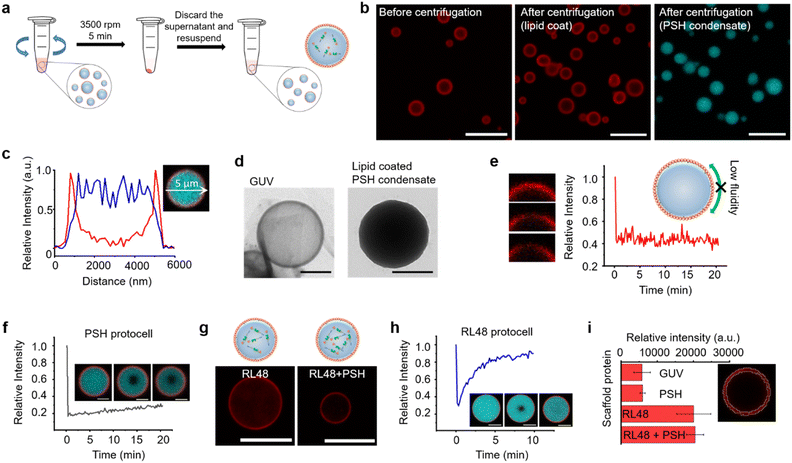
Compared to other phase separable biomolecules, most notably polycarbohydrates and nucleic acids, proteins offers extremely high diversities in attainable structures and functions. Therefore, various protein condensates have been reported to have a wide range of droplet properties such as interior rigidity.22 We envisioned that protocells formed with dense and rigid condensates might be able to withstand physical stresses such as centrifugation. We used previously developed metal ion-mediated LLPS of a 6His-tagged minimal scaffold protein consisting of proline-rich motif (PRM), a PRM binding SH3 protein domain, and a 6His tag (PRM-SH3-6His; PSH).24 The PSH phase separates into condensates by the simple addition of Ni2+, and various condensates of PSH variants with different droplet properties were also reported.24 More importantly, PSH condensates have extremely high (cytosolic-like) inside protein densities and high droplet rigidities.24Most coacervate protection to fabricate coacervate-based protocells has been induced by an electrostatic interaction-driven coating of amphiphilic lipids or polymers. For more stable and specific protection, we used a NTA-modified lipid, which can bind to abundant 6His tags on PSH condensates.24 Ni2+-containing PSH condensates were treated with a lipid mixture consisting of 94.5% DOPC (frame lipid), 5% DGS NTA (Ni), and 0.5% Liss Rhod PE (dye-lipid) in the presence of detergent octyl glucoside (OG) (Fig. S1†). Lipids were solubilized by OG (0.67%) in a PBS solution and incubated with condensates for 10 min to allow DGS-NTA binding to 6His tags on PSH condensates (Scheme 1). The solution OG percentage was decreased to 0.2% by rapid dilution, by which lipids can form more clustered structures on condensate surfaces. TEM images of lipid mixtures with different OG concentrations showed that 0.67% OG-solubilized lipids were well dispersed, while lipids formed larger clusters upon OG dilution to 0.2% (Fig. S2†). Lipid-covered condensates were centrifuged and resuspended in PBS to remove unbound proteins and lipids (Fig. 1a). Confocal images showed well-dispersed PSH condensates, which are thoroughly coated with lipids (Fig. 1b and c). We believe that this stable lipid coating prevents condensate merging/fusion during centrifugation. However, without the OG dilution (from 0.67% to 0.2%) step, condensates were fused into large coacervates by centrifugation (Fig. S3†), suggesting that clustered lipid structures on condensate surfaces are essential to prevent droplet fusion (Scheme 1). A recent study also reported that cellular condensate fusion was prevented by condensate surface-bound protein clusters, which act as stabilizing pickering agents.25 It is possible that our lipid-coated condensates are also close to these pickering emulsion structures. When the total lipid concentration was lowered, the lipid coating intensities clearly decreased, and a large portion of condensates agglomerated by centrifugation (Fig. S4a†), which also resulted in a low condensate recovery yield. When the lipid concentration was increased, however, a significant lipid signals were observed inside condensates. When OG was slowly lowered rather than being rapidly decreased, lipid signals were also heterogeneously distributed inside condensates (Fig. S4b†).
Specific binding of DGS-NTA to 6His on condensates was also important for stable coating since bound DGS-NTA could act as seeds for lipid cluster formation during subsequent OG dilution. In the absence of DGS-NTA, protein condensates were readily aggregated by centrifugation and could not be recovered. Lowering the DGS-NTA ratio resulted in less lipid coating and low recovery yields (Fig. S5a†). Excessively high DGS-NTA ratios, however, led to lipid aggregation around condensates (Fig. S5a†). The present method also allowed easy variation of protocell sizes. The size of the lipid-PSH condensate was readily increased by longer phase separation duration (Fig. S5b†). Lowered phase separation temperature also resulted in larger sizes but with irregular shapes (Fig. S5c†).
A high protein density inside the lipid-coated PSH condensates was observed in a TEM image, unlike liposomal giant unilamellar vesicles (GUVs) (Fig. 1d). The fluidity of the lipid coating was next monitored by fluorescence recovery after photobleaching (FRAP). Lipid coatings on PSH condensates were not fluid (Fig. 1e), whereas lipid layers on GUV were highly fluidic (Fig. S6†). PSH condensates were reported to be highly rigid,24 and we also observed that the fluidity of PSH proteins inside lipid-condensate protocells was extremely low (Fig. 1f). Strong 6His/Ni-NTA interaction and condensate rigidity might contribute to the static nature of the lipid coating. To vary the inside diffusivity of the protocells, we also used a PSH variant, PRM-SH3-RL48 H (RL48), which contains a rigid/long peptide linker between PRM-SH3 and 6His tag. Compared to PSH, RL48 forms similarly dense but more fluid protein condensates.24 Lipid-coated RL48 condensate protocells were also stably fabricated (Fig. 1g and S7a†). Compared to PSH protocells, however, slight penetration of lipids into condensates was observed (Fig. S7b and c†), likely due to the high fluidity of the RL48 condensates.24 RL48 proteins in protocells were significantly more mobile than PSH (Fig. 1h). On the other hand, the LLPS tendency of RL48 was weaker than that of PSH,24 and thereby RL48 produced fewer but larger condensate protocells (Fig. S7a†). Interestingly, adding a small portion of PSH (3%) to RL48 enhanced LLPS tendency and yielded more protocells, while maintaining condensate fluidity. In addition, lipid penetration into interior of this RL48 + PSH condensate was also reduced (Fig. 1g).
Average lipid signal intensities on the surface areas of various protocells were measured. Lipid coating signals on PSH condensates were similar to that of GUV (Fig. 1i). On the other hand, lipid signals for RL48 condensate protocells were several times higher than those of GUV and PSH protocells. Increased lipid signals likely originated from lipid penetration into more fluidic condensates. We further varied the protein condensates and applied them to protocell formation. Two PSH condensates were separately labeled, formed, and mixed with lipids. These PSH condensates were not miscible, forming janus-like structures, and lipids were stably coated on these heterogeneous condensates (Fig. S8†). The data suggest that additional compartmentalization is possible inside condensate-protocells. We also examined an intrinsically disordered protein, LAF, which is a well-studied LLPS scaffold protein both in vitro and in vivo.26 Again, lipids were specifically coated on LAF condensates (Fig. S9†). However, these LAF protocells were deformed and merged by centrifugation. LAF condensates are less dense and more fluidic than PSH condensates.27 Highly dense and rigid PSH condensate structures are likely key conditions for construction of stable protein condensate protocells.
Properties of lipid-coated protein condensates
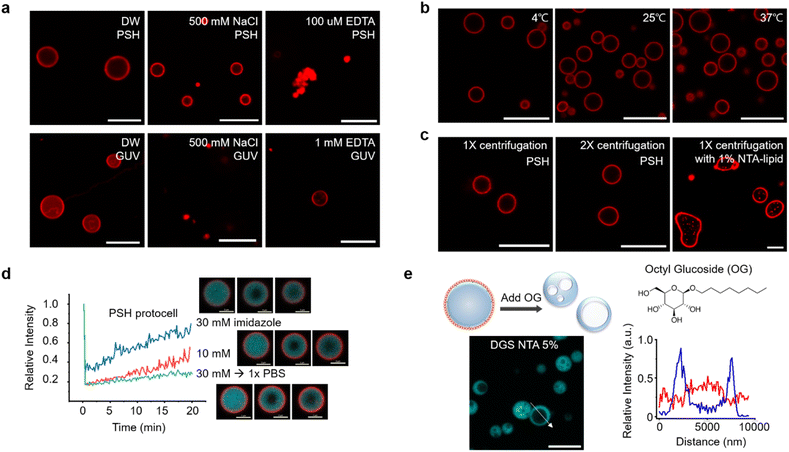
The stability of centrifuged lipid-protected PSH condensates was further examined against various environmental changes. Lipid-PSH condensates were stable in both desalted (DW) and high salt (500 mM NaCl) solutions (Fig. 2a). On the other hand, liposomal GUV collapsed quickly by 500 mM NaCl, likely due to osmotic pressure. Protein interaction networks inside the PSH condensates might contribute to this high protocell stability. When condensates were disassembled by Ni2+ chelation with EDTA and subsequent protein interaction disruption, the protocells shrunk and lipids agglomerated (Fig. 2a). In addition, lipid-PSH protocells were structurally unaffected and remained well dispersed without agglomeration after 1 h of incubation at low (4 °C) and high (37 °C) temperatures (Fig. 2b). We also examined whether the constructed protocells could be additionally centrifuged. Lipid-condensates were stable after a second centrifugation and resuspension (Fig. 2c). We expect that external substances can be repeatedly washed/exchanged, and the outer environments can be readily altered for the present lipid-condensate protocells, which will allow many complex protocell activities. For example, protocells can be used as bioreactors for multi-step biochemical reactions, and responses of artificial cells against sequential environmental changes can be more precisely monitored. More diffusive lipid-RL48 condensates could also be stably centrifuged and resuspended twice (Fig. S10†). Strong binding of NTA-lipid on 6His tags of condensates is important for this protocell stability against centrifugation given that condensates with a 1% (instead of 5%) NTA-lipid coating showed non-spherical irregular shapes after centrifugation (Fig. 2c).We next examined the effect of altering the condensate property on protocell structures. Although EDTA completely sequestered Ni2+ and disassembled condensates, imidazole moderately and concentration-dependently weakened the interaction between Ni2+ and 6His. When 10 mM or 30 mM imidazole was added to protocells, the inside condensate diffusivity was clearly (concentration-dependently) increased, likely by weakening protein interaction networks, whereas the lipid-coated condensate structures remained intact (Fig. 2d). In addition, imidazole removal by centrifugation restored the rigidity of the condensates. It has been reported that in-cell viscosity heavily influences many cellular activities inside cells.28 An ability to control the inside diffusivity of protocells will be a valuable asset when studying and utilizing artificial cell-like entities.
We also disrupted lipid structures by adding detergent OG to lipid-coated condensates. Upon OG addition, the outer lipid coatings completely disappeared. Interestingly, the resulting protein condensates showed cavity-containing hollow structures (Fig. 2e). Weak lipid signals were observed inside the cavities of these hollow condensates. Surface bound lipids are necessary for this cavity formation since this morphological change was not observed by adding OG to the protein condensates without a lipid coating (Fig. S11†). It is likely that with OG, a portion of condensate-bound lipids penetrated the protein condensate and formed a cavity. Even when condensates were covered with fewer lipids, the cavity formation was still observed (Fig. S11†). During OG-induced disruption of the lipid layers, some lipids might be uptaken by condensates, and amphiphilic lipids gather together to form interior cavities. In fact, the initially formed small cavities merged into larger ones inside the condensates (Fig. S12a†). During cavity formation, a small compound dye (calcein) in solution was included in the cavities, whereas larger proteins were mostly excluded from these cavities (Fig. S12b†). Although the underlying principles of the observed morphological changes are not fully understood, the present protocells offer another approach to create additional compartments inside condensates.
Biomolecule permeability of lipid-coated protein condensates
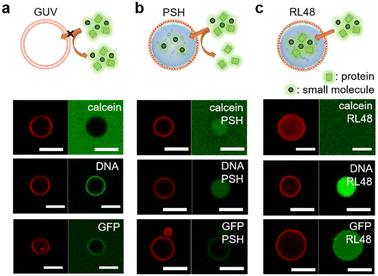
Natural cells precisely control the entry of outside substances through lipid bilayer membranes by using membrane embedded receptors and pores. This selective uptake, however, is still extremely difficult to achieve for artificial protocells. We first examined the permeability of lipid–protein condensates against a small compound dye (calcein), 21 base pair (bp) DNA, and a globular protein GFP. Liposomal GUV was not permeable to all these substances, although DNA was highly enriched around GUV surfaces (Fig. 3a and Table S1†). Interestingly, however, lipid-coated PSH condensates were permeable to calcein and DNA, while GFP was not observed inside the protocells (Fig. 3b and Table S1†). Importantly, DNA was strongly enriched inside the lipid-condensates, and even long (360 bp) DNA was also uptaken by the lipid-condensates (Fig. S13 and Table S1†). We envision that condensate surfaces might not be fully covered (or blocked) by lipid clusters, generating multiple entrances for external substances, while these lipid clusters still effectively stabilize coated condensates. Biomolecule uptake would then be largely governed by inside protein condensates. Previous studies reported that DNA and RNA molecules were well recruited into various protein condensates.27,29 More specifically, highly charged protein condensates (like PSH condensates) showed strong uptake abilities for nucleic acids, particularly for single stranded DNA and RNA. Unlike linear DNA, globular (6His tagged) GFP was localized only around PSH condensate surfaces, likely via 6His interaction with Ni2+ containing lipid-NTA or condensate 6His. On the other hand, GFP was permeable (even slightly enriched) to lipid-coated RL48 and RL48 + PSH condensates (Fig. 3c, S14, and Table S1†), which are significantly more fluidic than rigid PSH condensates. It would be easier for GFP to penetrate into more fluid condensates than into rigid condensates. In addition, the long linker on RL48 might generate more internal spaces in condensates for external proteins to enter.Unlike calcein, 5 kDa poly ethylene glycol (PEG) was slightly excluded from the all tested protocell condensates (Fig. S14 and Table S1†). Interestingly, all protocells were permeable to large cage proteins (ferritin30 ∼15 nm and mi331 ∼25 nm) (Fig. S14 and Table S1†). Uptake data indicate that the lipid coating stabilizes the protein condensates against physical (centrifugation) and chemical (buffer exchanges) stresses, but at the same time, this coating is permeable enough for condensates to uptake various substances. Uptake degrees then will depend on the properties of external substances such as size, charge, and shapes. The observed permeability data suggest that protein condensates can be engineered to manipulate their uptake abilities against diverse materials.
Selective biomolecule uptake by lipid-coated protein condensates
For more selective and controlled biomolecule uptake by protocells, we used specific interactions between protein condensates and external proteins (Fig. 4a). First, external GFP was fused with the condensate forming PSH, which can bind to highly dense PSH in protocells. A low concentration (0.1 μM) of GFP-PSH was treated to protocells to examine selective uptake strengths. GFP-PSH was strongly recruited and enriched into RL48 + PSH condensate protocells (Fig. 4b). The GFP-PSH concentration inside RL48-PSH protocells reached 30 μM, which is 300-fold enrichment from treated 0.1 μM (Fig. S15†). Real time imaging of this uptake process indicated that GFP-PSH was slowly recruited from outside to inside over 10 min (Fig. 4b and Video S1†).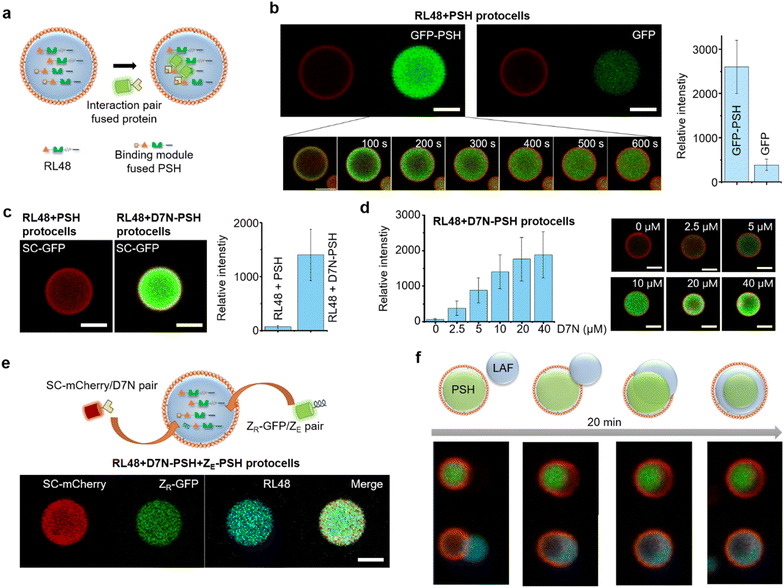 | ||
| Fig. 4 Selective protein uptake by functionalized protocells. (a) Schematics of interaction-driven selective uptake. (b) Confocal images of RL48 + PSH protocells treated with GFP-PSH (left) or free GFP (right). Relative intensities of recruited GFP signals are shown in the right graph. Error bars: 1 s. d. (n > 10). Real-time images of GFP-PSH uptake by a protocell are shown in the bottom. (c) Confocal images of RL48 + PSH (left) and RL48 + D7N-PSH (right) protocells treated with SC-GFP. Relative intensities of recruited GFP signals are shown in the right graph. Error bars: 1 s. d. (n > 10). (d) Relative SC-GFP intensities in RL48 + D7N-PSH protocells with different D7N-PSH portions inside condensates with representative confocal images (right). Error bars: 1 s. d. (n > 10). D7N 0 μM and 10 μM data are identical to those in Fig. 4c. (e) Simultaneous uptake of SC-mCherry (red) and ZR-GFP (green) by RL48 + D7N-PSH + ZE-PSH protocells. (f) Schematics and confocal images of LAF condensate (cyan) engulfing by a PSH protocell (red: lipid, green: PSH condensate). Scale bars 5 μm. | ||
We next implemented other interaction pairs for selective protein uptake to further diversify recruitment processes of protocells. A SpyTag variant D7N peptide, which specifically binds to SpyCatcher (SC) (KD ∼1 μM),32 was fused with PSH, and the resulting D7N-PSH was partly included in protocells for selective uptake of SC-fused proteins (Fig. 4a). Protocells with D7N-PSH behaved similar to other protocells (Fig. S16†). When 0.1 μM SC-GFP was treated, SC-GFP was strongly recruited into D7N-PSH-containing RL48 protocells but not to protocells without D7N-PSH (Fig. 4c). SC-GFP was enriched over 90 fold (∼9.2 μM) from initial 0.1 μM (Fig. S15†), indicating that a specific interaction with an only moderate affinity (KD ∼1 μM) could still facilitate strong protocell uptake. Furthermore, we also varied the functionalized scaffold (D7N-PSH) portion in protocells to control protocell uptake power against SC-fused external proteins. Recruited SC-GFP signals were clearly enhanced as D7N-PSH concentrations in protocell condensates increased (Fig. 4d and S15†).
Protocell-forming condensates were further functionalized with an additional binding module to induce simultaneous uptake of two external proteins via different interactions. A negative charged leucine zipper ZE, which forms a coiled coil structure with a positive ZR,33 was fused to PSH. ZR-GFP was effectively recruited into ZE-PSH-containing protocells, although enrichment degrees were notably lower than those by SC-GFP uptake (Fig. S17†), possibly due to repulsions between positively charged ZR and PSH condensates. Both D7N-PSH and ZE-PSH were included in the RL48 protocells, and SC-mCherry (red fluorescent protein) and ZR-GFP were treated. Both external proteins were effectively recruited into protocells (Fig. 4e). These data indicate that the present protein condensate-based protocells could uptake multiple external proteins with controlled recruitment powers by implementing multiple binding pairs and manipulating binding module portions in the condensates.
Many protein condensates can interact and fuse with other condensates both homogeneously (miscible) and heterogeneously (immiscible). Immiscible fusion produces various multiphase structures in cells also in model systems34 as we also demonstrated with immiscible PSH condensates (Fig. S8†). Therefore, we also examined external condensate fusion to our lipid-coated protein condensates. Less dense and fluidic LAF condensates (Fig. S9†) were treated to PSH protocells. Interestingly, PSH protocells slowly engulfed external LAF condensates (Fig. 4f), mimicking phagocytosis.35 The lipid coating was momentarily disrupted at the initial contact region but recovered around the protocell as engulfing was completed. Upon engulfing, a more fluidic LAF condensates were spread around more rigid and spherical PSH condensates in protocells. Similar engulfing processes and the resulting multiphase structures were observed with other protein condensates such as PSH condensate treatment to LAF protocells or RL48 condensate treatment to PSH protocells (Fig. S18†). This observation suggests that various external biomolecules can be condensed and engulfed by protocells for massive and highly concentrated biomolecular uptake.
Conclusions
We fabricated lipid-coated protein condensates as protocells with high stability and a selective uptake ability, which are two of the most challenging features to implement on artificial cell-like entities. Highly dense and rigid protein condensates offered unusual protocell stability against centrifugations and chemical stresses. His-Ni-NTA interaction-driven lipid coating was also crucial for protocell stability, importantly by preventing fusion between condensates. We suggest that condensates are coated by many lipid clusters to form pickering emulsion like structures, which allow entrance of external materials. With this permeability of lipid coats, protein condensate protocells showed strong uptake of target proteins via specific protein interactions and even engulfing of other condensates. As membrane-less organelles in cells, protein condensates can selectively uptake a specific set of biomolecules. By simple genetic engineering of condensate forming proteins, diverse binding modules can be included in protocells for selective uptake of many different biomolecules. However, the detail structure of lipid coating still remains to be determined. We will further modify the lipid coating method to apply for more diverse protein condensates with different fluidities. It will be also interesting to examine how lipid-coated protein condensates interact with natural cells. Our protocells will also offer great systems to study biological and chemical activities inside phase separated biomolecular condensates with their abilities to confine specific reactants (by selective uptake) and to be isolated for reliable analysis (by centrifugation).Data availability
All experimental supporting data and procedures are available in the ESI.†Author contributions
J. Son conducted all experiments. J. Son and Y. Jung designed the project and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by BioNano Health Guard Research Center funded by the Ministry of Science and ICT (MSIT) as Global Frontier Project (H-GUARD_2014M3A6B2060507 (1711073453)) and the KC30 grant funded by KAIST.References
- C. Guindani, L. C. da Silva, S. Cao, T. Ivanov and K. Landfester, Angew. Chem., Int. Ed., 2022, 61, e202110855 CrossRef CAS PubMed.
- K. Ruiz-Mirazo, C. Briones and A. de la Escosura, Chem. Rev., 2014, 114, 285–366 CrossRef CAS PubMed.
- P. Schwille, J. Spatz, K. Landfester, E. Bodenschatz, S. Herminghaus, V. Sourjik, T. J. Erb, P. Bastiaens, R. Lipowsky, A. Hyman, P. Dabrock, J. C. Baret, T. Vidakovic-Koch, P. Bieling, R. Dimova, H. Mutschler, T. Robinson, T. D. Tang, S. Wegner and K. Sundmacher, Angew. Chem., Int. Ed., 2018, 57, 13382–13392 CrossRef CAS PubMed.
- S. Mann, Angew. Chem., Int. Ed., 2013, 52, 155–162 CrossRef CAS PubMed.
- D. W. Green, J. A. Watson, B. Ben-Nissan, G. S. Watson and A. Stamboulis, Biomaterials, 2021, 276, 120941 CrossRef CAS.
- K. A. Podolsky and N. K. Devaraj, Nat. Rev. Chem., 2021, 5, 676–694 CrossRef CAS.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC.
- K. L. Thompson, M. Williams and S. P. Armes, J. Colloid Interface Sci., 2015, 447, 217–228 CrossRef CAS.
- D. van Swaay and A. deMello, Lab Chip, 2013, 13, 752–767 RSC.
- R. Rodríguez-García, M. Mell, I. López-Montero, J. Netzel, T. Hellweg and F. Monroy, Soft Matter, 2011, 7, 1532–1542 RSC.
- D. C. Dewey, C. A. Strulson, D. N. Cacace, P. C. Bevilacqua and C. D. Keating, Nat. Commun., 2014, 5, 4670 CrossRef CAS PubMed.
- N. N. Deng, Biomicrofluidics, 2020, 14, 051301 CrossRef CAS PubMed.
- R. J. Ellis, Trends Biochem. Sci., 2001, 26, 597–604 CrossRef CAS PubMed.
- C. A. Strulson, R. C. Molden, C. D. Keating and P. C. Bevilacqua, Nat. Chem., 2012, 4, 941–946 CrossRef CAS PubMed.
- E. Sokolova, E. Spruijt, M. M. Hansen, E. Dubuc, J. Groen, V. Chokkalingam, A. Piruska, H. A. Heus and W. T. Huck, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11692–11697 CrossRef CAS PubMed.
- Y. Zhang, Y. Chen, X. Yang, X. He, M. Li, S. Liu, K. Wang, J. Liu and S. Mann, J. Am. Chem. Soc., 2021, 143, 2866–2874 CrossRef CAS PubMed.
- S. Liu, Y. Zhang, M. Li, L. Xiong, Z. Zhang, X. Yang, X. He, K. Wang, J. Liu and S. Mann, Nat. Chem., 2020, 12, 1165–1173 CrossRef CAS PubMed.
- C. Zhao, J. Li, S. Wang, Z. Xu, X. Wang, X. Liu, L. Wang and X. Huang, ACS Nano, 2021, 15, 10048–10057 CrossRef PubMed.
- Y. Qiao, M. Li, R. Booth and S. Mann, Nat. Chem., 2017, 9, 110–119 CrossRef CAS PubMed.
- W. J. Altenburg, N. A. Yewdall, D. F. M. Vervoort, M. van Stevendaal, A. F. Mason and J. C. M. van Hest, Nat. Commun., 2020, 11, 6282 CrossRef CAS.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS.
- G. L. Dignon, R. B. Best and J. Mittal, Annu. Rev. Phys. Chem., 2020, 71, 53–75 CrossRef CAS PubMed.
- S. Alberti, A. Gladfelter and T. Mittag, Cell, 2019, 176, 419–434 CrossRef CAS PubMed.
- K. Hong, D. Song and Y. Jung, Nat. Commun., 2020, 11, 5554 CrossRef CAS PubMed.
- A. W. Folkmann, A. Putnam, C. F. Lee and G. Seydoux, Science, 2021, 373, 1218–1224 CrossRef CAS PubMed.
- S. Elbaum-Garfinkle, Y. Kim, K. Szczepaniak, C. C. Chen, C. R. Eckmann, S. Myong and C. P. Brangwynne, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 7189–7194 CrossRef CAS PubMed.
- Y. Jo and Y. Jung, Chem. Sci., 2019, 11, 1269–1275 RSC.
- L. B. Persson, V. S. Ambati and O. Brandman, Cell, 2020, 183, 1572–1585 CrossRef CAS PubMed.
- T. J. Nott, T. D. Craggs and A. J. Baldwin, Nat. Chem., 2016, 8, 569–575 CrossRef CAS PubMed.
- P. M. Harrison and P. Arosio, Biochim. Biophys. Acta, 1996, 1275, 161–203 CrossRef.
- Y. Hsia, J. B. Bale, S. Gonen, D. Shi, W. Sheffler, K. K. Fong, U. Nattermann, C. Xu, P.-S. Huang, R. Ravichandran, S. Yi, T. N. Davis, T. Gonen, N. P. King and D. Baker, Nature, 2016, 535, 136–139 CrossRef CAS PubMed.
- B. Zakeri, J. O. Fierer, E. Celik, E. C. Chittock, U. Schwarz-Linek, V. T. Moy and M. Howarth, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E690–E697 CrossRef CAS PubMed.
- W. H. Landschulz, P. F. Johnson and S. L. McKnight, Science, 1988, 240, 1759–1764 CrossRef CAS PubMed.
- Y. Shin and C. P. Brangwynne, Science, 2017, 357, eaaf4382 CrossRef PubMed.
- D. M. Richards, Adv. Exp. Med. Biol., 2020, 1246, 55–70 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc03123j |
| This journal is © The Royal Society of Chemistry 2022 |