 Open Access Article
Open Access ArticleEnzyme-powered micro- and nano-motors: key parameters for an application-oriented design
Xavier
Arqué
a,
Tania
Patiño
 ab and
Samuel
Sánchez
ab and
Samuel
Sánchez
 *ac
*ac
aInstitute for Bioengineering of Catalonia (IBEC), The Barcelona Institute of Science and Technology (BIST), Barcelona 08028, Spain. E-mail: ssanchez@ibecbarcelona.eu
bBio-Organic Chemistry, Institute for Complex Molecular Systems, Eindhoven University of Technology, 5600 MB Eindhoven, The Netherlands
cInstitució Catalana de Recerca i Estudis Avançats (ICREA), Barcelona 08010, Spain
First published on 21st July 2022
Abstract
Nature has inspired the creation of artificial micro- and nanomotors that self-propel converting chemical energy into mechanical action. These tiny machines have appeared as promising biomedical tools for treatment and diagnosis and have also been used for environmental, antimicrobial or sensing applications. Among the possible catalytic engines, enzymes have emerged as an alternative to inorganic catalysts due to their biocompatibility and the variety and bioavailability of fuels. Although the field of enzyme-powered micro- and nano-motors has a trajectory of more than a decade, a comprehensive framework on how to rationally design, control and optimize their motion is still missing. With this purpose, herein we performed a thorough bibliographic study on the key parameters governing the propulsion of these enzyme-powered devices, namely the chassis shape, the material composition, the motor size, the enzyme type, the method used to incorporate enzymes, the distribution of the product released, the motion mechanism, the motion media and the technique used for motion detection. In conclusion, from the library of options that each parameter offers there needs to be a rational selection and intelligent design of enzymatic motors based on the specific application envisioned.
1. Introduction: enzyme-powered micro- and nano-motors
Inspired by the active motion of certain biological systems, there has been increasing interest in the past few decades to engineer artificial motile systems, namely nanomachines or nanomotors. Among them, catalytic micro- and nano-motors self-propel by converting free chemical energy from in situ catalytic reactions.1 The field of self-propelled motors has gained interest from fundamental knowledge and basic science to their use in distinct applications. For instance, these biocompatible devices have a promising use in biomedicine exploiting enhanced penetration and targeting for visualization inside a tissue or as treatment through heat or drug delivery.2 Other fields of applications are taking advantage of the catalytic and micromixing effect like sensing of compounds, water decontamination and antimicrobial applications.3Although most of the reported systems have relied on the use of inorganic catalysts, enzymatic engines offer unique advantages such as a wide variety of catalytic reactions or fuel biocompatibility and bioavailability.4 Over the last few years, several improvements have been presented to optimize the performance of enzyme-powered motors for specific applications,5–8 but a comprehensive framework on their key parameters to rationally design, control and optimize motion is still missing. From the most fundamental aspects to the final applications there are features of each micro- and nano-motor reported that determine their motion performance. For instance, it is known that active motion of catalytic nano-/micro-motors requires an asymmetric release of products, which depends on the chassis design, material composition9 and the location of the catalyst in the chassis.10
The propulsion performance of enzyme-powered motors can be further improved based on the enzyme selected and the method for enzyme incorporation.11 These characteristics determine the mechanism of motion, which is still not clearly understood for most of the systems reported. Nonetheless, other aspects such as the composition of the fluid media and their size can also play a role in motion dynamics and performance and consequently in the detection technique chosen for motion analysis.
After more than a decade of trajectory of the field, in this review we compile and analyze the main parameters of enzyme-powered micro- and nano-motors to promote the development of the field and help to rationally optimize the motor design and implementation.
2. Materials for chassis composition
In the field of enzyme-powered micro- and nano-motors, the building base material is key for an easy surface modification, flexibility, degradability, porosity, size, or shape of the structure. Hence, the chemical composition of the chassis has a central role in the motor design. In Fig. 1A we depict a bibliographic study of the incidence of different chassis compositions. Among the materials used at least in 25% of reports each, polymers are distinctively appealing because of their flexibility, silica because of their porosity and metals because of their photo- and magnetic responsiveness and conductivity. Other chassis are built with less common materials that are currently emerging such as carbon, metal–organic frameworks and bio-inspired chassis made from liposomes or cells.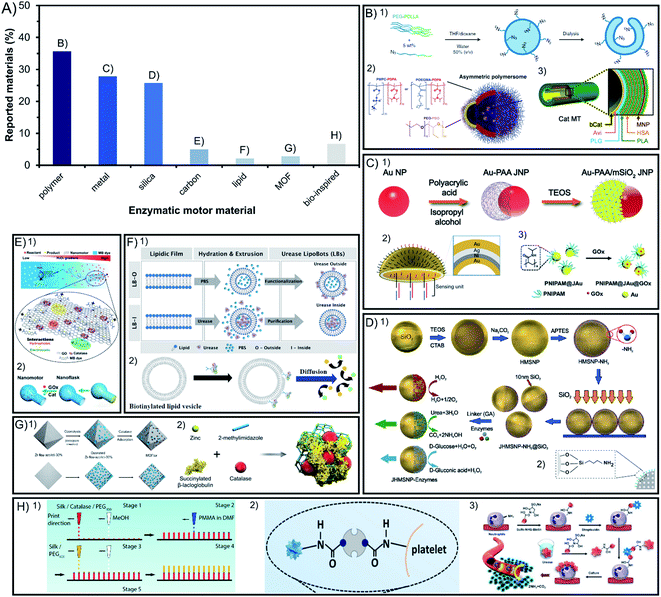 | ||
| Fig. 1 Chassis materials of enzyme-powered micro- and nano-motors. (A) Representation of each chassis material in the publications of the field. Examples of enzymatic motors made of (B) polymers,27,37,63 (C) metals,47,64,100 (D) silica,67,93 (E) carbon,103,104 (F) lipid vesicles,112,113 (G) MOFs107,110 and (H) bio-inspired materials.115–117 Panel (B) adapted with permission from (1) ref. 27 Copyright 2019 Springer Nature, (2) ref. 37 Copyright 2017 AAAS, and (3) ref. 63 Copyright 2021 Royal Society of Chemistry. Panel (C) adapted with permission from (1) ref. 100 Copyright 2021 American Chemical Society, (2) ref. 47 Copyright 2019 American Chemical Society, and (3) ref. 64 Copyright 2019 Wiley. Panel (D) adapted with permission from (1) ref. 67 Copyright 2015 American Chemical Society and (2) ref. 93 Copyright 2021 AAAS. Panel (E) adapted with permission from (1) ref. 104 Copyright 2021 Wiley and (2) ref. 103 Copyright 2019 American Chemical Society. Panel (F) adapted with permission from (1) ref. 112 Copyright 2020 Wiley and (2) ref. 113 Copyright 2019 American Chemical Society. Panel (G) adapted with permission from (1) ref. 110 Copyright 2020 American Chemical Society and (2) ref. 107 Copyright 2019 Wiley. Panel (H) adapted with permission from (1) ref. 117 Copyright 2016 Wiley, (2) ref. 115 Copyright 2020 AAAS, and (3) ref. 116 Copyright 2022 American Chemical Society. | ||
2.1. Polymers
Among the materials reported for enzymatic motors, 36% of them are polymeric materials (Fig. 1B). The enzymatic motors based on a single-polymer chassis commonly use polypyrrole (PPy),12,13 polystyrene (PS),14,15 poly(lactic-co-glycolic acid) (PLGA)16, poly(styrene-co-maleic anhydride) (PSMA),17,18 polycaprolactone (PCL),19 poly(lactic acid) (PLA)20 and polydiacetylene (PDA).21,22 These materials allow for an easy surface modification, degradability, and biocompatibility.Polymeric materials can be organized in multilayered combination of polymers, which was pioneered by Wilson and van Hest's groups, the most prominent users of polymeric chassis. The first combination used was poly(ethylene glycol) with polystyrene (PEG–PS)23–25, followed by poly(ethylene glycol) with poly-DL-lactide (PEG-PDLLA)26,27 and poly(ethylene glycol) diacrylate with dextran (PEGDA-dextran).28,29 This allows for different properties on each side of the structure that enable morphological flexibility to encapsulate enzymes or drugs and to generate an asymmetric shape. This is also the case for other examples that have used dual polymeric compounds like poly(ethylene oxide) with poly(lactic-co-glycolic acid) (PEO-PLGA),30 poly(ethylene glycol) with aggregation-induced emission polymers (PEG-P(AIE)),31,32 (poly(ethylene oxide-b-butadiene)) (PEO-PB),33 polystyrene with poly(glycidyl methacrylate) (PS-PGMA),15 polydopamine with human serum albumin (PDA-HSA)34 or poly(ethylene glycol) with poly(propylene glycol) (PEG-PPG-PEG)35. Even more complex structures are polymeric vesicles made of three-layered polymers such as poly(ethylene glycol) with polycaprolactone and poly(trimethylene carbonate) (PEG-PCL-PTMC)36 to allow for the mobility of enzymes on the coacervate surface. Vesicles can also combine different types of dual polymers to create asymmetric structures by using poly[(2-methacryloyl)ethyl phosphorylcholine] or poly[oligo(ethylene glycol) methyl methacrylate] with poly[2-(diisopropylamino)ethyl methacrylate] ((PMPC or POEGMA)-PDPA), and a part of the vesicle composed of poly(ethylene oxide) with poly(butylene oxide) (PEO-PBO)37 to allow only one side of the motor to permeate molecules through the membrane, named polymersomes. Polymeric materials have also been used to create tubular structures with multiple layers, for instance combining poly(L-arginine), human serum albumin and poly(L-glutamic acid) (PLA-HSA-PLG),38,39 or brush structures combining poly(glycidyl methacrylate), polyacrylic acid and poly(oligo(ethylene glycol) methyl ether acrylate) ((PGMA-g-PAA)-b-POEGA)40. Recently, an innovative chassis was constructed by accumulation of the aminoacidic sequence Ac-RLVFFAL-NH2 based on cross-β amyloid.41 The appealing properties of these materials are easy functionalization and biocompatibility, and that their structure flexibility can be used to encapsulate molecules inside a cavity in the chassis, justifying its wide use for biomedical applications. The chassis composition can be further expanded through combination with other materials, such as metals or silica.
2.2. Metals
Metals are the second most commonly used material, representing 28% of materials used for enzymatic motors (Fig. 1C). Enzymatic motors built solely with metals used gold (Au),42 iron oxide (Fe3O4),43 or a combination of different layered metals44 like titanium with gold (Ti–Au),45 titania with cadmium sulfide (TiO2–CdS),46 gold with silver (Au–Ag–Au),47,48 and gold with nickel and silver (Au–Ni–Ag).49 Metals are combined with silica and metals thanks to their appealing properties, such as responsiveness to light or magnetic fields, or high conductivity. Gold has been combined with polymers like polypyrrole (PPy) to exploit their electron transfer capacity,50–52 or to allow enzyme attachment,53 similarly applied with (poly(3,4-ethylenedioxythiophene)) polymer (PEDOT)54–58. To exploit the magnetic and photo-responsive properties, metals like Au, MnFe2O4, Fe3O4 or a combination of Ga–In–Sn have been incorporated inside or underneath polymeric structures like polylysine (PL),59 polydopamine (PDA),60 polycaprolactone (PCL),61 poly(ethylene glycol) (PEG)62 or PEG-PS multipolymers.63 Metals have also been used as a layer within the structure of (poly(3,4-ethylenedioxythiophene)), poly(sodium styrene sulfonate) (PSS-PEDOT-Au),64 within polystyrene with poly(diallyldimethylammonium chloride), poly(styrene sulfonate) and poly(L-lysine) (PS-PPDA-PSS-MnFe2O4-PLL)65 or within poly(L-arginine), human serum albumin, Fe3O4 and poly(L-glutamic acid) (PLG-HAS-Fe3O4-PLG).66 In some cases, polymers like PNIPAM (or other water-soluble polymers) have been attached on one side of a metal nanoparticle (Au or TiO2)67,68 to create asymmetry and take advantage of their photo-responsive properties.2.3. Silica
Silica (SiO2) is the third most commonly used material and represents 26% of the chassis compositions used for enzymatic motors (Fig. 1D), with Sánchez's group being the top user. Enzymatic motors are widely reported to be used in the forms of mesoporous silica69–79, core–shell structures80 or rigid silica.78,81–87 Silica allows for easy spherical formation and surface modification, as well as biocompatibility and molecule encapsulation inside mesopores making this material promising for clinical translation.88 Hence, it is combined with polymers to additionally confer these properties, for example silica with polystyrene (PS-SiO2)89, poly(sodium styrene sulfonate) (PSS-SiO2),57 PNIPAM (PNIPAM-SiO2)90 or other combinations of increasing complexity like silica with polydopamine, poly(L-lysine) and poly(ethylene glycol) (SiO2-PDA-PLL-PEG)91.Metals like Au and Fe3O4 have been also combined with silica to take advantage of the same magnetic and photo-reactive properties,92–95 to attach isotopes for visualization,96 to create an asymmetric incorporation of enzymes,97–101 or to synthesize an organoclay chassis.102 There are even examples of polymers combined with both silica and metals such as (1) MnFe2O4 added as a layer in the combination of silica with poly(L-lysine), poly(methacrylic acid) and poly(ethylene glycol) (SiO2-PLL-PMA-MnFe2O4-PEG)103 or (2) Au nanoparticles added between layers of poly(ethylene glycol), poly(acrylic acid) and mesoporous silica (PEG-Au-PAA-mSiO2),104 to offer magnetic guidance and to induce an asymmetric chassis structure, respectively.
2.4. Other chassis materials: carbon, MOFs, lipids and bio-inspired
Emerging materials used in other fields have been also implemented lately to build chassis for enzyme-powered motors, currently representing between 2 and 7% of the total motors. Carbonaceous materials are used in 5% of reports since the earliest times of this field105,106 (Fig. 1E) and have recently gained more presence to fabricate innovative chassis shapes,107 for instance using graphene oxide,108 combined with silica109 or PDA and PEI,110 or by adding Fe3O4 to convey magnetic responsiveness properties for biomedical applications.111Lately, some chassis of enzymatic motors have also used the combination of metals with organic molecules to form metal–organic frameworks (MOFs) (Fig. 1G) in 3% of reports, like ZIF-8 (ref. 112–114) and UiO types,115 taking advantage of the controllable porosity to encapsulate enzymes, drugs, or absorb contaminants.
Bio-inspired materials have also emerged as innovative chassis composition, the most popular of which are lipidic vesicles used in 2% of publications. Lipidic vesicles (Fig. 1F), a well-stablished structure in the field of nanomedicine,116 are attractive because of their biological characteristics such as flexibility, permeability and biocompatibility, as well as the possibility of incorporating enzymatic engines inside the cavity or transmembrane.117–119 Other recently used materials taken from nature (Fig. 1H) are platelet120 and neutrophile121 motors as well as previous examples of motors made of silk,122–124 cellulose combined with MOFs,125 or plant tissues like radish,126 carrot, potato or millet seeds.127
2.5. Conclusions on chassis materials
The development of the chassis composition points towards enlarging the versatility of materials and to rationally design them towards applications. The well stablished polymeric materials, silica, and metals will be further consolidated, as well as their combination to fuse their appealing properties in one system, namely easy fabrication, surface and shape modification and magnetic and photo-responsiveness. For biomedical applications, the biocompatibility of the material is essential and even biodegradability is required in scenarios where the devices cannot be extracted. Mesoporous silica holds great promise for its safety, effectivity, and viability in vivo. Bio-inspired materials like liposomes, biodegradable polymers or the usage of cells are especially appealing since they offer native-like properties that can be helpful inside an organism, like high biocompatibility, flexibility, solubility, and permeability. For applications outside living organisms, such as environmental applications or sensing, other properties from inorganic structures will be fostered like the highly controllable porosity of metal–organic frameworks (MOFs) or metals in general for their photo- and magnetic responsiveness.3. Chassis shape and product distribution
Symmetry breaking is essential for all chemically powered micro- and nano-motors in order to ensure the creation of a gradient of released products around the chassis. The shape of enzymatic micro- and nano-motors is key to promote this asymmetry, either physical or molecular, and the resulting active motion. Fig. 2A shows a summary of the field in terms of shapes reported, where spheres lead with vast predominance. The rest of the shapes reported are present in less than 14% of reports each. While some shapes have physical asymmetry such as internal and external different parts like tubes, stomatocytes, bottles, vesicles and shells, others need to have asymmetry forced on their exterior surface area like rods and crystals. However, lately the spontaneous generation of asymmetry for several of the mentioned shapes without extra fabrication steps required has been reported.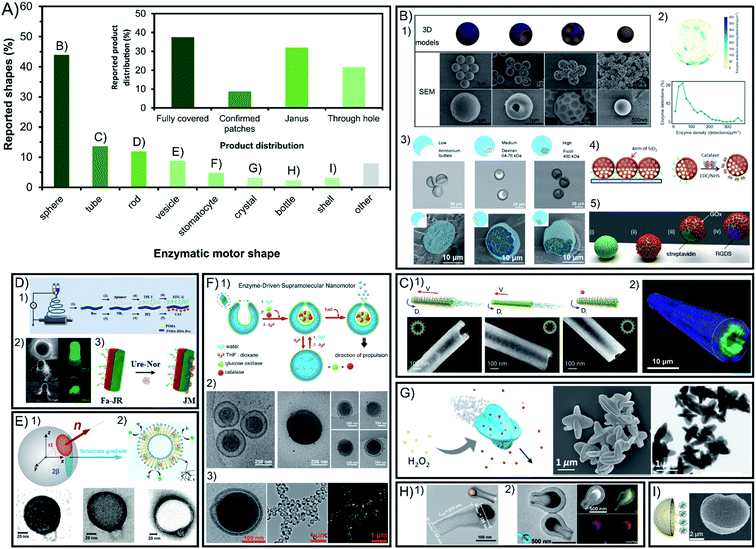 | ||
| Fig. 2 Chassis shape and product release distribution of enzyme-powered micro- and nano-motors. (A) Representation of each chassis shape in the publications of the field. Inset: representation of each configuration of product release in the publications of the field. Examples of enzymatic motors with shapes of (B) spheres,15,29,60,66,86 (C) tubes,30,83 (D) rods,17,20,117 (E) vesicles,37,113 (F) stomatocytes,23,24,27 (G) crystals,107 (H) bottles35,95 and (I) shells.46 Panel (B) adapted with permission from (1) ref. 15 Copyright 2019 Elsevier, (2) ref. 86 Copyright 2018 American Chemical Society, (3) ref. 29 Copyright 2019 Wiley, (4) ref. 66 Copyright 2017 Elsevier and (5) ref. 60 Copyright 2017 Royal Society of Chemistry. Panel (C) adapted with permission from (1) ref. 83 Copyright 2016 American Chemical Society and (2) ref. 30 Copyright 2016 Wiley. Panel (D) adapted with permission from (1) ref. 17 Copyright 2020 Elsevier, (2) ref. 117 Copyright 2016 Wiley, and (3) ref. 20 Copyright 2021 Elsevier. Panel (E) adapted with permission from (1) ref. 37 Copyright 2017 AAAS and (2) ref. 113 Copyright 2019 American Chemical Society. Panel (F) adapted with permission from (1) ref. 24 Copyright 2016 American Chemical Society, (2) ref. 27 Copyright 2019 Springer Nature and (3) ref. 23 Copyright 2016 American Chemical Society. Panel (G) adapted with permission from (1) ref. 107 Copyright 2019 Wiley. Panel (H) adapted with permission from (1) ref. 95 Copyright 2021 American Chemical Society and (2) ref. 35 Copyright 2022 Wiley. Panel (I) adapted with permission from (1) ref. 46 Copyright 2019 Wiley. | ||
3.1. Spheres
Spherical shapes can be easily generated spontaneously in solution and offer a wider range of sizes from hundreds of nanometers to a few micrometers, and hence represent 44% of all the shapes reported (Fig. 2B). However, it has been argued that the asymmetry of product release is needed for self-propulsion to take place,128 which is normally provided by adding fabrication steps to break the complete symmetry of spherical structures. For years, this has been solved by attaching enzymatic engines only on one side of the motor creating Janus structures through different methods such as synthesizing motors via a microfluidics system,28,29 growing different materials on each side of the particle,95,101,109 directly attaching two spherical particles together (each of them being a side of the Janus structure),97 or modifying one side of the particle after synthesis using the interfacial energy difference,104 through a sputtering process,44,69,70,92,99 by spin-coating,110 by using a Pickering emulsion,68,77,91,94,103 and modifying the spheres while attached on a bigger sphere67 or while magnetically attracted to a surface.43,62There are examples of spherical shapes where instead of forcing asymmetry, active motion was produced by fully covering with enzymes the surface of rigid spheres,13,14,16,31,34,57,60,65,85 porous spheres71–76,78,80,96,125 and hollow spheres,21,22,78,79,81–84,87,90,93 as well as embedding the enzymes inside the structure.61 However, the full coverage of most of these spherical structures was not analyzed in detail. Recent studies show that in some cases asymmetry may appear spontaneously through a patchy distribution of the enzymes on the particle's surface.89 This poses the relevant question of whether the so-claimed fully covered structures reported in fact present a given asymmetry of enzyme distribution on the surface. To elucidate that, the field would benefit from a thorough analysis of the enzyme distribution, as it has been reported using super resolution microscopy, contributing to a more controllable and rational design.71,87,89 Ultimately, the effect of the enzymatic coverage on the speed of spherical motors has been studied in multiple reports,15,29,36 indicating that the highest asymmetry maximizes the active motion, with Janus configurations being faster than patch configurations, while the slowest performance is obtained with full coverage and complete symmetry of product release.
3.2. Tubes
Other structures allow for a straightforward asymmetry of product release, such as the exploitation of the cavity to attach enzymes in tubular structures (Fig. 2C), being the second most commonly used shape with 14% of representation30,38,39,41,45,53–58,64,66. Tubular motors enable the release of the product through a hole, and the highest degree of asymmetry, even when the enzymes are embedded in the chassis material59. However, a recent example of fully covered tube bundles also shows active motion without confirmed asymmetry.46 The enzyme coverage on tubular structures has been studied by the Sánchez group, where tubes with enzymes covering the inner and outer surfaces showed the maximum speed, followed by only coverage of the inner cavity, while attaching enzymes only outside the tube produced almost no propulsion.863.3. Rods
Rod-like structures are almost 12% of chassis reported (Fig. 2D) and bear similarities to spheres since two different sides are often incorporated on the structure across the longitudinal section50–52,105 or the cross section,17,18,20,124 as shown by the pioneer work of Mano and Heller. However, the initial report of Feringa used the formation of spontaneous aggregates on the structure to power motion106 and latter reports also showed active motion without forcing any asymmetry.19,127 However, Janus conformations have proven to promote faster speeds for rod shapes as demonstrated in comparison studies.122,1263.4. Other chassis shapes: vesicles, stomatocytes, bottles, etc.
Beyond the main structures, other shapes used are present at a minimal percentage between 2 and 9% of total publications. Vesicle shapes represent almost 9% of reports (Fig. 2E) and can break symmetry by controlling their permeability while having the enzymes inside the lumen37 although most examples of vesicle motors with enzymes in the inner part or on the membrane release the product all around the spherical33,36,102,117–119 or elongated vesicle.26 The specific case of vesicle-like platelets and neutrophiles also supports the design of higher asymmetry for faster motion by comparing Janus and non-Janus conformations.120,121 Stomatocytes are used in more than 5% of the motors reports23–25,27,32,63 and are similar to vesicles but release the product through a hole created by the invagination of flexible polymers and the formation of a cavity where enzymes are encapsulated (Fig. 2F). Additionally, there have been few crystal-shaped motors (Fig. 2G) that contain the enzymes inside the structure and propel without forcing asymmetry.112–115 Similar structures to tubes are bottle-like shapes,35,98,107 which release the product through a single hole in the structure (Fig. 2H). These examples of apparently fully covered vesicles and crystals confirm that a spontaneous asymmetry can also drive motion in these shapes. Finally, shell shapes with Janus configuration have also been reported (Fig. 2I) but with one of the lowest representations in the field.47–49,100Other examples of shapes are rectangular prisms with enzymes attached under the structure to propel at the interface,12 as well as structures fully covered by enzymes like cube-like particles,111 elongated cucurbits that produce higher spontaneous asymmetry than spheres,31 2D sheets that increase the accessible area of the substrate for enzymes,108,129 zeta pentomino shapes,123 3- and 4-arm crosses,124 brush-shaped structures,67 or random organization of clusters,42 all of them with an inherent asymmetry of the product release.
3.5. Conclusions on the chassis shape
The shape of the chassis strongly determines the motion dynamics. The literature suggests that the shapes that facilitate the asymmetry of product release are beneficial for self-propulsion, since 62% of reports present motors with a certain degree of asymmetry through patches, Janus structures or release of the product through a hole (Fig. 2A inset). However, the remaining 38% of reports do not confirm the full coverage of the chassis with enzymes, and since asymmetry could also be present the field would benefit from improving the analysis techniques of enzyme distribution and quantification. Although the most effective asymmetry may be the release of the product through a hole and synthesis methods are becoming more efficient over the years, some fabrication processes still require extra steps to selectively incorporate the enzyme inside the motor. Spherical shapes tend to be applied for biomedical applications, while elongated structures of larger sizes are commonly used for sensing and decontamination applications thanks to their vigorous propulsion and micromixing effect. Lately, promising shapes are being tested as motor chassis because of their inherent conditions like vesicles and their spontaneous generation of asymmetry like elongated cucurbits and to increase the surface area in contact with media like 2D sheets. Further research needs to be pursued to conceive new materials and platforms that allow for shape tuning maintaining the desired material conditions.4. Enzyme incorporation method
Enzymatic engines are incorporated through different strategies that determine crucial aspects for active motion ranging from the stability of the modification to the stability of enzymes and the maintenance of their relevant properties. Fig. 3A presents the different incorporation methods which are divided into covalent bounds that increase stability but may hinder catalysis and the non-covalent methods of non-covalent interactions or encapsulation that keep the native-like conditions of enzymes but may make them more susceptible to detachment.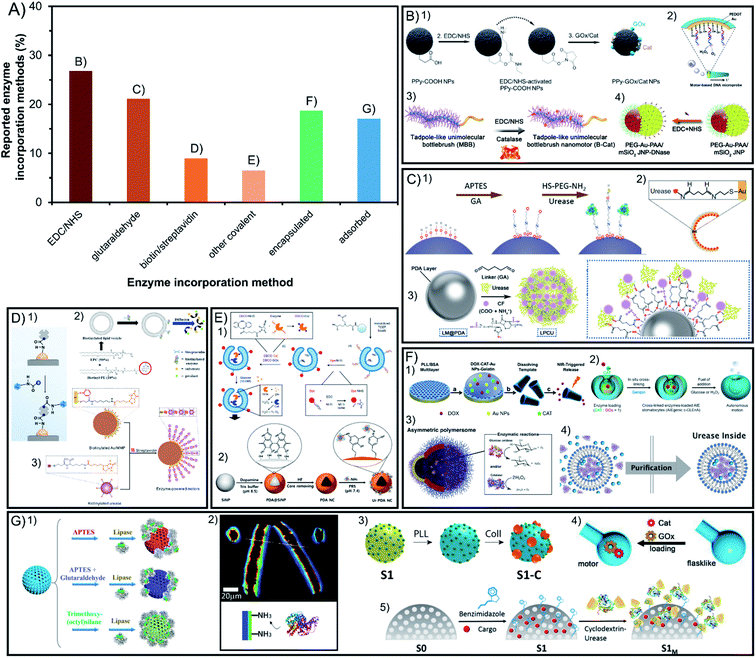 | ||
| Fig. 3 Enzyme incorporation method of enzyme-powered micro- and nano-motors. (A) Representation of each enzyme incorporation method in the publications of the field. Examples of enzymatic motors with enzymes attached by using covalent attachment like (B) EDC/NHS,13,40,56,100 (C) glutaraldehyde,58,70,97 (D) biotin/streptavidin43,113,115 or (E) other covalent methods,21,27 and non-covalent incorporation methods like (F) encapsulation32,37,57,112 and (G) non-covalent interaction.30,35,62,71,73 Panel (B) adapted with permission from (1) ref. 13 Copyright 2021 American Chemical Society, (2) ref. 56 Copyright 2017 Elsevier, (3) ref. 40 Copyright 2019 American Chemical Society, and (4) ref. 100 Copyright 2021 American Chemical Society. Panel (C) adapted with permission from (1) ref. 70 Copyright 2019 American Chemical Society, (2) ref. 97 Copyright 2021 American Institute of Physics and (3) ref. 58 Copyright 2021 American Chemical Society. Panel (D) adapted with permission from (1) ref. 115 Copyright 2020 AAAS, (2) ref. 113 Copyright 2019 American Chemical Society, and (3) ref. 43 Copyright 2020 American Chemical Society. Panel (E) adapted with permission from (1) ref. 27 Copyright 2019 Springer Nature and (2) ref. 21 Copyright 2022 Elsevier. Panel (F) adapted with permission from (1) ref. 57 Copyright 2015 American Chemical Society, (2) ref. 32 Copyright 2021 Multidisciplinary Digital Publishing Institute, (3) ref. 37 Copyright 2017 AAAS and (4) ref. 112 Copyright 2020 Wiley. Panel (G) adapted with permission from (1) ref. 73 Copyright 2020 Wiley, (2) ref. 30 Copyright 2016 Wiley, (3) ref. 62 Copyright 2019 American Chemical Society, (4) ref. 35 Copyright 2022 Wiley, and (5) ref. 71 Copyright 2019 American Chemical Society. | ||
4.1. Covalent incorporation in chassis
More than 63% of enzymatic motors are built by attaching the enzymes covalently to the chassis structure, which are divided between the linkers EDC/NHS and glutaraldehyde with more than 20% of representation each, biotin/streptavidin with less than 10% and other methods barely used (Fig. 3A). Glutaraldehyde is the most widely used linker in biocatalysis but causes crosslinking and poor control in enzyme orientation. However, systems like EDC/NHS, or especially biotin/streptavidin and the few methods using click chemistry, allow for a selecting linkage. Almost all these methods have been used to create Janus structures but also show motion when fully covering the chassis with enzymes, which suggest that a spontaneous asymmetry is generated.4.2. Non-covalent incorporation in chassis
The other almost 36% of enzymatic motors do not use covalent attachment and either encapsulate the enzymes inside a cavity of the chassis or the attachment interactions with the surface are non-covalent, with more than 19% and 17% of representation, respectively. While the encapsulation of enzymes benefits from native-like protection of the macromolecules from the exterior media, the incorporation of enzymes through non-covalent interactions is the easiest and fastest to apply.4.3. Conclusions on enzyme incorporation methods
The specific effect of different incorporation methods has been tested on the same structure, for instance, the enzyme stability and resulting speed obtained by tubular motors, supporting covalent binding as the best method for the fastest speed.45 The incorporation of enzymes into liposome-based nanomotors has been carried out using different strategies, including their surface binding through streptavidin–biotin approaches118,119 or encapsulating them inside the liposome cavity.117 The latter report demonstrated the advantages of compartmentalizing the enzymes when allowing a flow of molecules through a permeabilized membrane compared to covalently attached enzymes outside the vesicle. Additionally, for the specific case of the lipase enzyme on silica nanoparticles, covalent attachment showed improved active motion compared to non-covalent interactions. However, a specific covalent attachment through trimethoxy-(octyl)silane that optimized enzyme orientation demonstrated faster diffusion than the unspecific glutaraldehyde method, showing that a thorough understanding of the enzymatic structure and orientation of the active site outwards can be beneficial for both catalysis and active motion.76Overall, in the field of enzyme-powered micro- and nanomotors there is a marked inclination towards covalent immobilization of enzymes due to its resistance and the fine control of enzyme orientation, as well as the possibility to create multilayers of enzymes around the surface of the motors, which can boost the motion performance. However, due to the negative effects of covalent bonds on the enzyme structure and catalysis, as well as the need for protecting enzymes in all applications ranging from environmental to biomedical fields, the encapsulation of enzymes inside a compartment could gain more and more interest in the near future. This protective effect can also be achieved through polymeric coatings covering the enzymes. However, free enzymes in solution inside a cavity with a controlled environment represents a bio-inspired compartmentalization that may offer better catalysis under native-like conditions. This would also be the case for non-covalent interactions of transmembrane enzymes. Nevertheless, while more efficient methods are developed in this direction, covalent attachment will be applied because of its reliability and non-covalent interactions for being the easiest to apply.
5. Enzyme library and motion mechanism
Despite the apparent diversity of 23 different enzymes used to power active motion among all the reports of enzymatic micro- and nano-motors published over nearly two decades, more than 65% enzymatic engines used are either catalase or urease, as seen in Fig. 4A. These two enzymes share certain properties like a high turnover number and substrate/product molecules with a low molecular weight, which could be decisive for a proper motion. However, catalase has the distinctive property of generating oxygen bubbles as a product, while urease produces ionic products, which also play a key role in achieving propulsion. Other enzymes with around 10% or lower representation are glucose oxidase alone or combined with catalase and recently lipase and ATPase have gained more presence in the field to degrade undesired compounds. However, most enzymes have only been tested once either alone or in combination with other enzymes.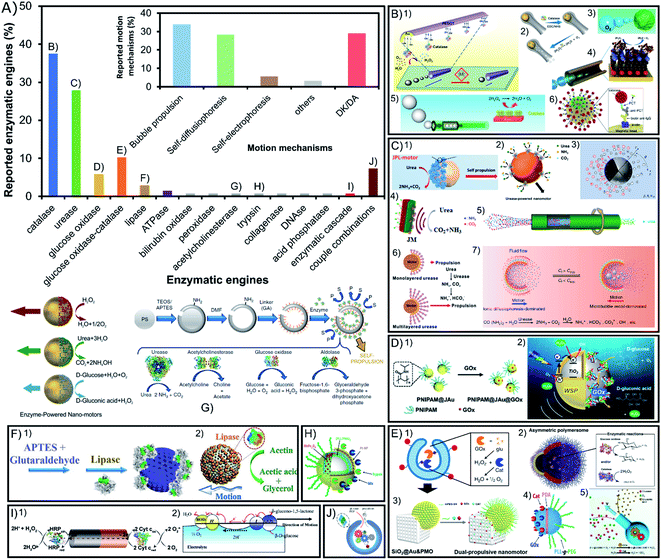 | ||
| Fig. 4 Enzyme type and motion mechanism of enzyme-powered micro- and nano-motors. (A) Representation of each enzyme type in the publications of the field. Inset: representation of each motion mechanism in the publications of the field. Examples of enzymatic motors powered with the enzymes (B) catalase,38,42,44,52,59,95 (C) urease,20,22,39,43,79,97,115 (D) glucose oxidase,64,65 (E) glucose oxidase and catalase,27,35,37,88,92 (F) lipase,72,73 (G) acetylcholinesterase,80 (H) trypsin,99 (I) enzyme combinations50,101 and (J) enzymatic pathway.23 Panel (B) adapted with permission from (1) ref. 52 Copyright 2013 American Chemical Society, (2) ref. 95 Copyright 2021 American Chemical Society, (3) ref. 59 Copyright 2019 American Chemical Society, (4) ref. 44 Copyright 2010 American Chemical Society, (5) ref. 38 Copyright 2018 American Chemical Society, and (6) ref. 42 Copyright 2019 Elsevier. Panel (C) adapted with permission from (1) ref. 115 Copyright 2020 AAAS, (2) ref. 22 Copyright 2020 American Chemical Society, (3) ref. 79 Copyright 2020 American Physical Society, (4) ref. 20 Copyright 2021 Elsevier, (5) ref. 39 Copyright 2019 Wiley, (6) ref. 43 Copyright 2020 American Chemical Society, and (7) ref. 97 Copyright 2021 American Institute of Physics. Panel (D) adapted with permission from (1) ref. 64 Copyright 2019 Wiley and (2) ref. 65 Copyright 2021 American Chemical Society. Panel (E) adapted with permission from (1) ref. 27 Copyright 2019 Springer Nature, (2) ref. 37 Copyright 2017 AAAS, (3) ref. 92 Copyright 2022 American Chemical Society, (4) ref. 88 Copyright 2015 American Chemical Society, and (5) ref. 35 Copyright 2022 Wiley. Panel (F) adapted with permission from (1) ref. 73 Copyright 2020 Wiley and (2) ref. 72 Copyright 2019 Wiley. Panel (G) adapted with permission from (1) ref. 80 Copyright 2019 Springer Nature and (2) ref. 67 Copyright 2015 American Chemical Society. Panel (H) adapted with permission from (1) ref. 99 Copyright 2017 American Chemical Society. Panel (I) adapted with permission from (1) ref. 23 Copyright 2016 American Chemical Society. Panel (J) adapted with permission from (1) ref. 50 Copyright 2014 Wiley and (2) ref. 101 Copyright 2005 American Chemical Society. | ||
5.1. Catalase
Catalase is the most common enzymatic engine, with 38% of motors using it (Fig. 4B) because of its high turnover number and the capacity to consume hydrogen peroxide (H2O2) and produce water (H2O) and oxygen bubbles (O2) that drive a jet-like motion mechanism16–19,28–30,38,45,48,49,53–55,57–59,61,63,64,66,77,85,108,115,122–124,126,127 or a buoyancy effect mechanism.102,114,125 Due to this powerful bubble propulsion, faster motion and micromixing of the fluid are desired for sensing17–19,43,49,55,57,58,64,77,126 and water decontamination.54,61,66,108,115 When the thrust of bubbles is not visible it has been proposed that either there are nanosized bubbles being produced or that active motion is generated by the products of the reaction creating phoretic fluctuations around the particle14,24–26,31,33,36,69,70,97,98,110,112,113,119. Additionally, a recent report presented bubble-propelled motor shells but when they are reduced to sizes smaller than 5 μm, the motors displayed motion without visible bubbles,47 which underlines the relevance of the motor size for catalase motion mechanisms.132 While multiple reports show enhanced diffusion of catalase-powered nanomotors, no reports claim active motion of micrometer-sized motors or bigger except by bubble propulsion but when the size of these motors is increased at the microscale. Wang, Tang, Ma and coworkers explained this by proposing a self-electrophoretic mechanism powering H2O2-consuming micromotors when there is no jet-like propulsion,132 supported by previous studies showing Pt-based micromotors reducing the speed in ionic media133,134 or when their surface roughness was altered.134,135 Hence, in comparison to the Pt film coverage of the motor, the enzymatic coverage is not enough to allow for electron transport through the surface and no bubble-free motion is visible for catalase micromotors. Only Janus particles half covered with a Pt film where the electrons can flow have shown H2O2-consuming active motion without bubble propulsion. However, as mentioned before, for smaller sizes we find catalase nanomotors able to navigate even in ionic media, so a different mechanism could be taking place, either (neutral or ionic) self-diffusiophoresis or nanobubble propulsion. Additionally, the main drawback of this enzyme is the high concentrations of toxic substrate (H2O2) needed for motion, although recently more motors used low substrate concentrations26,33,34,40,43,47,58,102,108 and for biomedical applications where this particular substrate is bioavailable,16,40,57,104 despite being originally used for environmental and sensing applications.3,545.2. Urease
Urease is the second most common enzyme with almost 28% of motors using it as engine (Fig. 4C) that consumes urea (CO(NH2)2) and releases ammonium ions (NH4+) and bicarbonate (CO32−).14,20–22,31,34,36,39,44,60,70–74,78–81,83,84,86,89,90,92,93,99,117–120,129 Interestingly, a study of urease-powered micromotors shows that the gradient of ionic products may be responsible for the self-propulsion,84 an ionic self-diffusiophoretic mechanism that has been further supported by theoretical models.82,136 This was proved by the decreasing speed of urease micromotors after exposing them to different ionic compounds. Interestingly, this net negative influence of ionic media is not observed for nanosized motors since they show an enhanced diffusion even in PBS solution,80 as well as one specific case of platelet micromotors.120 Hence, again properties like the size and chassis material may play an important role in the underlying mechanism of motion and its effectiveness. Recent reports also presented urease motors powered by bubble propulsion.100,121 A clear change from an ionic self-diffusiophoretic mechanism to a bubble thrust mechanism takes place when urea concentration reaches a certain threshold and CO2 and NH3 gases are generated. In Guan's work, this capacity to change the motion mechanism is possible given the shell cavity to nucleate bubbles and the localization of the enzyme in the inner part, highlighting again the importance of the motor shape and enzyme distribution to promote a specific mechanism. Additionally, so far this is the only enzyme that has been purified prior to functionalization, which proved to improve loading, self-propulsion, and endurance of motion.87 Beyond fundamental mechanistic aspects, this enzyme is gaining much interest to propel nanomotors for biomedical applications given its substrate bioavailability, biocompatibility, pH basification and high turnover rate. Urease-powered motors have been applied in different studies to change the media to basic pH81,84 and fight bacteria,46,72,78,93 and for treatments against bladder cancer,137 where urea is bioavailable.22,60,73,96 Due to their biocompatibility and efficient active motion, urease motors have been expanded to multiple other biomedical applications.17,21,34,74,80,99,1205.3. Glucose oxidase
Glucose oxidase is the third most commonly used single enzyme, which is present in more than 6% of reports (Fig. 4D) to power motion consuming glucose and producing gluconic acid and H2O2.67,68,70 This enzyme's biocompatibility and substrate bioavailability have made it gain interest for biomedical applications,37,62 especially recently to power drug delivery systems against breast cancer.111 However, the turnover rate of glucose oxidase is rather slow, and the mechanism of action has been understudied.5.4. Glucose oxidase and catalase
Glucose oxidase is notably used combined with catalase as 10% of the enzymes used in the field (Fig. 4E), even more often than glucose oxidase alone. This enzymatic tandem has the purpose to potentiate the appealing properties of both enzymes while avoiding their drawbacks. The motion mechanism is primarily related to catalase propulsion since it consumes the H2O2 produced by glucose oxidase to release oxygen bubbles that propel the motor. It was initially reported by the Feringa group106 and then followed by others23,24,27,35,47,91,102,107 with high motivation for biomedical applications given the consumption of glucose, high turnover rate of catalase effective for motion, and the biocompatible combination of enzymes.32,37,42,95 Recently, glucose oxidase-catalase motors have also been applied for water decontamination.13 Similarly, catalase has been substituted by Pt (glucose oxidase-Pt) to drive the same catalytic tandem.1035.5. Other enzymes: lipase, ATPase, etc.
Lipase degrades water-insoluble esters such as triglycerides and produces free fatty acids (FFA) and monoglycerides (Fig. 4F). This enzymatic reaction is the fourth most commonly used with almost 3% of representation in the field given the fundamental interest for enzyme attachment studies such as the effect on active motion depending on enzyme orientation76 or enzyme distribution on the motor.15 The consumption of the lipidic substrate in the form of fat degradation has also attracted attention for biomedical and environmental applications,75,109 since lipase is a widely consolidated enzyme in the industrial sector.138,139 Another example of a less frequently used enzyme is ATPase, which degrades adenosine triphosphate (ATP) into adenosine diphosphate (ADP) and a phosphate ion. Given its transmembrane nature it has been used on liposome motors for fundamental studies of enzyme attachment methods118 and to study the chemotactic behavior of lipidic vesicles.119 Other enzymes have been used only once like peroxidase for contaminant degradation,56 bilirubin oxidase for its electron uptake12 and acid phosphatase for the degradation of ATP.118 Substrate bioavailability is an appealing property for biomedical applications that has also motivated to test the power capacity of acetylcholinesterase83 (Fig. 4G), trypsin103 (Fig. 4H), DNAse104 and collagenase.655.6. Other enzyme combinations
Apart from the combination of catalase with glucose oxidase, 8% of motors are powered by a tandem of two enzymes with their reactions chained (Fig. 4I). Some scarce but remarkable combinations of enzymes are used for electron transfer between their catalyzed reactions, such as the pioneer work from Mano et al. with bilirubin oxidase–glucose oxidase,105 or others like peroxidase–catalase, peroxidase–cytochrome c and peroxidase–peroxidase.51,52 All these combinations rely on the transference of electrons through the motor structure to generate a self-electrophoretic mechanism that produces motion, but there are others that just exploit the tandem reactions to trigger self-diffusiophoresis such as glucose oxidase–horseradish peroxidase,94 sarcosine oxidase and catalase,41 or diamine oxidase and catalase.101 A similar designed has been reported by combining enzymatic engines with inorganic catalysts such as xanthin oxidase-Pt and glutamate oxidase-Pt.50 There is even one example of a whole enzymatic network (Fig. 4J) powering active motion by the chained reactions of 6 enzymes: hexokinase, pyruvate kinase, L-lactate dehydrogenase, L-lactate oxidase, glucose-6-phosphate dehydrogenase, and catalase.23 Finally, all the previous examples show a single substrate propulsion, but the Stadler group reported the combination of trypsin with glucose oxidase-Pt to create a double-fueled enzymatic motor,103 followed by recently reported motors combining Pt and urease79 or Pt and lipase.1095.7. Conclusions on enzymatic engines
Recently, multiple studies have compared different enzymatic engines on the same system,31,36,118,119 and the study of the enzymatic properties has provided information on the relevance of the enzyme type for active motion. These comparative studies point towards a central relevance of the turnover rate14,70,83 with recent work from Sánchez pointing towards large conformational changes of flexible enzymes as a precondition for optimal catalysis.83 Additionally, by observing the fastest enzymatic engines the relevance of small and charged product molecules with fast diffusion, as well as the exothermicity of the reaction, can also be considered.83Multiple insightful reports in the field have shed light on the mechanism of the most commonly used enzymes like catalase, urease or tandems of enzymes for electron transfer. These publications feature the relevance of the product properties in terms of asymmetric charge gradients (production of ions or electrons) or asymmetric bubble release (production of gas bubbles) (Fig. 4A inset). More specifically, catalase has been largely reported to power motion through either bubble propulsion or (ionic or neutral) self-diffusiophoresis, while urease propels mainly thanks to self-diffusiophoresis of ionic products and only a couple of reports show bubble propulsion. Hence, the mechanism of motion is strongly dependent on the nature of the product molecules. A third motion mechanism with significant representation is self-electrophoresis, commonly based on a tandem of enzymes that transfer electrons from one to the other (due to their complementary enzymatic reactions) though a conductive chassis. However, for most enzymes tested there is no clear indication for an individual mechanism motion either by: (1) a lack of study and mention in the publication probably argued to be out-of-scope (29% of the reports) or (2) an excessive complexity of the study that only leads to hypotheses of multiple mechanisms. This poses special difficulty on clarifying the exact mechanisms governing the active motion of each motor enzyme–chassis complex, and more efforts should be carried out in this direction. There are other aspects not yet considered that may contribute to the motion mechanism, such as the biological function and native-like conditions of enzymes, the need for a metal cofactor, the structural multimerism, or even aspects of the purity of the sample used. Additionally, the size of micromotors, chassis shape, surface morphology and enzyme distribution appear to play a relevant role in the motion mechanism140 as it has been mentioned for urease84,100 and catalase,47,132 where micromotors and nanomotors may be driven by different mechanisms or at least affected differently by the motor design or media composition.84
In conclusion, based on the whole library of enzymatic engines and the studies on the enzyme type, certain parameters play a central role in active motion such as the turnover rate of exothermic reactions, high structural flexibility and small and charged product molecules with fast diffusion. These properties are promoted in the field, but specific characteristics may be more attractive for particular applications, for instance, the degradation of contaminants and the generation of bubbles for micromixing both in sensing and decontamination applications, or high biocompatibility and substrate bioavailability for biomedical applications.
6. Motor size and motion detection technique
The sizes of enzyme-powered motors range from a few tens of nanometers up to centimeter-sized chassis but almost 77% of all enzymatic motors are between 0.3 or 100 micrometers, as depicted in the bibliographic summary of Fig. 5A. Regarding applications, there has been a tendency to reduce the size of motors given the increasing interest in biomedical applications, while motors of larger sizes have been normally used for either fundamental studies or applications in sensing or environmental applications.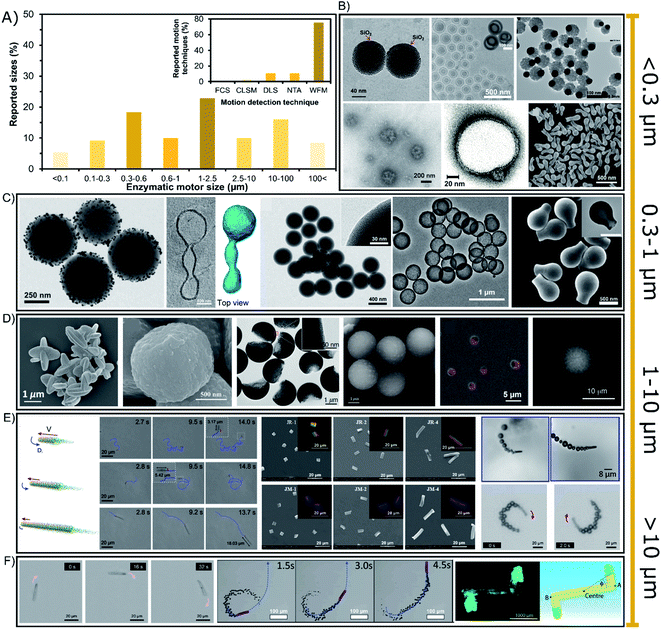 | ||
| Fig. 5 (A) Sizes of enzyme-powered micro- and nano-motors and motion detection technique. Representation the different motor sizes in the publications of the field. Inset: representation of each motion detection technique in the publications of the field. Examples of enzymatic motors powered with sizes of (B) < 0.3 (ref. 66, 25, 100, 41, 37 and 95), (C) 0.3–1 (ref. 93, 31, 73, 76 and 103), (D and E) 1–10 (ref. 107, 106, 97, 81, 115, 60, 83, 17 and 52) and (F) > 10 μm.30,39,63,118 Panel (B) < 0.3 μm adapted with permission from ref. 66 Copyright 2017 Elsevier, ref. 25 Copyright 2019 American Chemical Society, ref. 100 Copyright 2021 American Chemical Society, ref. 41 Copyright 2021 Elsevier, ref. 37 Copyright 2017 AAAS, and ref. 95 Copyright 2021 American Chemical Society. Panel (C) 0.3–1 μm adapted with permission from ref. 93 Copyright 2021 AAAS, ref. 31 Copyright 2021 American Chemical Society, ref. 73 Copyright 2020 Wiley, ref. 76 Copyright 2021 American Chemical Society, and ref. 103 Copyright 2019 American Chemical Society. Panels (D and E) 1–10 μm adapted with permission from ref. 107 Copyright 2019 Wiley, ref. 106 Copyright 2021 American Chemical Society, ref. 97 Copyright 2021 American Institute of Physics, ref. 81 Copyright 2020 AAAS, ref. 115 Copyright 2020 AAAS, ref. 60 Copyright 2017 Royal Society of Chemistry, ref. 83 Copyright 2016 American Chemical Society, ref. 17 Copyright 2020 Elsevier, and ref. 52 Copyright 2013 American Chemical Society. Panel (F) > 10 μm adapted with permission from ref. 63 Copyright 2021 Royal Society of Chemistry, ref. 39 Copyright 2019 Wiley, ref. 30 Copyright 2016 Wiley, and ref. 118 Copyright 2019 Wiley. | ||
6.1. Nano-sized enzymatic motors
Enzymatic micro- and nano-motors have sizes distributed from a few nanometers to centimeter-sized structures (Fig. 5), the smallest of which are gold spheres with a diameter of 25 nm synthesized by He's group.67 The next smallest enzymatic motors are CaCO3-based spheres of 60 nm110 and silica spheres of 90 nm,69 both motors used for fundamental studies of motion at the nanoscale. For larger motors, there is a special focus on sizes between 0.1 and 0.6 μm (Fig. 5B and C) appealing for biomedical applications, with special interest around 200 nm diameter for transport in the bloodstream. Such enzymatic motors are made of different materials like polymeric structures,13,23,37,40 liposomal vesicles,117–119 metal-based structures42,62,101,113 and silica-based particles,71,97,99,104,109 mainly applied as active drug delivery systems and chemotactic nanotools towards biologically relevant environments. Their small size promotes a proper navigation in complex media, and although only a few enzymes fit on the structure, it is enough to produce active motion. Since sizes below 300 nm (Fig. 5B) are not visible through widefield microscopy (WFM), the diffusion of nanomotors is studied through techniques like Dynamic Light Scattering (DLS) and NanoSight Tracking Analysis (NTA), each of which was used in almost 11% of the publications (Fig. 5A inset). These techniques are based on the measurement of collective diffusion though exposure with a laser beam. Other laser-based techniques have been rarely used to study the active motion of enzymatic nanomotors such as Fluorescence Correlation Spectroscopy (FCS)118 and confocal microscopy (CLSM).13,34,119 It is relevant to highlight the need for fluorescent dyes for techniques like NTA, FCS, CLSM or even stochastic optical reconstruction microscopy (STORM). However, the effect of these dyes on catalysis and active motion should be addressed in future work. However, more than 85% of the enzymatic motors are bigger than 300 nm, and hence, widefield microscopy has been used in almost 76% of the reports. Although this technique makes more difficult to study the collective active motion in 3D, it allows the detection of the trajectory of single enzymatic motors in 2D. Some nanomotors are above the threshold of visibility for widefield microscopy,24,27,31,41,63,70,76,79,98 suitable for biomedical applications based on active drug delivery or photo-responsive nanotools with chemotaxis, molecular targeting, bioavailable substrate consumption, bacteria killing and enhanced penetration.25,32,34,57,60,65,72–75,80,94–966.2. Micron-sized up to centimeter-sized enzymatic motors
Especially sizes of 0.6–10 μm diameter have attracted much attention since more than 42% of enzymatic motors are between these dimensions (Fig. 5D and E), with more than half of them in the range of 1 and 2.5 μm (Fig. 5D). These sizes are useful because more enzymes fit in the structure and their motion trajectory becomes clearly visible, showing a directional propulsive behavior that is suitable for fundamental studies14,15,26,29,35,36,44,48,51–53,68,81–87,89–91,100,103,107,118,131 and applications that take place outside the body, such as sensing,17,43,50,77 water remediation,54–56,108,126 and bacteria killing.78,93 However, in certain situations these sizes are also intended to be used for biomedical applications16,21,22,57,111,112,114,120 that need faster motion for a stronger micromixing or a clearly visible propulsion, as well as easier fabrication and less aggregation.Enzymatic motors of tens of micrometers are also suitable for bacteria capture and destruction,46,66 sensing17,19,49,58,64 and bioremediation61,115 or for fundamental studies,28,33,38,39,45,86,102,123 although there are a few examples for biomedical applications.59,121 Scaling up, there are two reports of motors ranging in the hundreds of microns30,122 (Fig. 5F) focusing on fundamental studies, as well as few millimeter/centimeter-sized motors105,106,125,127,129 that pioneered the field of enzyme-powered motors and have recently been applied for micro-stirring124 and to detect enantiomers in solution.12
6.3. Conclusions on the motor size and motion detection technique
Over the years, there has been a tendency of reducing the size of enzymatic motors, especially in response to their potential for biomedical applications that demand navigation inside the human body. Hence, while widefield microscopy is extensively used, novel techniques to detect the collective motion of swarms in 3D have gained relevance, especially DLS and NTA. Meanwhile, enzymatic motors at the microscale are intended for applications such as molecule sensing and water decontamination, for which widefield microscopy is fitting properly. Finally, some insightful reports compare a range of sizes for the same motor, which present an increase in the speed and rotation for smaller chassis dimensions.17,18,47,86,111,1187. Applications of enzymatic micro- and nano-motors
Beyond fundamental studies, one important motivation to build enzyme-powered micro- and nano-motors is to apply them as tools to solve specific problems, the reason for which is almost 48% of the publications aim at applications (56 out of 117). The most prominent benefit of using enzymes as engines for active motion instead of inorganic catalysts is the increase of the biocompatibility and versatility of reactions used, both appealing for their implementation in biomedicine. In Fig. 6A (green bars) it is shown that 52% of applied publications focus on the different approaches of biomedical applications such as active drug delivery, visualization of motors in tissue due to enhanced penetration and targeting, and thermal treatment by photo- or magnetic stimulation. Among these publications the most commonly used enzymes are urease, catalase, and few examples with glucose oxidase, which can exploit the bioavailability of substrates. The other 48% of applied publications focus on applications that are not taking place inside living organisms. Following the prominent activities of catalytic micro- and nano-motors in environmental applications, enzyme-powered motors have also been used to decontaminate water. In all cases, the same catalytic reaction is used to exploit the bubble micromixing effect but substituting the Pt catalyst by the catalase enzyme. This feature is also useful for the applications of sensing and antimicrobial effects in solution, which also tend to use catalase as a propelling system and recently some examples of urease have been reported.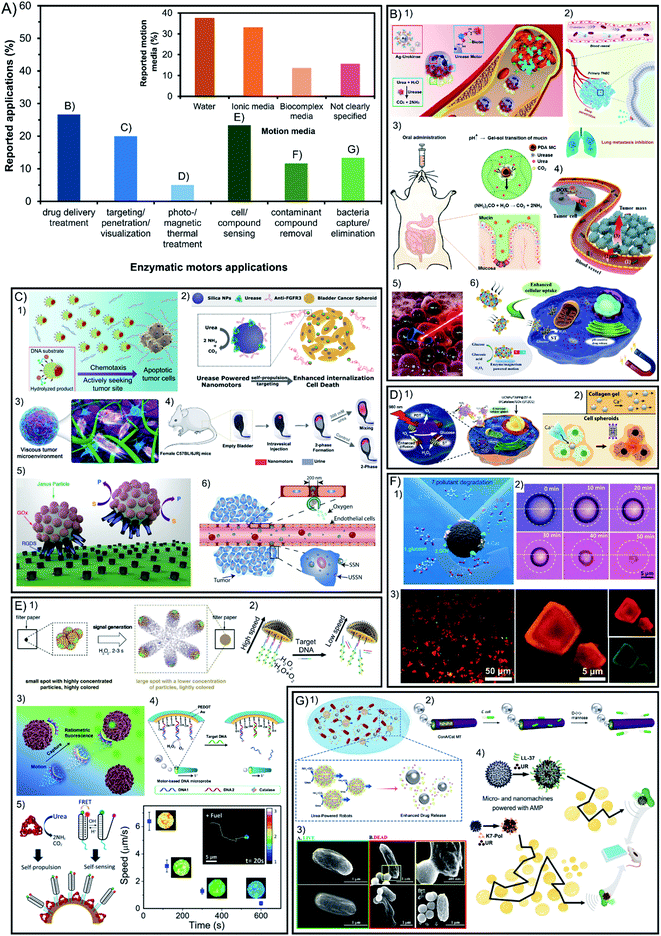 | ||
| Fig. 6 (A) Applications of enzyme-powered micro- and nano-motors and motion detection technique. Representation the different motor applications in the publications of the field. Inset: representation of each motion media in the publications of the field. Examples of enzymatic motors applied in (B) drug delivery treatment,20,21,41,57,106,116 (C) visualization inside organisms exploiting enhanced targeting and penetration,25,40,60,70,93,100 (D) photo- and magnetic thermal treatment,62,108 (E) cell and compound sensing,17,42,47,56,78 (F) contaminant compound removal13,72,110 and (G) bacteria capture or elimination.58,63,69,75 Panel (B) adapted with permission from (1) ref. 116 Copyright 2022 American Chemical Society, (2) ref. 41 Copyright 2021 Elsevier, (3) ref. 21 Copyright 2022 Elsevier, (4) ref. 20 Copyright 2021 Elsevier, (5) ref. 57 Copyright 2015 American Chemical Society, and (6) ref. 106 Copyright 2021 American Chemical Society. Panel (C) adapted with permission from (1) ref. 100 Copyright 2021 American Chemical Society, (2) ref. 70 Copyright 2019 American Chemical Society, (3) ref. 40 Copyright 2019 American Chemical Society, (4) ref. 93 Copyright 2021 AAAS, (5) ref. 60 Copyright 2017 Royal Society of Chemistry, and (6) ref. 25 Copyright 2019 American Chemical Society. Panel (D) adapted with permission from (1) ref. 108 Copyright 2019 Elsevier and (2) ref. 62 Copyright 2019 American Chemical Society. Panel (E) adapted with permission from (1) ref. 42 Copyright 2019 Elsevier, (2) ref. 47 Copyright 2019 American Chemical Society, (3) ref. 17 Copyright 2020 Elsevier, (4) ref. 56 Copyright 2017 Elsevier, and (5) ref. 78 Copyright 2019 American Chemical Society. Panel (F) adapted with permission from (1) ref. 13 Copyright 2021 American Chemical Society, (2) ref. 72 Copyright 2019 Wiley, and (3) ref. 110 Copyright 2020 American Chemical Society. Panel (G) adapted with permission from (1) ref. 58 Copyright 2021 American Chemical Society, (2) ref. 63 Copyright 2021 Royal Society of Chemistry, (3) ref. 69 Copyright 2021 American Chemical Society, and (4) ref. 75 Copyright 2022 American Chemical Society. | ||
7.1. Biomedical applications
Within the reported studies on the biomedical application of enzyme nanomotors, 27% is devoted to the development of drug delivery systems for disease treatment16,20,34,42,59,74,80,97,99,110–112,114,120,121 (Fig. 6B). Another 20% of the applications are focused on the visualization of micro- and nano-motors inside living organisms exploiting their capacities of enhanced targeting and penetration21,22,25,37,40,57,60,62,73,95,96,99,104 (Fig. 6C). The remaining 5% of publications applied on biomedicine focus on disease treatment through photo-responsive or magnetic thermal treatment32,34,65,113 (Fig. 6D).As observed in each of the previous sections, the different parameters and optimal design of enzymatic micro- and nano-motors are determined by the final application for that specific motor. For the future of biomedical applications, spherical shapes created from materials with biological-like properties such as biocompatibility, biodegradability, flexibility, solubility, and permeability will be promoted to offer native-like properties, which points towards bio-inspired materials like liposomes or biodegradable polymers. Silica poses promising features as a chassis material for biomedical nanomotors given its biocompatibility, and easy modification of the shape, size, and surface functionalization.60 Interestingly, inorganic materials could be exploited for their magnetic and photo-responsiveness, aiming at the minimum amount to be effective yet not affecting the biological-like properties mentioned above. These nanomotors tend to be powered through enzymes that also offer high biocompatibility and substrate bioavailability. Sizes from 100 to 800 nm are optimal for biomedical applications, with special interest between 100 and 400 nm, given the positive results in circulating times, extravasation and cell interaction,141 and their suitable diffusive motion for complex media. Moreover, these sizes are in the observable limit of widefield microscopy and since there is a need to detect collective motion of swarms in 3D, other methods like DLS and NTA are gaining more relevance.
7.2. Other applications: sensing, environmental and antimicrobial
The sensing of cellular or molecular compounds represents almost 23% of the applied publications12,17,19,43,49,52,54,55,58,64,77,81,94 (Fig. 6E). Additionally, lipase has been recently included in that field of applications thanks to the appeal of lipid degradation in solution. Another 12% of applied publications target the field of water remediation and decontamination from molecular pollutants, which can be removed by adsorption or degradation13,56,61,75,108,115,126 (Fig. 6F). Moreover, 13% of applied reports are based on the capture and elimination of pathogenic bacteria, which in fact can take place outside and inside living organisms18,46,60,66,72,78,93,120 (Fig. 6G).For the above-mentioned, the latter share common features such as elongated tubular structures with sizes ranging from 1 μm to 100 μm (even up to centimeter-sized structures), which can be visualized though widefield microscopy and show a clearly directional propulsion. It is important that the materials do not release toxic products, are able to be recovered and have specific reactivity, which are common features of inorganic materials that are magnetic and photo-responsive. The enzymes suitable for environmental applications tend to prioritize the degradation of a specific molecule to decontaminate or sense, as well as generate a thrust of bubbles to power motion and create a micromixing effect, such as the catalase enzyme. Specifically for environmental applications, it is appealing to have a chassis composition with highly controllable porosity, shape diversity and reactivity to stimuli, which are more frequent in inorganic compounds. Apart from water decontamination, other applications can benefit from specific enzymatic reactions. For instance, the basification of the media through urease catalysis has been used to produce an antimicrobial effect,72,78 or the degradation of phenolic compounds and production of color and light through horseradish peroxidase has been used for sensing and environmental applications.56
8. Discussion
In conclusion, this manuscript reviews all the publications in the field of enzymatic micro- and nano-motors and provides an overview to help rationally design motors tailored to specific applications. However, there are fundamental aspects in each report that can be beneficial for any kind of application and more in-depth basic science studies should be pursued for a greater impact in the field. For instance, material and shape preferences tend to be aligned with easy fabrication, modification, and scalability. Regarding the improvement of active motion in general, it is highly desired to have faster propulsion and navigation even at low concentrations of substrate. It is also interesting to have shapes and materials (like the well stablished polymeric materials, silica, and metals, and their combinations) that facilitate the asymmetry of product release and provide easy fabrication, surface and shape modification and magnetic and photo-responsiveness. This would also depend on promoting the analysis of the quantity and distribution of enzymes on the structure, for which the most effective configuration tends to be the release of the product through a hole when enzymes are kept inside a cavity. Enzymes could be incorporated through covalent attachment (with their final orientation optimized) for more resistance and with minimal loss of catalytic activity. Covalent binding has also presented an appealing configuration by creating multilayers of the enzyme, which has proven to significantly boost active motion. However, if the cavity is close enough to encapsulate the enzymes free in solution and release the product through a hole, a more native-like environment can be achieved by controlling the inner media and membrane permeability, thus promoting both the active motion and protection of enzymes from external conditions. For the same purpose, different polymeric coatings may be applied to cover the enzyme area and catalysis itself also contributes to decrease the protein corona blocking the particle surface. However, since these configurations may currently require extra steps of fabrication, in general the most cost-effective choice will be direct adsorption of the enzyme through non-covalent interactions and/or spontaneous generation of asymmetry. There are also enzymatic properties that could be appealing for any purpose of enzyme-powered motors given their central role in propulsion performance. Generally, these properties could be summarized as a high turnover rate of exothermic reactions, high structural flexibility and small and charged product molecules with fast diffusion. Motion dynamics can vary based on the enzyme type and the mechanisms of motion. This latter aspect is relevant to consider as it has been observed how the size and shape can have a clear effect on the motion speed86,142 even by means of changing the motion mechanism. For instance, catalase motors show faster motion when an oxygen bubble thrust is produced (bubble propulsion mechanism or buoyancy effect) rather than through (neutral or ionic) self-diffusiophoresis or self-electrophoresis. Bubble propulsion can be triggered by increasing the motor size or modifying the surface morphology,47 as it was also observed for Pt-based motors.134,143 Other aspects may trigger a bubble-propelled mechanism, such as the presence of bubble nucleation points, which are promoted by containing the enzymes in a cavity and releasing the product through a hole144–146 or the presence of a surfactant in the media can also allow for bubble formation.147,148 Hence, the influence of the surrounding media is also key to the motion mechanism, as it is also observed for ion-dependent mechanisms (self-electrophoresis and ionic self-diffusiophoresis), like motors powered by electron transfer and urease motors, which show faster propulsion in the absence of ions in the media. Recently, urease motors have shown a bubble-propelled mechanism by increasing the fuel concentration.100,121 There is even the possibility of coexisting mechanisms,149 and although several key properties (shape, surface, size, enzyme type and location and media composition) appear to interact with each other and have a relevant role in the motion mechanisms, they are heavily understudied. To fully understand how all these parameters affect each other and the generation of a motion mechanism out of them, it is necessary that the field devotes special attention, curiosity and study to the most fundamental aspects of these navigating systems while pursuing the final applications. For instance, more than 29% of publications do not mention or specify a motion mechanism for the motors reported (Fig. 5A inset), and almost 16% of reports lack a clear indication of the media in which motion was tested (Fig. 6A inset). However, over the years more publications show propulsion in ionic media (specially in PBS) and in more complex environments like cell medium, serum or urine, a crucial step for the implementation of these devices. Since most of the biomedical applications take place in buffered solutions and enzymes require specific pH for their optimal catalysis, a more in-depth study of the effect of buffers in active motion is needed in the field. Regarding these understudied media effects, a recent report studied the formation of a protein corona around urease nanomotors in biological media, showing that catalysis and active motion reduce the presence of proteins around the particle.719. Outlook
As a final point, special emphasis is needed towards the exploration of novel options in each of the design parameters, to push for a wider library and open new avenues, and the possibility of advantages yet to be discovered. Some recently featured examples with novel properties are (1) highly porous materials like metal–organic frameworks or bio-inspired cell-based chassis; (2) shapes with an increased surface area like 2D sheets or enhanced spontaneous asymmetry generated with cucurbit shapes; (3) intelligent attachment methods that are resistant and interchangeable by using click chemistry or azobenzene/benzimidazole interaction with β-CD-modified enzymes; (4) innovative enzymes for their application-oriented substrate consumption and attachment studies for enzyme orientation and transmembrane attachment, as it is the case for lipase and ATPase, respectively. Additionally, multiple understudied enzymological aspects could be key to the induced faster propulsion, such as the biological function and native-like conditions of enzymes, the need for a metal cofactor, and the structural multimerism. Enzyme purification has proven to improve active motion, so the field would benefit from in-house synthesis and on-demand modification of enzymes to optimize their catalysis and structure for a better propulsion. Direct communication with other fields will also be decisive to break new ground, an example of which could be the incorporation of nanozymes. Overall, this is a review of the explored parameters of enzyme-powered micro- and nano-motors, their diversity of options and advantages, and the outlook on motor design optimization and exploration of novel features.Author contributions
X. A. wrote the manuscript. T. P. and S. S. revised and supervised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Spanish MINECO (project RTI2018-095622-B-I00), the Catalan AGAUR (project 2017 SGR 238), and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No 866348; i-NanoSwarms) and from the “la Caixa” Foundation under the grant agreement LCF/PR/HR21/52410022 (BLADDEBOTS project). It was also funded by the grant RTI2018-098164-B-I00 from the MICIN/AEI/10.13039/5011000110333 and the “FEDER Una manera de hacer Europa” (BOTSinFluids project), the CERCA program by the Generalitat de Catalunya, the Secretaria d'Universitats i Recerca del Departament d'Empresa i Coneixement de la Generalitat de Catalunya through the project 2017 SGR 1148 and the “Centro de Excelencia Severo Ochoa”, funded by the Agencia Estatal de Investigación (CEX2018-000789-S). X. A. thanks the Spanish MINECO for the Severo Ochoa program (SEV-2014-0425) for the PhD fellowship (PRE2018-083712).References
- Y. Zhang and H. Hess, Nat. Rev. Chem., 2021, 5, 500–510 CrossRef CAS.
- M. Mathesh, J. Sun and D. A. Wilson, J. Mater. Chem. B, 2020, 8, 7319–7334 RSC.
- J. Parmar, D. Vilela, K. Villa, J. Wang and S. Sánchez, J. Am. Chem. Soc., 2018, 140, 9317–9331 CrossRef CAS PubMed.
- Z. Guo, J. Liu, D.-W. Wang, J. Xu and K. Liang, Biophys. Rep., 2020, 6, 179–192 CrossRef.
- G. Salinas, S. M. Beladi-Mousavi and A. Kuhn, Curr. Opin. Electrochem., 2022, 32, 100887 CrossRef CAS.
- H. Yuan, X. Liu, L. Wang and X. Ma, Bioact. Mater., 2021, 6, 1727–1749 CrossRef CAS PubMed.
- X. Zhao, K. Gentile, F. Mohajerani and A. Sen, Acc. Chem. Res., 2018, 51, 2373–2381 CrossRef CAS PubMed.
- T. Patino, X. Arqué, R. Mestre, L. Palacios and S. Sánchez, Acc. Chem. Res., 2018, 51, 2662–2671 CrossRef CAS PubMed.
- F. Soto, E. Karshalev, F. Zhang, B. Esteban Fernandez De Avila, A. Nourhani and J. Wang, Chem. Rev., 2021, 112, 5365–5403 Search PubMed.
- S. Hermanová and M. Pumera, Chem.–Eur. J., 2020, 26, 11085–11092 CrossRef PubMed.
- Q. Yang, Y. Gao, L. Xu, W. Hong, Y. She and G. Yang, Int. J. Biol. Macromol., 2021, 167, 457–469 CrossRef CAS PubMed.
- S. Arnaboldi, G. Salinas, A. Karajić, P. Garrigue, T. Benincori, G. Bonetti, R. Cirilli, S. Bichon, S. Gounel, N. Mano and A. Kuhn, Nat. Chem., 2021, 13, 1241–1247 CrossRef CAS PubMed.
- E. M. Kutorglo, R. Elashnikov, S. Rimpelova, P. Ulbrich, J. Říhová Ambrožová, V. Svorcik and O. Lyutakov, ACS Appl. Mater. Interfaces, 2021, 13, 16173–16181 CrossRef CAS PubMed.
- K. K. Dey, X. Zhao, B. M. Tansi, W. J. Méndez-Ortiz, U. M. Córdova-Figueroa, R. Golestanian and A. Sen, Nano Lett., 2015, 15, 8311–8315 CrossRef CAS PubMed.
- Y. Hu and Y. Sun, Biochem. Eng. J., 2019, 149, 107242 CrossRef CAS.
- J. Wang, B. J. Toebes, A. S. Plachokova, Q. Liu, D. Deng, J. A. Jansen, F. Yang and D. A. Wilson, Adv. Healthcare Mater., 2020, 9, 1–8 Search PubMed.
- L. Zhao, Y. Liu, S. Xie, P. Ran, J. Wei, Q. Liu and X. Li, Chem. Eng. J., 2020, 382, 123041 CrossRef CAS.
- L. Zhao, S. Xie, Y. Liu, Q. Liu, X. Song and X. Li, Nanoscale, 2019, 11, 17831–17840 RSC.
- M. Liu, Y. Sun, T. Wang, Z. Ye, H. Zhang, B. Dong and C. Y. Li, J. Mater. Chem. C, 2016, 4, 5945–5952 RSC.
- Z. Zhang, H. Yan, S. Li, Y. Liu, P. Ran, W. Chen and X. Li, Chem. Eng. J., 2021, 404, 127073 CrossRef CAS.
- H. Choi, S. H. Jeong, T. Y. Kim, J. Yi and S. K. Hahn, Bioact. Mater., 2022, 9, 54–62 CrossRef CAS PubMed.
- H. Choi, S. H. Cho and S. K. Hahn, ACS Nano, 2020, 14, 6683–6692 CrossRef CAS PubMed.
- M. Nijemeisland, L. K. E. A. Abdelmohsen, W. T. S. Huck, D. A. Wilson and J. C. M. Van Hest, ACS Cent. Sci., 2016, 2, 843–849 CrossRef CAS PubMed.
- L. K. E. A. Abdelmohsen, M. Nijemeisland, G. M. Pawar, G. J. A. Janssen, R. J. M. Nolte, J. C. M. Van Hest and D. A. Wilson, ACS Nano, 2016, 10, 2652–2660 CrossRef CAS PubMed.
- J. Sun, M. Mathesh, W. Li and D. A. Wilson, ACS Nano, 2019, 13, 10191–10200 CrossRef CAS PubMed.
- B. J. Toebes, L. K. E. A. Abdelmohsen and D. A. Wilson, Polym. Chem., 2018, 9, 3190–3194 RSC.
- B. J. Toebes, F. Cao and D. A. Wilson, Nat. Commun., 2019, 10, 1–6 CAS.
- S. Keller, S. P. Teora, G. X. Hu, M. Nijemeisland and D. A. Wilson, Angew. Chem., Int. Ed., 2018, 57, 9814–9817 CrossRef CAS PubMed.
- S. Keller, G. X. Hu, M. I. Gherghina-Tudor, S. P. Teora and D. A. Wilson, Adv. Funct. Mater., 2019, 29, 1–6 CrossRef.
- A. Sitt, J. Soukupova, D. Miller, D. Verdi, R. Zboril, H. Hess and J. Lahann, Small, 2016, 12, 1432–1439 CrossRef CAS PubMed.
- S. Cao, H. Wu, I. A. B. Pijpers, J. Shao, L. K. E. A. Abdelmohsen, D. S. Williams and J. C. M. Van Hest, ACS Nano, 2021, 15, 18270–18278 CrossRef CAS PubMed.
- J. Shao, S. Cao, H. Wu, L. K. E. A. Abdelmohsen and J. C. M. van Hest, Pharmaceutics, 2021, 13, 1–13 Search PubMed.
- W. S. Jang, H. J. Kim, C. Gao, D. Lee and D. A. Hammer, Small, 2018, 14, 1–8 Search PubMed.
- M. Wu, S. Liu, Z. Liu, F. Huang, X. Xu and Q. Shuai, Colloids Surf., B, 2022, 212, 112353 CrossRef CAS PubMed.
- C. Zhou, C. Gao, Y. Wu, T. Si, M. Yang and Q. He, Angew. Chem., Int. Ed., 2022, 61, e202116013 CAS.
- S. Song, A. F. Mason, R. A. J. Post, M. De Corato, R. Mestre, N. A. Yewdall, S. Cao, R. W. van der Hofstad, S. Sanchez, L. K. E. A. Abdelmohsen and J. C. M. van Hest, Nat. Commun., 2021, 12, 1–9 CrossRef PubMed.
- A. Joseph, C. Contini, D. Cecchin, S. Nyberg, L. Ruiz-Perez, J. Gaitzsch, G. Fullstone, X. Tian, J. Azizi, J. Preston, G. Volpe and G. Battaglia, Sci. Adv., 2017, 3, e1700362 CrossRef PubMed.
- N. Sugai, Y. Nakai, Y. Morita and T. Komatsu, ACS Appl. Nano Mater., 2018, 1, 3080–3085 CrossRef CAS.
- N. Sugai, Y. Morita and T. Komatsu, Chem.–Asian J., 2019, 14, 2953–2957 CrossRef CAS PubMed.
- H. Li, Z. Sun, S. Jiang, X. Lai, A. Böckler, H. Huang, F. Peng, L. Liu and Y. Chen, Nano Lett., 2019, 19, 8749–8757 CrossRef CAS PubMed.
- A. Chatterjee, S. Ghosh, C. Ghosh and D. Das, Angew. Chem., Int. Ed., 2022, 1–6, DOI:10.1002/anie.202201547.
- W. Yu, R. Lin, X. He, X. Yang, H. Zhang, C. Hu, R. Liu, Y. Huang, Y. Qin and H. Gao, Acta Pharm. Sin. B, 2021, 11, 2924–2936 CrossRef CAS PubMed.
- S. M. Russell, A. Alba-Patiño, M. Borges and R. de la Rica, Biosens. Bioelectron., 2019, 140, 111346 CrossRef CAS PubMed.
- M. Luo, S. Li, J. Wan, C. Yang, B. Chen and J. Guan, Langmuir, 2020, 36, 7005–7013 CAS.
- S. Sanchez, A. A. Solovev, Y. F. Mei and O. C. Schmidt, J. Am. Chem. Soc., 2010, 132, 13144–13145 CrossRef CAS PubMed.
- K. Villa, H. Sopha, J. Zelenka, M. Motola, L. Dekanovsky, D. C. Beketova, J. M. Macak, T. Ruml and M. Pumera, Small, 2022, 2106612 CrossRef PubMed.
- C. Chen, Z. He, J. Wu, X. Zhang, Q. Xia and H. Ju, Chem.–Asian J., 2019, 14, 2491–2496 CrossRef CAS PubMed.
- D. Wang, C. Chen, J. Sun, H. Ao, W. Xiao, H. Ju and J. Wu, ACS Appl. Mater. Interfaces, 2022, 14, 27074–27082 CrossRef CAS PubMed.
- X. Zhang, C. Chen, J. Wu and H. Ju, ACS Appl. Mater. Interfaces, 2019, 11, 13581–13588 CrossRef CAS PubMed.
- A. I. Bunea, I. A. Pavel, S. David and S. Gáspár, Biosens. Bioelectron., 2015, 67, 42–48 CrossRef CAS PubMed.
- A. I. Bunea, I. A. Pavel, S. David and S. Gáspár, Chem. Commun., 2013, 49, 8803–8805 RSC.
- I. A. Pavel, A. I. Bunea, S. David and S. Gáspár, ChemCatChem, 2014, 6, 866–872 CrossRef CAS.
- W. Gao, S. Sattayasamitsathit, A. Uygun, A. Pei, A. Ponedal and J. Wang, Nanoscale, 2012, 4, 2447–2453 RSC.
- J. Orozco, V. García-Gradilla, M. D'Agostino, W. Gao, A. Cortés and J. Wang, ACS Nano, 2013, 7, 818–824 CrossRef CAS PubMed.
- V. V. Singh, K. Kaufmann, B. Esteban-Fernández De Ávila, M. Uygun and J. Wang, Chem. Commun., 2016, 52, 3360–3363 RSC.
- M. Bayraktaroğlu, B. Jurado-Sánchez and M. Uygun, J. Hazard. Mater., 2021, 418, 126268 CrossRef PubMed.
- E. S. Olson, J. Orozco, Z. Wu, C. D. Malone, B. Yi, W. Gao, M. Eghtedari, J. Wang and R. F. Mattrey, Biomaterials, 2013, 34, 8918–8924 CrossRef CAS PubMed.
- Y. Xie, S. Fu, J. Wu, J. Lei and H. Ju, Biosens. Bioelectron., 2017, 87, 31–37 CrossRef CAS PubMed.
- Z. Wu, X. Lin, X. Zou, J. Sun and Q. He, ACS Appl. Mater. Interfaces, 2015, 7, 250–255 CrossRef CAS PubMed.
- D. Xu, J. Hu, X. Pan, S. Sánchez, X. Yan and X. Ma, ACS Nano, 2021, 15, 11543–11554 CrossRef CAS PubMed.
- D. Wang, G. Zhao, C. Chen, H. Zhang, R. Duan, D. Zhang, M. Li and B. Dong, Langmuir, 2019, 35, 2801–2807 CrossRef CAS PubMed.
- G. Rucinskaite, S. A. Thompson, S. Paterson and R. De La Rica, Nanoscale, 2017, 9, 5404–5407 RSC.
- J. Shao, S. Cao, H. Che, M. T. De Martino, H. Wu, L. K. E. A. Abdelmohsen and J. C. M. Van Hest, J. Am. Chem. Soc., 2022, 1–6 Search PubMed.
- S. Fu, X. Zhang, Y. Xie, J. Wu and H. Ju, Nanoscale, 2017, 9, 9026–9033 RSC.
- M. A. Ramos-Docampo, M. Fernández-Medina, E. Taipaleenmäki, O. Hovorka, V. Salgueirinõ and B. Städler, ACS Nano, 2019, 13, 12192–12205 CrossRef CAS PubMed.
- M. Umebara, N. Sugai, K. Murayama, T. Sugawara, Y. Akashi, Y. Morita, R. Kato and T. Komatsu, Mater. Adv., 2021, 2, 6428–6438 RSC.
- Y. Ji, X. Lin, Z. Wu, Y. Wu, W. Gao and Q. He, Angew. Chem., Int. Ed., 2019, 58, 12200–12205 CrossRef CAS PubMed.
- W. Noh, S. Jo, J. Kim and T. S. Lee, Langmuir, 2021, 37, 6301–6310 CrossRef CAS PubMed.
- X. Ma and S. Sánchez, Tetrahedron, 2017, 73, 4883–4886 CrossRef CAS.
- X. Ma, A. Jannasch, U. R. Albrecht, K. Hahn, A. Miguel-López, E. Schäffer and S. Sánchez, Nano Lett., 2015, 15, 7043–7050 CrossRef PubMed.
- T. Patiño, J. Llacer-wintle, S. Pujals, L. Albertazzi and S. Sánchez, ChemRxiv, 2021, 1–10 Search PubMed.
- D. Vilela, N. Blanco-Cabra, A. Eguskiza, A. C. Hortelao, E. Torrents and S. Sanchez, ACS Appl. Mater. Interfaces, 2021, 13, 14964–14973 CrossRef CAS PubMed.
- A. C. Hortelao, R. Carrascosa, N. Murillo-Cremaes, T. Patino and S. Sánchez, ACS Nano, 2019, 13, 429–439 CrossRef CAS PubMed.
- A. Llopis-Lorente, A. Garciá-Fernández, N. Murillo-Cremaes, A. C. Hortelaõ, T. Patinõ, R. Villalonga, F. Sancenón, R. Martínez-Máñez and S. Sánchez, ACS Nano, 2019, 13, 12171–12183 CrossRef CAS PubMed.
- L. Wang, A. C. Hortelão, X. Huang and S. Sánchez, Angew. Chem., 2019, 131, 8076–8080 CrossRef.
- L. Wang, M. Marciello, M. Estévez-Gay, P. E. D. Soto Rodriguez, Y. Luengo Morato, J. Iglesias-Fernández, X. Huang, S. Osuna, M. Filice and S. Sánchez, Angew. Chem., 2020, 132, 21266–21273 CrossRef.
- J. Simmchen, A. Baeza, D. Ruiz, M. J. Esplandiu and M. Vallet-Regí, Small, 2012, 8, 2053–2059 CrossRef CAS PubMed.
- X. Arqué, M. D. T. Torres, T. Patiño, A. Boaro, S. Sánchez and C. de la Fuente-Núñez, ACS Nano, 2022, 1–12 Search PubMed.
- Z. Ye, Y. Wang, S. Liu, D. Xu, W. Wang and X. Ma, J. Am. Chem. Soc., 2021, 143, 15063–15072 CrossRef CAS PubMed.
- A. C. Hortelão, T. Patiño, A. Perez-Jiménez, À. Blanco and S. Sánchez, Adv. Funct. Mater., 2018, 28, 1–10 CrossRef.
- T. Patino, A. Porchetta, A. Jannasch, A. Lladó, T. Stumpp, E. Schäffer, F. Ricci and S. Sánchez, Nano Lett., 2019, 19, 3440–3447 CrossRef CAS PubMed.
- M. De Corato, X. Arqué, T. Patinõ, M. Arroyo, S. Sánchez and I. Pagonabarraga, Phys. Rev. Lett., 2020, 124, 1–6 CrossRef PubMed.
- X. Arqué, A. Romero-Rivera, F. Feixas, T. Patiño, S. Osuna and S. Sánchez, Nat. Commun., 2019, 10, 2826 CrossRef PubMed.
- X. Arqué, X. Andrés, R. Mestre, B. Ciraulo, J. Ortega Arroyo, R. Quidant, T. Patiño and S. Sánchez, Research, 2020, 2020, 1–14 CrossRef PubMed.
- J. Simmchen, A. Baeza, D. Ruiz-Molina and M. Vallet-Regí, Nanoscale, 2014, 6, 8907–8913 RSC.
- X. Ma, A. C. Hortelao, A. Miguel-López and S. Sánchez, J. Am. Chem. Soc., 2016, 138, 13782–13785 CrossRef CAS PubMed.
- M. Valles, S. Pujals, L. Albertazzi and S. Sánchez, ACS Nano, 2019, 19, 6019–6026 Search PubMed.
- T. I. Janjua, Y. Cao, C. Yu and A. Popat, Nat. Rev. Mater., 2021, 6, 1072–1074 CrossRef CAS PubMed.
- T. Patiño, N. Feiner-Gracia, X. Arqué, A. Miguel-López, A. Jannasch, T. Stumpp, E. Schäffer, L. Albertazzi and S. Sánchez, J. Am. Chem. Soc., 2018, 140, 7896–7903 CrossRef PubMed.
- S. Wang, D. Xu, X. Liu, Y. Wang, H. Ye and X. Ma, ChemNanoMat, 2022, e202100447 CAS.
- P. Schattling, B. Thingholm and B. Städler, Chem. Mater., 2015, 27, 7412–7418 CrossRef CAS.
- X. Ma, X. Wang, K. Hahn and S. Sánchez, ACS Nano, 2016, 10, 3597–3605 CrossRef CAS PubMed.
- D. Xu, C. Zhou, C. Zhan, Y. Wang, Y. You, X. Pan, J. Jiao, R. Zhang, Z. Dong, W. Wang and X. Ma, Adv. Funct. Mater., 2019, 29, 1–11 Search PubMed.
- H. Shen, X. Zheng, Z. Zhou, W. He, M. Li, P. Su, J. Song and Y. Yang, J. Mater. Chem. B, 2020, 8, 8467–8475 RSC.
- M. Liu, L. Chen, Z. Zhao, M. Liu, T. Zhao, Y. Ma, Q. Zhou, Y. S. Ibrahim, A. A. Elzatahry, X. Li and D. Zhao, J. Am. Chem. Soc., 2022, 144, 3892–3901 CrossRef CAS PubMed.
- A. C. Hortelao, C. Simó, M. Guix, S. Guallar-Garrido, E. Julián, D. Vilela, L. Rejc, P. Ramos-Cabrer, U. Cossío, V. Gómez-Vallejo, T. Patiño, J. Llop and S. Sánchez, Sci. Robot., 2021, 6, 1–13 Search PubMed.
- A. Llopis-Lorente, A. García-Fernández, E. Lucena-Sánchez, P. Díez, F. Sancenón, R. Villalonga, D. A. Wilson and R. Martínez-Máñez, Chem. Commun., 2019, 55, 13164–13167 RSC.
- M. Yan, L. Xie, B. Qiu, S. Zhou, T. Liu, J. Zeng, Q. Liang, J. Tang, K. Liang, D. Zhao and B. Kong, ACS Nano, 2021, 15, 11451–11460 CrossRef CAS PubMed.
- Z. Chen, T. Xia, Z. Zhang, S. Xie, T. Wang and X. Li, Chem. Eng. J., 2019, 375, 122109 CrossRef CAS.
- Y. Feng, Y. Yuan, J. Wan, C. Yang, X. Hao, Z. Gao, M. Luo and J. Guan, Appl. Phys. Rev., 2021, 8, 011406 CAS.
- D. Fu, Y. Ye, C. Gao, D. Xie and F. Peng, ChemNanoMat, 2022, 1–7, DOI:10.1002/cnma.202200152.
- B. V. V. S. P. Kumar, A. J. Patil and S. Mann, Nat. Chem., 2018, 10, 1154–1163 CrossRef CAS PubMed.
- P. S. Schattling, M. A. Ramos-Docampo, V. Salgueiriño and B. Städler, ACS Nano, 2017, 11, 3973–3983 CrossRef CAS PubMed.
- Y. Ye, F. Tong, S. Wang, J. Jiang, J. Gao, L. Liu, K. Liu, F. Wang, Z. Wang, J. Ou, B. Chen, D. A. Wilson, Y. Tu and F. Peng, Nano Lett., 2021, 21, 8086–8094 CrossRef CAS PubMed.
- N. Mano and A. Heller, J. Am. Chem. Soc., 2005, 127, 11574–11575 CrossRef CAS PubMed.
- D. Pantarotto, W. R. Browne and B. L. Feringa, Chem. Commun., 2008, 1533–1535 RSC.
- C. Gao, C. Zhou, Z. Lin, M. Yang and Q. He, ACS Nano, 2019, 13, 12758–12766 CrossRef CAS PubMed.
- M. Mathesh, E. Bhattarai and W. Yang, Angew. Chem., 2022, 61, e202113801 CAS.
- Y. Xing, X. Du, T. Xu and X. Zhang, Soft Matter, 2020, 16, 9553–9558 RSC.
- J. Ren, P. Hu, E. Ma, X. Zhou, W. Wang, S. Zheng and H. Wang, Appl. Mater. Today, 2022, 27, 101445 CrossRef.
- J. Wu, S. Ma, M. Li, X. Hu, N. Jiao, S. Tung and L. Liu, ACS Appl. Mater. Interfaces, 2021, 13, 31514–31526 CrossRef CAS PubMed.
- S. Gao, J. Hou, J. Zeng, J. J. Richardson, Z. Gu, X. Gao, D. Li, M. Gao, D. W. Wang, P. Chen, V. Chen, K. Liang, D. Zhao and B. Kong, Adv. Funct. Mater., 2019, 29, 1–10 Search PubMed.
- Y. You, D. Xu, X. Pan and X. Ma, Appl. Mater. Today, 2019, 16, 508–517 CrossRef.
- Z. Guo, T. Wang, A. Rawal, J. Hou, Z. Cao, H. Zhang, J. Xu, Z. Gu, V. Chen and K. Liang, Mater. Today, 2019, 28, 10–16 CrossRef CAS.
- Y. Yang, X. Arqué, T. Patiño, V. Guillerm, P. R. Blersch, J. Pérez-Carvajal, I. Imaz, D. Maspoch and S. Sánchez, J. Am. Chem. Soc., 2020, 142, 20962–20967 CrossRef CAS PubMed.
- A. C. Apolinário, L. Hauschke, J. R. Nunes and L. B. Lopes, Prog. Lipid Res., 2021, 82, 101096 CrossRef PubMed.
- A. C. Hortelão, S. García-Jimeno, M. Cano-Sarabia, T. Patiño, D. Maspoch and S. Sanchez, Adv. Funct. Mater., 2020, 30, 2002767 CrossRef.
- S. Ghosh, F. Mohajerani, S. Son, D. Velegol, P. J. Butler and A. Sen, Nano Lett., 2019, 19, 6019–6026 CrossRef CAS PubMed.
- A. Somasundar, S. Ghosh, F. Mohajerani, L. N. Massenburg, T. Yang, P. S. Cremer, D. Velegol and A. Sen, Nat. Nanotechnol., 2019, 14, 1129–1134 CrossRef CAS PubMed.
- S. Tang, F. Zhang, H. Gong, F. Wei, J. Zhuang, E. Karshalev, B. E. F. De vila, C. Huang, Z. Zhou, Z. Li, L. Yin, H. Dong, R. H. Fang, X. Zhang, L. Zhang and J. Wang, Sci. Robot., 2020, 5, 41–43 Search PubMed.
- J. Zheng, R. Qi, C. Dai, G. Li and M. Sang, ACS Nano, 2022, 16, 2330–2344 CrossRef CAS PubMed.
- D. A. Gregory, Y. Zhang, P. J. Smith, X. Zhao and S. J. Ebbens, Small, 2016, 12, 4048–4055 CrossRef CAS PubMed.
- Y. Zhang, D. A. Gregory, Y. Zhang, P. J. Smith, S. J. Ebbens and X. Zhao, Small, 2019, 15, 1–9 Search PubMed.
- P. Kumar, Y. Zhang, S. J. Ebbens and X. Zhao, J. Colloid Interface Sci., 2022, 623, 96–108 CrossRef CAS PubMed.
- M. Hosseini, F. Babayekhorasani, Z. Guo, K. Liang, V. Chen and P. T. Spicer, arXiv, 2022, 1–29.
- S. Sattayasamitsathit, K. Kaufmann, M. Galarnyk, R. Vazquez-Duhalt and J. Wang, RSC Adv., 2014, 4, 27565–27570 RSC.
- Y. Gu, S. Sattayasamitsathit, K. Kaufmann, R. Vazquez-Duhalt, W. Gao, C. Wang and J. Wang, Chem. Commun., 2013, 49, 7307–7309 RSC.
- R. Golestanian, T. B. Liverpool and A. Ajdari, Phys. Rev. Lett., 2005, 94, 1–4 CrossRef PubMed.
- Y. Xu, L. Ji, S. Izumi and S. Nakata, Chem.–Asian J., 2021, 16, 1762–1766 CrossRef CAS PubMed.
- O. Barbosa, C. Ortiz, Á. Berenguer-Murcia, R. Torres, R. C. Rodrigues and R. Fernandez-Lafuente, RSC Adv., 2014, 4, 1583–1600 RSC.
- X. Ma, X. Wang, K. Hahn and S. Sánchez, ACS Nano, 2016, 10, 3597–3605 CrossRef CAS PubMed.
- X. Lyu, X. Liu, C. Zhou, S. Duan, P. Xu, J. Dai, X. Chen, Y. Peng, D. Cui, J. Tang, X. Ma and W. Wang, J. Am. Chem. Soc., 2021, 143, 12154–12164 CrossRef CAS PubMed.
- A. Brown and W. Poon, Soft Matter, 2014, 10, 4016–4027 RSC.
- S. Ebbens, D. A. Gregory, G. Dunderdale, J. R. Howse, Y. Ibrahim, T. B. Liverpool and R. Golestanian, Epl, 2014, 106, 58003 CrossRef.
- Y. Ibrahim, R. Golestanian and T. B. Liverpool, J. Fluid Mech., 2017, 828, 318–352 CrossRef CAS.
- E. S. Asmolov, T. V. Nizkaya and O. I. Vinogradova, arXiv, 2022, 1–8.
- J. Shi, Sci. Robot., 2020, 5, 1–3 Search PubMed.
- A. Houde, A. Kademi and D. Leblanc, Appl. Biochem. Biotechnol., 2004, 118, 155–170 CrossRef CAS PubMed.
- P. Chandra, Enespa, R. Singh and P. K. Arora, Microbial lipases and their industrial applications: A comprehensive review, BioMed Central, 2020, vol. 19 Search PubMed.
- S. Meng, Y. Zhang, Y. Liu, Z. Zhang, K. Ma, X. Chen, Q. Gao, X. Ma, W. Wang and H. Feng, JCIS Open, 2022, 5, 100046 CrossRef.
- N. Hoshyar, S. Gray, H. Han and G. Bao, Nanomedicine, 2016, 11, 673–692 CrossRef CAS PubMed.
- T. Patino, X. Arqué, R. Mestre, L. Palacios and S. Sánchez, Acc. Chem. Res., 2018, 51, 2662–2671 CrossRef CAS PubMed.
- X. Ma, S. Jang, M. N. Popescu, W. E. Uspal, A. Miguel-López, K. Hahn, D. P. Kim and S. Sánchez, ACS Nano, 2016, 10, 8751–8759 CrossRef CAS PubMed.
- Y. Lin, X. Geng, Q. Chi, C. Wang and Z. Wang, Micromachines, 2019, 10, 415 CrossRef PubMed.
- G. Gallino, F. Gallaire, E. Lauga and S. Michelin, Adv. Funct. Mater., 2018, 28, 1–10 CrossRef.
- U. Choudhury, L. Soler, J. G. Gibbs, S. Sanchez and P. Fischer, Chem. Commun., 2015, 51, 8660–8663 RSC.
- J. Simmchen, V. Magdanz, S. Sanchez, S. Chokmaviroj, D. Ruiz-Molina, A. Baeza and O. G. Schmidt, RSC Adv., 2014, 4, 20334–20340 RSC.
- H. Wang, G. Zhao and M. Pumera, J. Phys. Chem. C, 2014, 118, 5268–5274 CrossRef CAS.
- D. A. Wilson, B. De Nijs, A. Van Blaaderen, R. J. M. Nolte and J. C. M. Van Hest, Nanoscale, 2013, 5, 1315–1318 RSC.
| This journal is © The Royal Society of Chemistry 2022 |
