Open Access Article
Open Access ArticleAn N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction†
Victoria N.
Drago
ab,
Steven
Dajnowicz
ab,
Jerry M.
Parks
 c,
Matthew P.
Blakeley
c,
Matthew P.
Blakeley
 d,
David A.
Keen
d,
David A.
Keen
 e,
Nicolas
Coquelle
d,
Kevin L.
Weiss
b,
Oksana
Gerlits
f,
Andrey
Kovalevsky
e,
Nicolas
Coquelle
d,
Kevin L.
Weiss
b,
Oksana
Gerlits
f,
Andrey
Kovalevsky
 *b and
Timothy C.
Mueser
*b and
Timothy C.
Mueser
 *a
*a
aDepartment of Chemistry and Biochemistry, University of Toledo, Toledo, OH 43606, USA. E-mail: timothy.mueser@utoledo.edu
bNeutron Scattering Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, USA. E-mail: kovalevskyay@ornl.gov
cBiosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, USA
dLarge Scale Structures Group, Institut Laue–Langevin, 71 Avenue des Martyrs, 38000 Grenoble, France
eISIS Facility, Rutherford Appleton Laboratory, Harwell Campus, Didcot, OX11 0QX, UK
fDepartment of Natural Sciences, Tennessee Wesleyan University, Athens, TN 37303, USA
First published on 17th August 2022
Abstract
Pyridoxal 5′-phosphate (PLP)-dependent enzymes have been extensively studied for their ability to fine-tune PLP cofactor electronics to promote a wide array of chemistries. Neutron crystallography offers a straightforward approach to studying the electronic states of PLP and the electrostatics of enzyme active sites, responsible for the reaction specificities, by enabling direct visualization of hydrogen atom positions. Here we report a room-temperature joint X-ray/neutron structure of aspartate aminotransferase (AAT) with pyridoxamine 5′-phosphate (PMP), the cofactor product of the first half reaction catalyzed by the enzyme. Between PMP NSB and catalytic Lys258 Nζ amino groups an equally shared deuterium is observed in an apparent low-barrier hydrogen bond (LBHB). Density functional theory calculations were performed to provide further evidence of this LBHB interaction. The structural arrangement and the juxtaposition of PMP and Lys258, facilitated by the LBHB, suggests active site preorganization for the incoming ketoacid substrate that initiates the second half-reaction.
Introduction
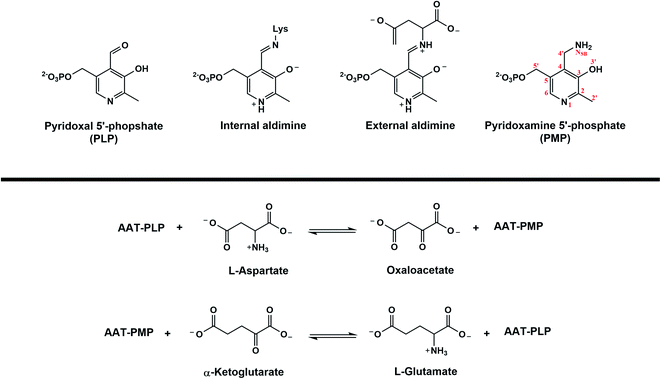
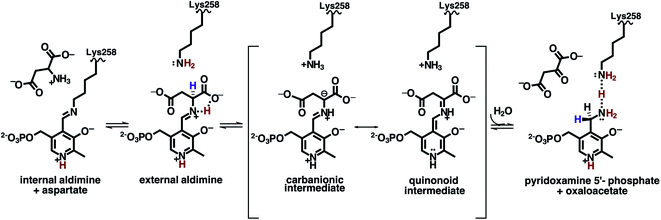
The family of enzymes utilizing biologically active phosphorylated derivatives of vitamin B6, pyridoxal 5′-phosphate (PLP) and pyridoxamine 5′-phosphate (PMP), have long been studied for their chemical diversity. A genomic study of PLP-dependent enzymes revealed that ∼4% of all classified activities can be attributed to this ubiquitous family of enzymes, which include transamination, β- and γ-elimination, decarboxylation, phosphorylation, aldol cleavage, racemization, and replacement reactions.1,2 Owing to their involvement in amino acid metabolism and synthesis of amino acid derivatives, many PLP-dependent enzymes have been identified as potential drug targets.3 Detailed understanding of PLP-facilitated catalysis has therefore become necessary for specific inhibitor design.PLP-dependent enzymes are unified by the conserved active site lysine and the formation of internal and substrate-bound external aldimine structures (Fig. 1a). From the external aldimine onward, the reaction specificity arises. How PLP-mediated catalysis is achieved is governed by two primary concepts. Bond cleavage in PLP-dependent enzymes is promoted by the arrangement of the scissile bond perpendicular to the conjugated system of PLP, known as the Dunathan hypothesis.4 The PLP resonance then acts as an electron sink to withdraw electrons from the scissile bond into the pyridine ring to stabilize intermediates and promote bond cleavage and rearrangements.5 These basic concepts explain how PLP can promote catalysis, but they do not describe the diversity of PLP-dependent enzyme activity. Selective protonation of the PLP cofactor directed by the active site environment is considered the most likely explanation for how PLP-dependent enzymes promote specific chemistries.6 Hence, PLP-dependent catalysis and the enzyme specificities cannot be thoroughly described without knowing the positions of hydrogen atoms that determine the protonation states in the active sites. As a means of directly visualizing hydrogen (and deuterium) positions within a protein, neutron diffraction has proven to be an invaluable technique for our exploration of PLP-driven catalysis.7
Neutron diffraction is a non-destructive technique that allows for the direct observation of hydrogen, H, (or deuterium, D) positions in a protein because the coherent neutron scattering cross-sections of H and D are comparable to those of C, N and O. Unlike X-ray diffraction, which requires ultra-high resolution data to visualize hydrogens, H and D positions are visible in neutron scattering length (or nuclear) maps even at moderate resolutions of ∼2.5 Å.8,9 Moreover, cold neutrons with the wavelengths >2 Å normally used in macromolecular neutron crystallography experiments do not cause radiation damage to protein crystals; thus, radiation damage-free structures can be obtained at near-physiological, ambient temperature. Knowledge of protonation states gained from neutron diffraction is vital in understanding catalysis and ligand binding, can be used to gain further information on protein dynamics, and can help generate accurate starting models for quantum chemical calculations.10–14 Visualization of protonation states is essential for understanding the electronic modulation of PLP, which is necessary to discern enzymatic mechanisms.
Aspartate aminotransferase (AAT), a model system for fold type I PLP-dependent enzymes, is a biological homodimer that catalyzes the reversible conversion between aspartate and oxaloacetate, and α-ketoglutarate and glutamate in an ordered, sequential (ping-pong) mechanism (Fig. 1b).15 In the first half-reaction, substrate aspartate reacts with the internal aldimine to free catalytic Lys258 to produce the external aldimine through a gem-diamine intermediate, setting the stage for a series of proton transfers between substrate-conjugated PLP and Lys258 that ultimately result in the generation of intermediate products PMP and oxaloacetate. In the second, less well-studied half-reaction, PMP facilitates conversion of α-ketoglutarate to glutamate going backward along the reaction path described above to regenerate the internal aldimine.
The AAT monomer is described as having a large, fixed domain and a small, flexible domain. The physiological assembly of AAT is a homodimer with the intermonomer contacts primarily between the large domains. The first segment of the N-terminal residues tethers the two domains and the next segment contacts the C-terminal region to form the small flexible domain. The active site residues are positioned between the small and large domains (Fig. 2a). The small domain undergoes conformational change upon substrate binding, switching between the substrate-free internal aldimine open-state to the substrate-bound external aldimine closed-state. The open conformation allows the incoming substrate to bind as a Michaelis complex, inducing a hydrophobic patch of residues to close over the active site (Fig. 2b). In the crystal structures of AAT, crystal packing holds the small domain of one monomer (chain B in Fig. 2a) open, providing a unique opportunity to view the active sites in two different forms.16,17 Thus, we have exploited the uniqueness of AAT crystal packing to study the enzyme's catalytic mechanism using neutron crystallography. We previously obtained a neutron structure that contained both internal and external aldimines after the reaction with α-methylaspartate substrate analogue because it reacts only with the PLP located in the monomer capable of the open–close conformational change in the crystal (chain A in Fig. 2a).17 We thus directly determined the protonation states of the PLP in both the internal and external aldimine states, which were similar to those previously obtained using NMR.18,19
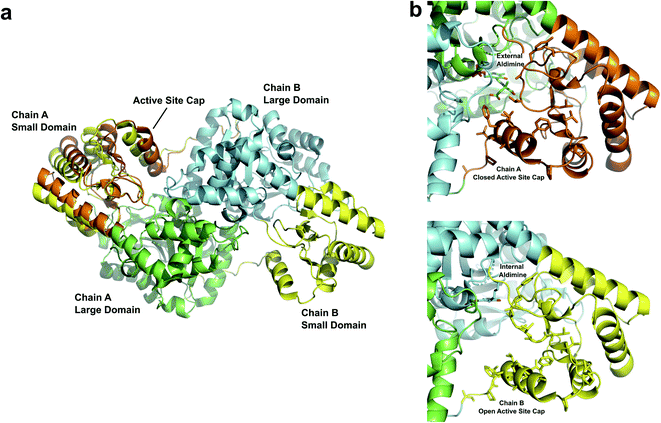 | ||
| Fig. 2 (a) Overview of AAT homodimer with domain labels (PDB 5VJZ). Chain A (large domain in green) is shown with the small domain in the open (yellow) and closed (orange) conformations. Both positions of the small domain are shown to demonstrate the movement of the active site cap. Chain A (large domain in cyan), which has crystal contacts preventing the closure of the active site cap, is depicted with the small domain in an open (yellow) conformation. (b) Zoomed-in views of the active site opening with external (top) and internal (bottom) aldimines bound. The internal aldimine state adopts an open conformation. The external aldimine becomes closed off by hydrophobic residues in the active site cap. | ||
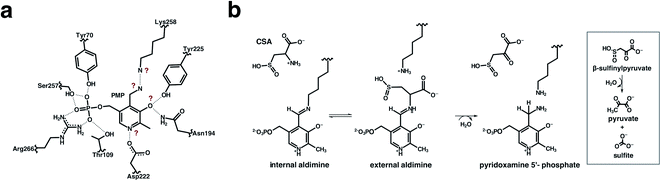
Here we describe a 2.2 Å joint X-ray/neutron (XN) structure of AAT in which we captured the stable pyridoxamine 5′-phosphate (PMP), the product of the first half-reaction. By directly observing D atoms in the AAT-PMP complex, we determined the protonation states of the chemical groups on the PMP and active site residues specifically indicated with question marks in Fig. 3a that are of particular interest for mapping the enzyme's catalytic mechanism, allowing visualization of hydrogen bonding interactions in the active site. To generate the AAT-PMP complex, we used a crystal in which α-methylaspartate is already bound in the active site of chain A.17 This crystal was treated with a solution of L-cysteinesulfinic acid (CSA) which, instead of displacing the 2-methylasparate in chain A, reacted with the internal aldimine PLP located in the “locked open” active site of chain B and the reaction proceeded all the way to PMP. In the normal aspartate reaction, the Schiff base nitrogen (NSB) of the internal or external aldimines becomes the primary amine of PMP when oxaloacetate is released by hydrolysis. When CSA reacts, deamination releases an unstable product, β-sulfinylpyruvate, which hydrolyzes to pyruvate and sulfite, thereby abrogating the reverse reaction (Fig. 3b).20 The PMP nitrogen rotates to the si face, forming a close hydrogen bond with the ζ-amine of Lys-258. Nuclear density maps revealed the presence of a potential low-barrier hydrogen bond (LBHB) between the amino groups of the catalytic Lys258 and PMP. To our knowledge, this is the first time an N⋯H⋯N LBHB has been experimentally detected in any protein structure, with H observed as D in the joint XN structure. Quantum chemical calculations on a cluster model derived from the neutron structure provided additional evidence for the LBHB. Our neutron structure contributes to the overall understanding of the reaction mechanism of AAT by mapping exact locations of hydrogen atoms in the AAT active site at the stage of the first half-reaction product.
Results and discussion
Joint XN structure
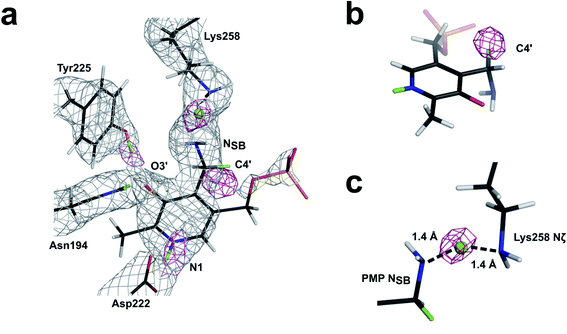
Room-temperature neutron diffraction data to 2.2 Å resolution were collected from a crystal of AAT in complex with PMP. Joint XN refinement was performed using the 1.7 Å room-temperature X-ray data collected from the same crystal to solve the structure and discern the protonation states of the PMP, Lys258 and surrounding ionizable residues (Fig. 3a and Table S1†). Deuterated AAT was purified and crystallized in hydrogenated solvents and the crystal was H/D exchanged by vapor diffusion prior to data collection. Importantly, we have not been able to co-crystallize AAT with substrate analogues, resorting instead to crystal soaking to obtain the AAT-PMP neutron structure. In the AAT crystal, the crystal packing holds the small domain of one chain (chain B) in the homodimer in an open position, resulting in the two active sites possessing different in crystallo activities. In the oxidative catabolism of cysteine, the intermediary cysteine sulfinic acid (CSA) may undergo deamination by AAT (Fig. 3b). This reaction yields β-sulfinylpyruvate, which rapidly degrades to sulfite and pyruvate. In an isolated reaction, the CSA substrate can be utilized to form a stable PMP-bound AAT. In this study, we reused the crystal that was previously reacted with α-methylaspartate to exhibit the external aldimine in chain A and internal aldimine in chain B. By soaking this crystal with CSA, reaction in chain B converted the internal aldimine entirely to PMP, the final product of the first half-reaction even though the chain is locked open. For chain A, two scenarios are possible: (1) CSA displaces α-methylaspartate forming PMP or (2) no reaction with CSA occurs because the small domain is already in the closed conformation. Interestingly, CSA partially reacts with the external aldimine producing uninterpretable mixed electron and nuclear density maps. As a result, the active site in chain A will not be further discussed.One hypothesis for understanding PLP-dependent catalysis suggests that the PLP cofactor is electronically modulated through selective protonation driven by the active site environment. Therefore, knowledge of the hydrogen atom positions on the cofactor, the active site residues, and hydrogen atom movements during catalysis are critically important for fully understanding PLP-dependent enzymes. In the AAT-PMP neutron structure, we observed no nuclear density peak on the phenolic oxygen O3′ of PMP (see Fig. 1 for atom labels), indicating a deprotonated, negatively charged, state. The charge on the deprotonated O3′ is stabilized by the partial C–O double bond character provided through electron resonance with the pyridine ring and additionally by two hydrogen bonding interactions with Tyr225 and Asn194 side chains. The pyridine N1 nitrogen of PMP is found in the protonated, positively charged state, having a positive nuclear density peak for a D atom near N1 (Fig. 4a). Protonation of N1 is proposed to increase the charge delocalization by resonance. The same charge-separated state of PLP was observed in the internal and external aldimines.17 Thus, this aromatic part of PLP remains a zwitterion starting from the reactant (internal aldimine) and through the product (PMP). The hydrogen-bonding network consisting of Asp222, His143, and His189 connects PLP N1 to the bulk solvent and has been shown to influence cofactor electrophilicity. In AAT-PMP, His143 is singly protonated on Nε2, and thus neutral, whereas His189 is doubly protonated and thus positively charged. In the internal aldimine, His189 is singly protonated on Nε2,17 whereas same protonation states for these two histidine residues were observed in the external aldimine as in AAT-PMP. Hence, no protons were transferred to or from the bulk solvent for the reaction to proceed beyond the external aldimine. A singly protonated His193 located near His189 is locked in hydrogen bonding interactions with Ser136 and Thr196 and maintains its protonation state throughout the reaction. A direct visualization of the 1,3-proton shift is present in this new structure. A positive nuclear density peak was observed next to the aminomethyl C4′ of PMP, indicating that one of its hydrogens is a deuterium, i.e., C4′ is in fact a –C(H)D group (Fig. 4b). In this crystal, the protein is deuterated, the PLP cofactor is hydrogenated, and the C4′ is not an exchangeable site. Detection of a D atom on C4′ can be readily explained as follows. The proton on the substrate Cα is first abstracted by the Lys258 amino group, leading to the formation of a quinonoid intermediate whose lifetime is long enough for H/D exchange to occur. Subsequently, a D atom is delivered to the C4′ position by the protonated Lys258. Therefore, the 1,3-proton transfer through the si face stereochemical movement predicted by Dunathan4,21 and facilitated by Lys258 has occurred.
In the AAT-PMP neutron structure the amine NSB of PMP is positioned 2.8 Å away from the Lys258 amine Nζ, indicating a hydrogen bond. The nuclear density map revealed a D atom located halfway between the two amino nitrogens (Fig. 4c). Therefore, this hydrogen bond can potentially be a low-barrier hydrogen bond (LBHB). LBHBs are a type of short hydrogen bonds characterized by a delocalized proton shared equally by the two hydrogen bond acceptor atoms. In an LBHB, the zero-point vibrational frequency of the shared proton is close to or greater than the potential energy barrier, allowing the proton to move freely between the donor and acceptor atoms with the average position in the center.22–24 Although LBHBs have previously been found in the neutron structures of various enzymes, including an N⋯H⋯O LBHB detected in the external aldimine of AAT between the substrate carboxylate oxygen and NSB atoms,17,25,26 an N⋯H⋯N LBHB has not been observed previously. However, we note that in a neutron structure of cholesterol oxidase enzyme a short hydrogen bond between the main chain amide nitrogen of Gly residue and a flavin cofactor nitrogen was characterized as having an elongated amide N–H bond.27 The aminomethyl group of PMP in the AAT-PMP neutron structure is not coplanar with the pyridine, with the C4′–NSB bond rotated by 78° above the plane of pyridine (si face) to allow for the formation of the hydrogen bond with Lys258 (Fig. 5). The conformation of PMP in the active site, facilitated by the interaction with Lys258, is similar to that of the internal aldimine, which adopts a C3–C4–C4′–NSB torsion angle of 46° for the Schiff base. Conversely, in the external aldimine the C4′–NSB bond is moved below the pyridine plane (re face) possibly to avoid clash with the freed Lys258. LBHBs, with a high covalent character, are known to be stronger than conventional hydrogen bonds.28 As the aminomethyl group is free to rotate around the single C4–C4′ bond and PMP-NSB serves as a primary amine to react with ketoacids, the formation of a strong N⋯H⋯N LBHB would ensure a proper positioning of the PMP amine for the subsequent attack on the carbonyl of α-ketoglutarate, facilitating catalysis along the second half-reaction.
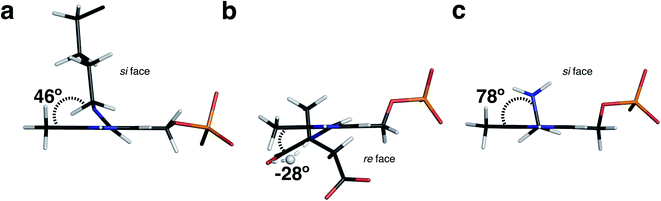 | ||
| Fig. 5 C3–C4–C4′-NSB torsion angles for (a) internal aldimine (PDB 5VJZ), (b) external aldimine (PDB 5VJZ), and (c) PMP. In both the internal aldimine and PMP, NSB is on the si face with C3–C4–C4′-NSB torsion angles of 46° and 78°, respectively. In the external aldimine NSB is on the re face with a C3–C4–C4′-NSB torsion angle of −28°. Active site preorganization holds the incoming substrate on the re face between Arg292 and Arg386 while Lys258 is held in the internal aldimine on the si face. When the substrate displaces Lys258 to form the external aldimine, the C3–C4–C4′-NSB torsion rotates so that NSB is on the re face. NSB returns to the si face upon formation of PMP and product release, suggesting that the active site is at a resting state when NSB is on the si face whereas NSB on the re face is active. | ||
Quantum chemical calculations
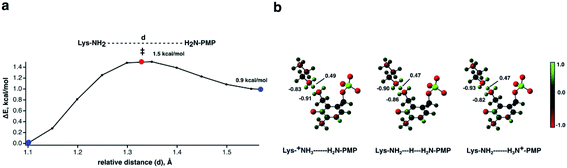
To provide further evidence that an LBHB is likely formed between PMP and Lys258 we performed density functional theory (DFT) calculations at the SMD/ωB97XD/Def2-SVP level of theory using a 151-atom cluster model derived from the neutron structure of AAT-PMP. Importantly, protonation states of the AAT amino acid residues and the PMP were kept as observed in the neutron structure, with the D atoms reverted to H. We placed the LBHB proton on the Lys258 amine, referred to as the reactant state, or on the PMP amine, referred to as the product state, and performed geometry optimizations of the two stationary points. The product state was found to be +0.9 kcal mol−1 higher in energy than the reactant. Thus, protonated PMP is slightly less stable than protonated Lys258 in the simplified cluster model of AAT (Fig. 6a). Next, we generated a potential energy profile for the proton transfer from the reactant to the product state. The calculated potential energy barrier heights for the forward and reverse proton transfers were very low at 1.5 kcal mol−1 for proton transfer from PMP to Lys258 and 0.6 kcal mol−1 for the reverse proton transfer. We emphasize that the calculations were performed at 0 K, with no zero-point energy corrections applied, and H nuclei were treated as classical, not quantum, particles. Therefore, the results should be considered as qualitative for comparison with the neutron structure in which the nuclei were observed directly from the experiment. At 300 K, the thermal energy kT would increase the energies of the reactant and product states relative to the transition state by ∼0.6 kcal mol−1, lowering the proton transfer barrier accordingly and indicating possible formation of an LBHB. In the DFT calculation the N⋯N separation is 2.7 Å in both the reactant and product states, and 2.6 Å in the transition state where H is located at the midpoint between the N atoms. The distances are close to the 2.8 Å N⋯N distance observed in the joint XN structure (Table S2†). Although the N⋯N separation appears shorter in the DFT calculations and may indicate an H/D isotopic effect leading to the N⋯N distance elongation in the joint XN structure, as was shown by NMR studies on small molecules,29–31 the difference of 0.1–0.2 Å is negligible. The reason is that the atomic position error for the heavy atoms in AAT-PMP complex based on the X-ray data is 0.17 Å, about the same as the N⋯N distance elongation. Thus, potential geometric H/D isotopic effects are not resolved within the margin of error in the joint XN structure. In the reactant and product states the respective amines are protonated ammonium ions (–NH3+). Natural population analysis (NPA) partial charges revealed that the partial charge of the shared proton is largely unaffected by its position in the hydrogen bond, with partial charges of +0.49, +0.47, and +0.47 in the reactant, transition state, and product, respectively (Fig. 6b). This result suggests that the proton is delocalized and that the hydrogen bond is also assisted by charge. In addition, considering the role of nuclear quantum effects such as zero-point motion, we conclude that the N⋯H⋯N hydrogen bond observed in the AAT-PMP neutron structure can be classified both as an LBHB and as a charge-assisted hydrogen bond.32,33Conclusion
Together with our previous neutron structures of AAT,17 we have now mapped out the positions of hydrogen atoms in the internal aldimine, external aldimine, and PMP, the three major intermediate states along the AAT-catalyzed transamination reaction pathway.3,15,34 Neutron diffraction has thus afforded us the opportunity to further clarify the mechanism of the first half-reaction in which Asp is converted to oxaloacetate (Fig. 7). As found in the internal and external aldimines, the PMP pyridine N1 is protonated and the PMP phenolic O3′ is deprotonated in the AAT-PMP neutron structure. A nuclear density peak on C4′ provides evidence of the fundamental 1,3-proton transfer in which the catalytic Lys258 facilitates proton abstraction from the substrate Cα and its subsequent placement on C4′’. PMP-NSB and Lys258-Nζ are primary amines locked in an apparent LBHB, orienting NSB on the si face of the active site above the plane of the PMP pyridine ring, ready to attack the carbonyl of an incoming α-ketoglutarate, which signifies the start of the second half-reaction. The LBHB appears to be important for catalysis as it preorganizes the active site for the incoming substrate preventing the highly dynamic aminomethyl group of PMP and the side chain of Lys258 from freely rotating in the active site cavity. DFT calculations provided evidence that an N⋯H⋯N LBHB is formed between PMP-NSB to Lys258-Nζ, in agreement with the observed position of the hydrogen atom in the AAT-PMP neutron structure.Materials and methods
Deuteration, purification and crystallization of AAT
E. coli T7 Express LacIQ cells carrying the gene for AAT were grown in deuterated minimal media prepared using hydrogenous glycerol and 99.8% D2O as described in detail previously.2,17,35 Overexpression of deuterated AAT was achieved in a BIOFLO 310 (Eppendorf) bioreactor, growing cells at 30 °C to an OD600 of 10.0. The cells were induced with 1 mM isopropyl β-D-thiogalactopyranoside at 20 °C and fed glycerol for 24 h. Purification was carried out as previously described using Ni-NTA affinity chromatography.35 The deuterated AAT crystal was grown in 40 mM NaOAc (pH 5.4), 2 mM PLP, 9% polyethylene glycol (PEG) 6000, and 10% glycerol using batch method crystallization. The PMP form of the cofactor was obtained by soaking the crystal with 50 mM Tris–HCl (pH 7.5), 5% PEG 6000, 5% glycerol, and 300 mM L-cysteinesulfinate in D2O for 24 h. The soaking was performed by directly adding liquid to the quartz capillary containing the crystal.Neutron diffraction data collection
Neutron quasi-Laue diffraction data from a 0.65 mm3 crystal of the AAT-PMP complex were collected at room temperature using the LADI-III diffractometer36 at the Institut Laue–Langevin (ILL) in Grenoble. A neutron wavelength range (Δλ/λ ∼ 30%) of 3.10–4.15 Å was used for data collection with diffraction data extending to 2.2 Å resolution. The crystal was held stationary at different φ (vertical rotation axis) for each exposure. A total of 32 images were recorded with an exposure time of 18 hours per image from 4 different crystal orientations. The neutron data were processed using the Daresbury Laboratory LAUE suite program LAUEGEN modified to account for the cylindrical geometry of the detector.37,38 The program LSCALE39 was used to determine the wavelength–normalization curve using the intensities of symmetry-equivalent reflections measured at different wavelengths. No explicit absorption corrections were applied. The data were merged and scaled using SCALA.40 Neutron data collection can be found in Table S1.†Room temperature X-ray diffraction data collection and structure refinement
After neutron diffraction data collection, room temperature X-ray diffraction data were collected on the same deuterated AAT crystal on beamline FIP-BM30A at the European Synchrotron Radiation Facility (ESRF) in Grenoble. X-ray diffraction data were integrated with XDS and reduced and scaled using XSCALE of the XDS program suite.41 The structure was refined against the room temperature X-ray diffraction data in Phenix42 to produce an input model for joint XN refinement. X-ray data collection statistics are shown in Table S1.†Joint X-ray/neutron (XN) refinement
Joint X/N refinement was carried out with the patch, nCNS,43 in the structure solution program, CNS44,45 using room temperature X-ray and neutron diffraction data. Following a single rigid body refinement, rounds of atomic position, individual atomic displacement, and occupancy refinements were performed with manual adjustments made to the structure in Coot.46 Joint XN refinement statistics are given in Table S1.† Based on the D incorporation into C–H bonds during protein expression and H/D exchange with D2O estimated using refined D atom occupancies, AAT is ∼85% deuterated.Quantum chemical calculations
A 151-atom cluster model was generated from the neutron crystal structure of AAT-PMP consisting of ten active site residues (Tyr70, Thr109, Trp140, N194, Asp222, Tyr225, Ser255, Ser257, Lys258, and Arg266), PMP, and two crystallographic waters (ESI Fig. S1†). The residues were truncated, and the free valences on terminal carbons were saturated with hydrogens. The terminal carbons were constrained to their crystallographic positions for all residues, as well as the oxygen positions of the two crystallographic waters. Tyr225, Tyr70, and Trp140 required additional atoms to be constrained to prevent internal rotations during the geometry optimizations.Geometry optimizations were performed at the SMD47/ωB97XD/Def2-SVP48,49 level of theory. Zero-point energies and other thermodynamic corrections were not included because the geometric constraints led to inaccuracies in the vibrational frequencies. A dielectric constant of 4.0 was used for the SMD solvent model to mimic the surrounding enzyme environment. The ωB97XD density functional approximation was chosen for its ability to describe hydrogen bonding interactions and barrier heights accurately.50 Vibrational frequency calculations were used to confirm energy minima and transition states. All calculations were performed with Gaussian 16.51 A rigid potential energy scan was used to map the potential energy pathway for the proton transfer between Lys258-Nζ and PMP-NSB. Ten steps were taken with a step size of 0.1 Å. The resulting profile provided an initial guess for the transition state, which was subsequently used to perform a transition state optimization. Natural population analysis (NPA) charges of the stationary points were calculated with the NBO program in Gaussian16.52
Data availability
The structure and corresponding structure factors have been deposited into the protein data bank with the PDB accession code 7TUR. ESI† is available online.Author contributions
SD, AK and TCM designed the study. VND and SD expressed and purified the protein. VND, SD and KLW performed protein deuteration. SD and AK crystallized the protein and generated the complex. NC collected X-ray data and VND refined the structure. VND and JMP performed DFT calculations. DAK, OG and MPB collected, reduced and analyzed neutron diffraction data. VND and AK performed joint X-ray/neutron structure refinement. VND, AK and TCM wrote original draft, and all authors edited and reviewed the paper.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research used resources at the Spallation Neutron Source and the High Flux Isotope Reactor, which are DOE Office of Science User Facilities operated by the Oak Ridge National Laboratory. The Office of Biological and Environmental Research supported research at ORNL's Center for Structural Molecular Biology (CSMB), a DOE Office of Science User Facility. ORNL is managed by UT-Battelle LLC for DOE's Office of Science, the single largest supporter of basic research in the physical sciences in the United States. The authors thank the Institut Laue Langevin (beamline LADI-DALI) for awarded neutron beamtime. This work was also supported by the National Institute of General Medical Sciences (NIGMS), the National Institutes of Health (NIH), grant number R01GM137008-01A1.Notes and references
- R. Percudani and A. Peracchi, EMBO Rep., 2003, 4, 850–854 CrossRef CAS PubMed.
- T. C. Mueser, V. Drago, A. Kovalevsky and S. Dajnowicz, Methods Enzymol., 2020, 634, 333–359 CAS.
- A. C. Eliot and J. F. Kirsch, Annu. Rev. Biochem., 2004, 73, 383–415 CrossRef CAS PubMed.
- H. C. Dunathan, Proc. Natl. Acad. Sci. U. S. A., 1966, 55, 712–716 CrossRef CAS PubMed.
- J. P. Richard, T. L. Amyes, J. Crugeiras and A. Rios, Curr. Opin. Chem. Biol., 2009, 13, 475–483 CrossRef CAS PubMed.
- M. D. Toney, Biochim. Biophys. Acta, 2011, 1814, 1407–1418 CrossRef CAS PubMed.
- M. P. Blakeley and A. D. Podjarny, Emerging Top. Life Sci., 2018, 2, 39–55 CrossRef CAS PubMed.
- D. W. Kneller, G. Phillips, K. L. Weiss, S. Pant, Q. Zhang, H. M. O'Neill, L. Coates and A. Kovalevsky, J. Biol. Chem., 2020, 295, 17365–17373 CrossRef CAS PubMed.
- O. Gerlits, K. L. Weiss, M. P. Blakeley, G. Veglia, S. S. Taylor and A. Kovalevsky, Sci. Adv., 2019, 5, eaav0482 CrossRef CAS PubMed.
- N. Niimura, M. Takimoto-Kamimura and I. Tanaka, Encyclopedia of analytical chemistry: applications, theory and instrumentation, 2006, pp. 1–30 Search PubMed.
- O. Gerlits, T. Wymore, A. Das, C. H. Shen, J. M. Parks, J. C. Smith, K. L. Weiss, D. A. Keen, M. P. Blakeley, J. M. Louis, P. Langan, I. T. Weber and A. Kovalevsky, Angew. Chem., Int. Ed. Engl., 2016, 55, 4924–4927 CrossRef CAS PubMed.
- D. W. Kneller, G. Phillips, K. L. Weiss, Q. Zhang, L. Coates and A. Kovalevsky, J. Med. Chem., 2021, 64, 4991–5000 CrossRef CAS PubMed.
- L. Gajdos, M. P. Blakeley, M. Haertlein, V. T. Forsyth, J. M. Devos and A. Imberty, Nat. Commun., 2022, 13, 194 CrossRef CAS PubMed.
- K. Koruza, B. P. Mahon, M. P. Blakeley, A. Ostermann, T. E. Schrader, R. McKenna, W. Knecht and S. Z. Fisher, J. Struct. Biol., 2019, 205, 147–154 CrossRef CAS PubMed.
- M. D. Toney, Arch. Biochem. Biophys., 2014, 544, 119–127 CrossRef CAS PubMed.
- S. Rhee, M. M. Silva, C. C. Hyde, P. H. Rogers, C. M. Metzler, D. E. Metzler and A. Arnone, J. Biol. Chem., 1997, 272, 17293–17302 CrossRef CAS PubMed.
- S. Dajnowicz, R. C. Johnston, J. M. Parks, M. P. Blakeley, D. A. Keen, K. L. Weiss, O. Gerlits, A. Kovalevsky and T. C. Mueser, Nat. Commun., 2017, 8, 955 CrossRef PubMed.
- H. H. Limbach, M. Chan-Huot, S. Sharif, P. M. Tolstoy, I. G. Shenderovich, G. S. Denisov and M. D. Toney, Biochim. Biophys. Acta, 2011, 1814, 1426–1437 CrossRef CAS PubMed.
- M. Chan-Huot, A. Dos, R. Zander, S. Sharif, P. M. Tolstoy, S. Compton, E. Fogle, M. D. Toney, I. Shenderovich, G. S. Denisov and H.-H. Limbach, J. Am. Chem. Soc., 2013, 135, 18160–18175 CrossRef CAS PubMed.
- N. C. Furumo and J. F. Kirsch, Arch. Biochem. Biophys., 1995, 319, 49–54 CrossRef CAS PubMed.
- H. C. Dunathan, Vitam. Horm., 1970, 28, 399–414 CrossRef CAS.
- J. A. Gerlt, M. M. Kreevoy, W. Cleland and P. A. Frey, Chem. Biol., 1997, 4, 259–267 CrossRef CAS PubMed.
- B. Schiott, B. B. Iversen, G. K. Madsen, F. K. Larsen and T. C. Bruice, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 12799–12802 CrossRef CAS PubMed.
- W. W. Cleland, Arch. Biochem. Biophys., 2000, 382, 1–5 CrossRef CAS PubMed.
- P. Kumar, P. K. Agarwal and M. J. Cuneo, ChemBioChem, 2021, 22, 288–297 CrossRef CAS PubMed.
- O. Gerlits, D. A. Keen, M. P. Blakeley, J. M. Louis, I. T. Weber and A. Kovalevsky, J. Med. Chem., 2017, 60, 2018–2025 CrossRef CAS PubMed.
- E. Golden, L. J. Yu, F. Meilleur, M. P. Blakeley, A. P. Duff, A. Karton and A. Vrielink, Sci. Rep., 2017, 7, 40517 CrossRef CAS PubMed.
- S. J. Grabowski, Chem. Rev., 2011, 111, 2597–2625 CrossRef CAS PubMed.
- S. N. Smirnov, N. S. Golubev, G. S. Denisov, H. Benedict, P. Schah-Mohammedi and H.-H. Limbach, J. Am. Chem. Soc., 1996, 118, 4094–4101 CrossRef CAS.
- H. Benedict, H.-H. Limbach, M. Wehlan, W.-P. Fehlhammer, N. S. Golubev and R. Janoschek, J. Am. Chem. Soc., 1998, 120, 2939–2950 CrossRef CAS.
- H.-H. Limbach, P. M. Tolstoy, N. Pérez-Hernández, J. Guo, I. G. Shenderovich and G. S. Denisov, Isr. J. Chem., 2009, 49, 199–216 CrossRef CAS.
- A. Cahlík, J. Hellerstedt, J. I. Mendieta-Moreno, M. Švec, V. M. Santhini, S. Pascal, D. Soler-Polo, S. I. Erlingsson, K. Výborný, P. Mutombo, O. Marsalek, O. Siri and P. Jelínek, ACS Nano, 2021, 15, 10357–10365 CrossRef PubMed.
- S. J. Grabowski, Annu. Rep. Prog. Chem., Sect. C: Phys. Chem., 2006, 102, 131–165 RSC.
- P. Christen and P. K. Mehta, Chem. Rec., 2001, 1, 436–447 CrossRef CAS PubMed.
- S. Dajnowicz, J. M. Parks, X. Hu, K. Gesler, A. Y. Kovalevsky and T. C. Mueser, J. Biol. Chem., 2017, 292, 5970–5980 CrossRef CAS PubMed.
- M. P. Blakeley, S. C. Teixeira, I. Petit-Haertlein, I. Hazemann, A. Mitschler, M. Haertlein, E. Howard and A. D. Podjarny, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 1198–1205 CrossRef CAS PubMed.
- J. Campbell, J. Appl. Crystallogr., 1995, 28, 228–236 CrossRef CAS.
- J. W. Campbell, Q. Hao, M. M. Harding, N. D. Nguti and C. Wilkinson, J. Appl. Crystallogr., 1998, 31, 496–502 CrossRef CAS.
- S. Arzt, J. W. Campbell, M. M. Harding, Q. Hao and J. R. Helliwell, J. Appl. Crystallogr., 1999, 32, 554–562 CrossRef CAS.
- M. Weiss, J. Appl. Crystallogr., 2001, 34, 130–135 CrossRef CAS.
- W. Kabsch, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 125–132 CrossRef CAS PubMed.
- D. Liebschner, P. V. Afonine, N. W. Moriarty, P. Langan and P. D. Adams, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2018, 74, 800–813 CrossRef CAS PubMed.
- M. Mustyakimov and P. Langan, C-06, 104 Patch for CNS: nCNS. An open source distribution patch for CNS for macromolecular structure refinement, Los Alamos National Security, Los Alamos, NM, USA, 2007 Search PubMed.
- A. T. Brunger, P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson and G. L. Warren, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1998, 54, 905–921 CrossRef CAS PubMed.
- P. D. Adams, M. Mustyakimov, P. V. Afonine and P. Langan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 567–573 CrossRef CAS PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS PubMed.
- F. Weigend, Phys. Chem. Chem. Phys., 2006, 8, 1057–1065 RSC.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- N. Mardirossian and M. Head-Gordon, Mol. Phys., 2017, 115, 2315–2372 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16, Revision A.03, Gaussian Inc., Wallingford, CT, 2016 Search PubMed.
- A. E. Reed, R. B. Weinstock and F. Weinhold, J. Chem. Phys., 1985, 83, 735–746 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc02285k |
| This journal is © The Royal Society of Chemistry 2022 |