Open Access Article
Open Access ArticleMetal-mediated DNA base pairing of easily prepared 2-oxo-imidazole-4-carboxylate nucleotides†
Lingyun
Hu
 ,
Yusuke
Takezawa
,
Yusuke
Takezawa
 * and
Mitsuhiko
Shionoya
* and
Mitsuhiko
Shionoya
 *
*
Department of Chemistry, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan. E-mail: takezawa@chem.s.u-tokyo.ac.jp; shionoya@chem.s.u-tokyo.ac.jp
First published on 23rd March 2022
Abstract
Metal-mediated DNA base pairs, which consist of two ligand-type artificial nucleobases and a bridging metal ion, have attracted increasing attention in recent years as a different base pairing mode from natural base pairing. Metal-mediated base pairing has been extensively studied, not only for metal-dependent thermal stabilisation of duplexes, but also for metal assembly by DNA templates and construction of functional DNAs that can be controlled by metals. Here, we report the metal-mediated base paring properties of a novel 2-oxo-imidazole-4-carboxylate (ImOC) nucleobase and a previously reported 2-oxo-imidazole-4-carboxamide (ImOA) nucleobase, both of which can be easily derived from a commercially available uridine analogue. The ImOC nucleobases were found to form stable ImOC–CuII–ImOC and ImOC–HgII–ImOC base pairs in the presence of the corresponding metal ions, leading to an increase in the duplex melting temperature by +20 °C and +11 °C, respectively. The ImOC bases did not react with other divalent metal ions and showed superior metal selectivity compared to similar nucleobase design reported so far. The ImOC–CuII–ImOC base pair was much more stable than mismatch pairs with other natural nucleobases, confirming the base pair specificity in the presence of CuII. Furthermore, we demonstrated the quantitative assembly of three CuII ions inside a DNA duplex with three consecutive ImOC–ImOC pairs, showing great potential of DNA-template based CuII nanoarray construction. The study of easily-prepared ImOC base pairs will provide a new design strategy for metal-responsive DNA materials.
Introduction
The high versatility and programming capabilities of DNA structures have served as a bridge from fundamental concepts to practicality in the field of DNA nanotechnology, which is a successful example of molecule-based bottom-up self-assembly processes.1–6 Much attention has been focused on the construction of modified DNA monomers to further enrich the diversity of DNA structure and function.7–9 In particular, a great deal of effort has been devoted to the development of artificial nucleobase pairs, which are increasingly being recognised as highly effective.10,11 Among them, metal-mediated base pairing, which consists of two ligand-type artificial nucleobases and a bridging metal ion, has attracted increasing interest due to its unique coordination binding mode.12–17 The newly introduced metal-mediated base pairs impart unique properties to the modified DNA, depending on the inherent nature of the metal. In addition to metal-dependent thermal stabilisation of duplexes, DNA-templated metal assembly,18–23 modulation of charge transfer,24–27 molecular sensing28–30 and metal-dependent control of DNAzymes and aptamers31–37 have been reported. Therefore, finding appropriate ligand-type nucleobases that can specifically and strongly bind to a certain metal ion and form metal-mediated base pairs is important to achieve these functions.A variety of ligand-type artificial nucleobases have been developed so far by modifying simple ligand scaffolds such as pyridine,38–40 maltol41,42 and pyrimidine.43–48 Among them, the imidazole ligand is one of the smallest scaffolds. It has been reported that most of the modified imidazole nucleobases form AgI-mediated base pairs by N–AgI–N coordination.49–51 This metal binding property can be altered by introducing an additional coordination site.52,53 For example, imidazole-4-carboxylate (ImC) nucleobases form a base pair via CuII- and AgI-mediated coordination, and the negatively charged carboxylates neutralise the positive charge of the bridging metal ion.52 The ImC–CuII–ImC pair is one of the most stabilising artificial base pairs developed so far,35 but ImC retains its binding affinity for other metals such as NiII and CoII. Furthermore, we have successfully applied ImC–CuII–ImC pairing to the metal-dependent functional regulation of DNAzymes.35
With these examples as a starting point, we sought to expand the structural diversity of imidazole-like scaffolds that can form metal-mediated base pairs. Based on imidazole-4-carboxylate (ImC), a novel ligand-type nucleobase, 2-oxo-imidazole-4-carboxylate (ImOC), was designed by modifying the C-2 position (Fig. 1). Unlike conventional examples, ImOC nucleosides can be easily derived from commercially available nucleosides,54,55 reducing the need for laborious synthesis. The ImOC nucleobase was predicted to have the following characteristics: (1) In contrast to the ImC nucleobase, the N3 atom of ImOC is protonated under neutral conditions.56 Since metal coordination requires deprotonation or amide–iminol tautomerisation, ImOC was expected to show a metal binding affinity different from the ImC nucleobase. (2) As the ImC nucleoside has a 2-carbonyl group, it preferentially adopts an anti-conformation suitable for metal-mediated base pairing, as reported for the 2-oxo-imidazole-4-carboxamide (ImOA) nucleoside.55 (3) The 2-carbonyl group may also function as a hydrogen bond acceptor for certain amino acids in DNA polymerases.57,58 Therefore, such a structure may be advantageous for future polymerase incorporation studies.
In this study, DNA duplexes containing 2-oxo-imidazole-4-carboxylate (ImOC) and 2-oxo-imidazole-4-carboxamide (ImOA) were synthesised and their ability to form metal-mediated base pairs was investigated. Heterologous base pairing of ImOC with other natural or unnatural nucleobases was also analysed in the presence and absence of certain metal ions. Furthermore, we investigated the construction of consecutive metal-mediated base pairs and explored the possibility of one-dimensional metal assembly inside DNA duplexes.
Results and discussion
Synthesis of DNA strands containing ImOC or ImOA nucleosides
The synthetic route for DNA strands containing ImOC or ImOA nucleosides is shown in Scheme 1. The 2-oxo-imidazole-4-carboxlate nucleoside (2) was synthesised by ring contraction of commercially available 5-bromo-2′-deoxyuridine (1).54 Its protected derivative 3 was prepared by the reported procedure.55 Phosphoramidite 4 was then synthesised without the N3 protection because the 3′-NH group was found to be intact during DNA synthesis just like thymidine. The resulting phosphoramidite was immediately used for solid-phase DNA synthesis.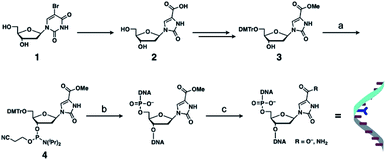 | ||
| Scheme 1 Synthesis of DNA strands containing ImOC or ImOA nucleotides. The DMTr-modified nucleoside 3 was synthesised by the reported procedure.55 (a) 2-Cyanoethyl N,N-diisopropylchlorophosphoramidite, DIPEA, CH2Cl2, rt, 0.5 h (78%); (b) DNA synthesiser; (c) 0.3 M NaOH aq., 37 °C (for ImOC) or 25% ammonia aq., 55 °C (for ImOA). DMTr = 4,4′-dimethoxytrityl. | ||
Since the 4′-substituent was protected as a methyl ester, the nucleobase moiety can be converted to either a carboxylate (ImOC) or a carboxamide (ImOA) depending on the deprotection conditions. To confirm this, we synthesised a trimer DNA strand with one unnatural nucleotide (5′-TXT-3′) and deprotected it with sodium hydroxide or ammonia solution. The deprotected products were analysed by HPLC (Fig. S1†) and ESI mass spectrometry (Fig. S2†). When the trinucleotide was treated with 0.3 M NaOH aq. at 37 °C, an almost complete conversion to the carboxylate ImOC was observed. When incubated in 25% ammonia solution, the artificial nucleobase was converted to the desired amide ImOA in over 90% yield. The strands containing ImOC and ImOA were easily isolated by reverse-phase HPLC.
To investigate the metal-mediated base-pairing properties of ImOC and ImOA nucleotides, 15-mer DNA strands containing one or three ImOC/ImOA nucleotides in the central position were synthesised (Table 1). All the strands were deprotected with NaOH or ammonia solution to produce ImOC or ImOA nucleobases, which were purified by HPLC (Fig. S3†). The DNA oligomers with the desired artificial nucleotides were characterised by ESI mass spectrometry (see ESI†).
| DNA | Sequences (5′ to 3′)a |
|---|---|
| a ImC: imidazole-4-carboxylate. | |
| 1 | CAC ATT AImOCT GTT GTA |
| 2 | TAC AAC AImOCT AAT GTG |
| 2N (N = A, T, G, C) | TAC AAC ANT AAT GTG |
| 3 | CAC ATT AImOAT GTT GTA |
| 4 | TAC AAC AImOAT AAT GTG |
| 5 | CAC ATT ImOCImOCImOC GTT GTA |
| 6 | TAC AAC ImOCImOCImOC AAT GTG |
| 7 | CAC ATT AImCT GTT GTA |
| 8 | TAC AAC AImCT AAT GTG |
CuII-mediated base pairing of ImOC nucleobases
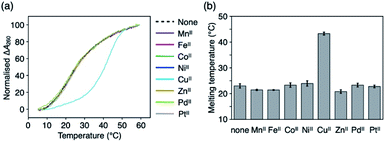
Metal-mediated base pairing was first examined in a duplex containing a single artificial base pair, ImOC–ImOC or ImOA–ImOA. Thermal melting analysis of duplexes 1·2 and 3·4 was performed in the absence and presence of various metal ions (1.0 equiv.), including first-row transition metal ions and square-planar PdII and PtII ions (Fig. 2, S4, and Table S1†). In the absence of metal ions, the melting temperatures (Tm) of the ImOC-modified duplex 1·2 and ImOA-modified duplex 3·4 were 23.0 °C and 34.5 °C, respectively. Both duplexes were significantly less stable than a fully matched duplex containing an A–T pair in the middle (Tm = 44.2 °C),33 indicating that both ImOC–ImOC and ImOA–ImOA behave as mismatch pairs. The Tm value of duplex 1·2 is about 12 °C lower than that of duplex 3·4. This may be due to the coulombic repulsion between the negatively charged carboxylates of the ImOC–ImOC base pair. A similar phenomenon was also observed in the DNA duplex containing a pair of imidazole-4-carboxylate nucleobases (ImC), which showed a Tm value (22.5 °C) similar to that of the ImOC-containing duplex 1·2.35 Among the metal ions tested, only the CuII ion showed significant stabilisation of duplex 1·2 (Tm = 43.3 °C, ΔTm = +20.3 °C). On the other hand, none of these metal ions caused a significant change in the melting profile of duplex 3·4 (−1 °C ≤ ΔTm ≤ +1 °C). The CuII-dependent stabilisation of duplex 1·2 suggests the CuII-mediated formation of the ImOC–CuII–ImOC base pair, which crosslinks the duplex via metal coordination bonds.Fig. 3a shows the melting curves of duplex 1·2 in the presence of various amounts of CuII ions. In the presence of 0.5 equiv. of CuII ions, a two-step transition was observed, indicating the presence of both metal-free and CuII-bound DNA duplexes. The addition of more than one equivalent of CuII did not cause any obvious change in the melting behaviour. In addition, CuII-dependent stabilisation was not observed in the fully matched duplex or in the duplex containing a T–T mismatch.33,41 These results proved that the metal-mediated base pair was formed by the binding of a single CuII ion, and also indicated that there is a high binding affinity between ImOC and CuII. The stoichiometry of the ImOC–CuII–ImOC base pair was further confirmed by ESI-TOF mass spectrometry (found: 1838.41 (z = 5); calcd. for [1·2 + CuII − 7H]5−: 1838.47; Fig. 3b and S5†).
 | ||
| Fig. 3 (a) Melting curves of the DNA duplex 1·2 containing an ImOC–ImOC base pair in the presence of different concentrations of CuII ions. [duplex] = 2.0 μM, [CuII]/[duplex] = 0, 0.5, 1 (solid lines), 2, and 3 (dashed lines) in 10 mM HEPES buffer (pH 7.0), 100 mM NaCl, 0.2 °C min−1 (b) ESI mass spectrum of the duplex 1·2 with 1 equiv. of CuII (negative mode). Small signals were attributed to the sodium and potassium adducts. See also Fig. S5.† (c) CD spectra of the duplex 1·2 in the absence (dotted lines) and presence (solid lines) of 1 equiv. of CuII. [duplex] = 2.0 μM in 10 mM HEPES buffer (pH 7.0), 100 mM NaCl, l = 0.5 cm, 4 °C. | ||
It is suggested that the ImOC–CuII–ImOC pair is formed by the coordination of both the N3 atoms and the carboxylate groups in a square planar geometry. CuII-mediated base pairing with similar coordination structures has been reported with imidazole-4-carboxylate nucleobases (ImC)52 and with 6-carboxypurines.59 The carboxylate group of the ImOC nucleobases not only coordinates with the CuII ion, but also neutralises the positive charge of the metal, thus maintaining a neutral environment within the DNA duplex. However, in the case of the ImOA nucleobase, the carboxamide group is a weak neutral ligand and cannot neutralise the resulting complex. This is the main reason why metal-mediated base pairing with ImOA is unfavourable.
The circular dichroism (CD) spectra of duplex 1·2 were also measured in the absence and presence of CuII ions (Fig. 3c). The spectra showed Cotton effects characteristic of right-handed B-DNA, indicating that the introduction of the ImOC–CuII–ImOC base pair did not alter the typical duplex structure. Due to the small size of the ImOC base, the π–π stacking interaction with the neighbouring base pairs may be reduced, making the duplex less stable. However, this destabilisation effect can be compensated for by the coordination bonds of ImOC–CuII–ImOC. Overall, the stability of the duplex containing an ImOC–CuII–ImOC pair (1·2·CuII, Tm = 43.3 °C) was completely comparable to that of the natural DNA duplex (44.2 °C).
It should be emphasised that ImOC showed better metal selectivity for CuII compared to the structurally relevant imidazole-4-carboxylate (ImC) nucleobase.35,52 A DNA duplex with an ImC–ImC base pair was stabilised by the addition of not only CuII (ΔTm = +35.2 °C) but also many transition metal ions such as NiII and CoII (+14.0 °C and + 11.3 °C, respectively).35 In contrast to ImC, the N3 atom of ImOC needs to be deprotonated for metal complexation. Thus, ImOC showed excellent metal selectivity in that it forms metal-mediated base pairs with only the most suitable CuII.
HgII-mediated base pairing of ImOC/ImOA nucleobases
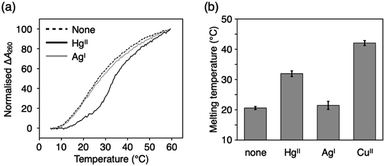
HgII and AgI ions are often involved in metal-mediated base pairing in two-coordinate geometry, such as the classical T–HgII–T60,61 and C–AgI–C62 base pairs. Since the 2-oxo-imidazole ring is derived from uridine/thymine nucleobases, ImOC and ImOA were expected to form a metal-mediated base pair similar to T–HgII–T. To investigate this, we conducted melting analysis of duplexes 1·2 and 3·4 in the presence of HgII or AgI ions (Fig. 4, S6 and Table S2†). In these measurements, sodium chloride in the buffer solution was replaced by sodium nitrate to prevent precipitation of metal chlorides.The addition of one equivalent of HgII ions markedly increased the stability of both ImOC- and ImOA-containing duplexes (ΔTm = +11.4 °C and +6.2 °C for duplexes 1·2 and 3·4, respectively). The addition of excess HgII did not stabilise the duplexes anymore (Fig. S7†). This suggests that the formation of ImOC–HgII–ImOC and ImOA–HgII–ImOA base pairs is mediated by a single HgII ion. The HgII-mediated base pairing was further confirmed by ESI-MS measurements of the ImOC-containing duplex (found: 2342.26 (z = 4); calcd for [1·2 + HgII + KI − 7H]4−: 2342.11; Fig. S8†) and the ImOA-containing one (found: 2332.08 (z = 4); calcd. for [3·4 + HgII − 6H]4−: 2332.13; Fig. S9†). ImOC and ImOA are thought to form a linear complex with HgIIvia coordination of the deprotonated N3 atom, similar to the T–HgII–T base pair.63,64 It is noteworthy that in the case of ImOC, the degree of HgII-dependent duplex stabilisation is comparable to that observed for the T–HgII–T base pair (ΔTm = +10.9 °C) under the same conditions.33
Notably, the addition of AgI did not stabilise the duplexes containing ImOC and ImOA. On the other hand, previous studies have shown that the duplex with an imidazole-4-carboxylate base pair (ImC–ImC) is stabilised by both HgII and AgI ions, with a slightly higher preference for AgI over HgII.35,52 These results indicate that ImOC and ImOA nucleobases improve the metal selectivity, which can be attributed to the protonated N3 atoms. Therefore, ImOC and ImOA bases are more suitable for the construction of complex molecular systems using multiple types of metal ions.
Heterologous base pairing of ImOC with other natural and unnatural nucleobases
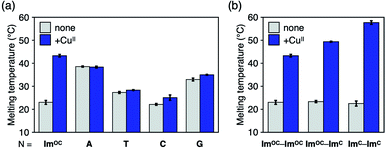
Apart from the homologous base pairing, base pairing between ImOC and other nucleobases was also examined to clarify the specificity. First, mismatch base pairing with four natural nucleobases was evaluated by duplex melting analysis (Fig. 5a and Table S3†). The Tm values of the duplexes with an ImOC–N pair (N = A, T, C and G) were different, and the pyrimidine mismatches (ImOC–T and ImOC–C) were found to be less stable than the purine mismatches (ImOC–A and ImOC–G). This trend can be well explained by the fact that the size of the ImOC–purine pairs is comparable to that of the natural base pairs. Similar results were also obtained in previous studies examining ImOA–N base pairing.55 The addition of one equivalent of CuII ions did not cause an obvious Tm change, indicating that ImOC does not form CuII-mediated base pairs with natural nucleobases. It is important to note that the Tm value of the duplex containing ImOC–CuII–ImOC is higher than all other duplexes with an ImOC–N pair. This result shows the specificity of the ImOC–CuII–ImOC base pairing over pairing with natural nucleobases in the presence of CuII ions.Next, we investigated the possibility of heterologous base pairing between ImC and ImOC in relation to the previously studied ImC–CuII–ImC base pair (Fig. 5b, S10 and Table S4†). As expected, the duplex melting analysis showed that the ImOC–CuII–ImC heterologous base pair was formed in the presence of CuII ions (ΔTm = +26.1 °C). The ΔTm value was intermediate between those of the duplexes containing the ImOC–CuII–ImOC and ImC–CuII–ImC pairs. This result reflects the CuII binding affinity of ImOC and ImC described above.
DNA-templated metal assembly using ImOC nucleobases
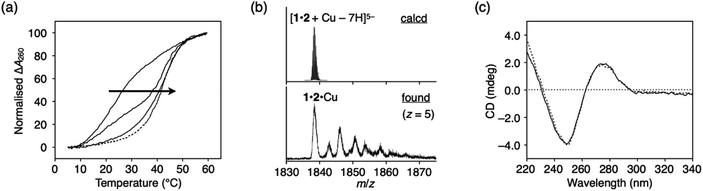
The construction of one-dimensional metal arrays using DNA as a template is one of the promising applications of metal-mediated base pairing.18–23 To investigate the possibility of DNA-templated metal assembly using ImOC nucleobases, a 15-mer duplex with three consecutive ImOC–ImOC base pairs in the centre (duplex 5·6) was prepared.Duplex melting experiments were conducted in the presence of different amounts of CuII ions (Fig. 6a). In the absence of CuII ions, duplex 5·6 was highly unstable, and the Tm value could not be determined. As the amount of CuII ions increased, the melting curves gradually shifted. When 3 equiv. of CuII ions were added, the Tm value was almost at its maximum (Tm = 51.7 °C). The melting curves were hardly changed by the addition of excess CuII. These results show that three base pairs of ImOC–CuII–ImOC were quantitatively formed in duplex 5·6. The Tm value of duplex 5·6 with 3 equiv. of CuII is about 10 °C higher than that of duplex 1·2 with equimolar CuII. This indicates that the incorporation of multiple ImOC–CuII–ImOC base pairs enhances the duplex stability more efficiently.
 | ||
| Fig. 6 (a) Melting curves of DNA duplex 5·6 containing three ImOC–ImOC base pairs in the presence of different concentrations of CuII ions. [duplex] = 2.0 μM, [CuII]/[duplex] = 0, 1, 2, 3 (solid lines), and 4, 5, and 6 (dashed lines). (b) CD spectra of duplex 5·6 in the presence of different concentrations of CuII ions. [CuII]/[duplex] = 0, 1, 2, 3 (solid lines) and 4, 5, and 6 (dotted lines). The changes in intensity at 250 and 275 nm are plotted in the inset. (c) ESI mass spectrum of duplex 5·6 with 3 equiv. of CuII (negative mode). See also Fig. S11.† The conditions are the same as in Fig. 3. | ||
The stoichiometric formation of the ImOC–CuII–ImOC base pairs was further examined by circular dichroism (CD) measurements at 4 °C (Fig. 6b). In contrast to the CD spectra of duplexes 1·2 and 3·4, the CD spectrum of duplex 5·6 showed a dramatic change upon CuII addition. As the amount of CuII ions increased, the CD intensity at 275 nm gradually decreased. The spectra changed linearly in the range of [CuII]/[5·6] = 0 to 3 and did not change with the addition of more than 3 equiv. of CuII. The result of the CD measurements were in excellent agreement with the behaviours of the melting curves, indicating that three CuII ions were bound within duplex 5·6. Such stoichiometry was also confirmed by ESI-MS measurements, where a duplex containing three CuII ions was mainly observed (found: 1860.92 (z = 5); calcd. for [5·6 + 3CuII − 11H]5−: 1860.84; Fig. 6c and S11†). These results suggest that the three CuII ions were quantitatively assembled inside the duplex due to the formation of the ImOC–CuII–ImOC base pairs. Thus, it was shown that a certain number of CuII ions can be aligned according to the number of ImOC bases in the template DNA duplex.
Conclusions
In this study, we have shown that a novel 2-oxo-imidazole-4-carboxylate (ImOC) nucleobase undergoes metal-mediated base pairing in DNA duplexes. The previously reported synthetic route for the corresponding phosphoramidite55 was shortened by removing one unnecessary protection step. The protected artificial nucleobase(s) in DNA oligomers can be easily converted to two types of nucleobases, ImOC and the previously reported carboxamide (ImOA), depending on the deprotection conditions. Other than the natural T and C bases, ImOC and ImOA are among the most easily prepared nucleobases that can form metal-mediated base pairs.The metal-mediated base pairing of both nucleobases was studied by duplex melting analysis, CD spectrometry and mass spectrometry. It was found that the ImOC homologous base pair forms stable metal-mediated base pairs with both CuII and HgII (i.e., ImOC–CuII–ImOC and ImOC–HgII–ImOC), and ΔTm was +20.0 °C and +11.4 °C, respectively. In contrast, ImOA showed only a slight increase in Tm in the presence of HgII (ΔTm = +6.2 °C) due to the formation of the ImOA–HgII–ImOA pair. This difference in metal ion affinity is mainly due to the substituent at the 4-position, since the negative carboxylate group can form a stronger coordination bond with CuII. Probably due to the protonated N3 atom, ImOC was found to exhibit superior metal selectivity compared to the structurally related imidazole-4-carboxylate (ImC) nucleobase.35,52
Further investigation of the possibility of heterologous base pairing in the absence and presence of metal ions revealed that the ImOC–CuII–ImOC base pair was more stable than heterologous base pairing with natural nucleobases (i.e.ImOC–N), suggesting the specificity of metal-mediated homologous base pairing among the possible base pairing patterns. When paired with the previously reported ImC, the resulting ImOC–CuII–ImC heterologous base pair showed intermediate stability (ΔTm = +26.1 °C) between ImOC–CuII–ImOC (+20.3 °C) and ImC–CuII–ImC (+35.2 °C). This result indicates that the metal-mediated base pair stabilisation effect can be rationally tuned by replacing the ligand-type nucleobases.
The stable ImOC–CuII–ImOC base pairing is expected to have applications such as the construction of DNA-templated metal arrays. In the preliminary experiments using a duplex containing three ImOC–ImOC base pairs, we succeeded in quantitatively assembling three CuII ions. Since the ImOC nucleoside can be prepared from a commercially available nucleoside by a short-step synthesis, ImOC oligomers are considered suitable for constructing one dimensional metal wires. In addition, the significant increase in duplex stability and high metal selectivity brought by the formation of the ImOC–CuII–ImOC base pair makes it a promising candidate for constructing metal-responsive DNA nanodevices and nanomachines. The newly introduced 2-oxo functional group is expected to play an important role in recognition by DNA polymerases. Applications such as enzymatic synthesis65,66 and PCR amplification67 are highly promising and will be probed in the future. Thus, metal-mediated base pairing with ImOC is believed to have high potential applications in DNA supramolecular chemistry and nanotechnology.
Data availability
All the data are shown in the ESI.†Author contributions
Y. T. and M. S. conceived and directed the study. L. H. performed the experiments and analysed the data with the aid of Y. T. All the authors prepared the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI grant numbers JP18H02081, JP21H02055, JP21H00384 (Molecular Engine) and JP21H05866 (Molecular Cybernetics) (to Y. T.), JP16H06509 (Coordination Asymmetry) and JP21H05022 (to M. S.) and JP21J11332 (Grant-in-Aid for JSPS Fellows; to L. H.). Y. T. also acknowledges financial support from the Iketani Science and Technology Foundation.Notes and references
- N. C. Seeman and H. F. Sleiman, Nat. Rev. Mater., 2017, 3, 17068–17091 CrossRef.
- M. Madsen and K. V. Gothelf, Chem. Rev., 2019, 119, 6384–6458 CrossRef CAS PubMed.
- P. Chidchob and H. F. Sleiman, Curr. Opin. Chem. Biol., 2018, 46, 63–70 CrossRef CAS PubMed.
- S. Nummelin, B. X. Shen, P. Piskunen, Q. Liu, M. A. Kostiainen and V. Linko, ACS Synth. Biol., 2020, 9, 1923–1940 CrossRef CAS PubMed.
- Y. G. Ke, C. Castro and J. H. Choi, Annu. Rev. Biomed. Eng., 2018, 20, 375–401 CrossRef CAS PubMed.
- E. Stulz and G. H. Clever, DNA in supramolecular chemistry and nanotechnology, Wiley, Chichester, 2015 Search PubMed.
- K. Nakatani and Y. Tor, Modified Nucleic Acids, Springer, 2016 Search PubMed.
- L. K. McKenzie, R. El-Khoury, J. D. Thorpe, M. J. Damha and M. Hollenstein, Chem. Soc. Rev., 2021, 50, 5126–5164 RSC.
- V. B. Pinheiro and P. Holliger, Trends Biotechnol., 2014, 32, 321–328 CrossRef CAS PubMed.
- A. W. Feldman and F. E. Romesberg, Acc. Chem. Res., 2018, 51, 394–403 CrossRef CAS PubMed.
- M. Kimoto and I. Hirao, Chem. Soc. Rev., 2020, 49, 7602–7626 RSC.
- Y. Takezawa and M. Shionoya, Acc. Chem. Res., 2012, 45, 2066–2076 CrossRef CAS PubMed.
- Y. Takezawa, J. Müller and M. Shionoya, Chem. Lett., 2017, 46, 622–633 CrossRef CAS.
- Y. Takezawa, M. Shionoya and J. Müller, in Comprehensive Supramolecular Chemistry II, ed. J. L. Atwood, Elsevier, Oxford, 2017, vol. 4, pp. 259–293 Search PubMed.
- S. Naskar, R. Guha and J. Müller, Angew. Chem., Int. Ed., 2020, 59, 1397–1406 CrossRef CAS PubMed.
- J. Müller, Coord. Chem. Rev., 2019, 393, 37–47 CrossRef.
- D. Ukale and T. Lönnberg, ChemBioChem, 2021, 22, 1733–1739 CrossRef CAS PubMed.
- K. Tanaka, A. Tengeiji, T. Kato, N. Toyama and M. Shionoya, Science, 2003, 299, 1212–1213 CrossRef CAS PubMed.
- K. Tanaka, G. H. Clever, Y. Takezawa, Y. Yamada, C. Kaul, M. Shionoya and T. Carell, Nat. Nanotechnol., 2006, 1, 190–194 CrossRef CAS PubMed.
- Y. Takezawa, W. Maeda, K. Tanaka and M. Shionoya, Angew. Chem., Int. Ed., 2009, 48, 1081–1084 CrossRef CAS PubMed.
- G. H. Clever and T. Carell, Angew. Chem., Int. Ed., 2007, 46, 250–253 CrossRef CAS PubMed.
- J. Kondo, Y. Tada, T. Dairaku, Y. Hattori, H. Saneyoshi, A. Ono and Y. Tanaka, Nat. Chem., 2017, 9, 956–960 CrossRef CAS PubMed.
- Y. Takezawa and M. Shionoya, in Biomimetics Bioinspired Materials, Mechanics, and Dynamics, Handbook of Biomimetics and Bioinspiration, ed. E. Jabbari, D.-H. Kim, L. P. Lee, A. Ghaemmaghami and A. Khademhosseini, World Scientific Publishing, Singapore, 2014, vol. 1, pp. 217–245 Search PubMed.
- S. Liu, G. H. Clever, Y. Takezawa, M. Kaneko, K. Tanaka, X. Guo and M. Shionoya, Angew. Chem., Int. Ed., 2011, 50, 8886–8890 CrossRef CAS PubMed.
- J. Joseph and G. B. Schuster, Org. Lett., 2007, 9, 1843–1846 CrossRef CAS PubMed.
- T. Ehrenschwender, W. Schmucker, C. Wellner, T. Augenstein, P. Carl, J. Harmer, F. Breher and H.-A. Wagenknecht, Chem.–Eur. J., 2013, 19, 12547–12552 CrossRef CAS PubMed.
- S. Hensel, K. Eckey, P. Scharf, N. Megger, U. Karst and J. Müller, Chem.–Eur. J., 2017, 23, 10244–10248 CrossRef CAS PubMed.
- A. Ono and H. Togashi, Angew. Chem., Int. Ed., 2004, 43, 4300–4302 CrossRef CAS PubMed.
- B. Jash and J. Müller, Eur. J. Inorg. Chem., 2017, 33, 3857–3861 CrossRef.
- S. Taherpour, O. Golubev and T. Lönnberg, Inorg. Chim. Acta, 2016, 452, 43–49 CrossRef CAS.
- J. Liu and Y. Lu, Angew. Chem., Int. Ed., 2007, 46, 7587–7590 CrossRef CAS PubMed.
- S. Shimron, J. Elbaz, A. Henning and I. Willner, Chem. Commun., 2010, 46, 3250–3252 RSC.
- Y. Takezawa, T. Nakama and M. Shionoya, J. Am. Chem. Soc., 2019, 141, 19342–19350 CrossRef CAS PubMed.
- T. Nakama, Y. Takezawa, D. Sasaki and M. Shionoya, J. Am. Chem. Soc., 2020, 142, 10153–10162 CrossRef CAS PubMed.
- Y. Takezawa, L. Hu, T. Nakama and M. Shionoya, Angew. Chem., Int. Ed., 2020, 59, 21488–21492 CrossRef CAS PubMed.
- M. H. Heddinga and J. Müller, Beilstein J. Org. Chem., 2020, 16, 2870–2879 CrossRef CAS PubMed.
- T. Nakama, Y. Takezawa and M. Shionoya, Chem. Commun., 2021, 57, 1392–1395 RSC.
- E. Meggers, P. L. Holland, W. B. Tolman, F. E. Romesberg and P. G. Schultz, J. Am. Chem. Soc., 2000, 122, 10714–10715 CrossRef CAS.
- N. Zimmermann, E. Meggers and P. G. Schultz, Bioorg. Chem., 2004, 32, 13–25 CrossRef CAS PubMed.
- K. Tanaka, Y. Yamada and M. Shionoya, J. Am. Chem. Soc., 2002, 124, 8802–8803 CrossRef CAS PubMed.
- K. Tanaka, A. Tengeiji, T. Kato, N. Toyama, M. Shiro and M. Shionoya, J. Am. Chem. Soc., 2002, 124, 12494–12498 CrossRef CAS PubMed.
- Y. Takezawa, K. Tanaka, M. Yori, S. Tashiro, M. Shiro and M. Shionoya, J. Org. Chem., 2008, 73, 6092–6098 CrossRef CAS PubMed.
- C. Switzer and D. Shin, Chem. Commun., 2005, 41, 1342–1344 RSC.
- Y. Takezawa, K. Nishiyama, T. Mashima, M. Katahira and M. Shionoya, Chem.–Eur. J., 2015, 21, 14713–14716 CrossRef CAS PubMed.
- K. Nishiyama, Y. Takezawa and M. Shionoya, Inorg. Chim. Acta, 2016, 452, 176–180 CrossRef CAS.
- K. Nishiyama, K. Mori, Y. Takezawa and M. Shionoya, Chem. Commun., 2021, 57, 2487–2490 RSC.
- Y. Takezawa, A. Suzuki, M. Nakaya, K. Nishiyama and M. Shionoya, J. Am. Chem. Soc., 2020, 142, 21640–21644 CrossRef CAS PubMed.
- D. Ukale, V. S. Shinde and T. Lönnberg, Chem.–Eur. J., 2016, 22, 7917–7923 CrossRef CAS PubMed.
- S. Johannsen, N. Megger, D. Böhme, R. K. O. Sigel and J. Müller, Nat. Chem., 2010, 2, 229–234 CrossRef CAS PubMed.
- K. Petrovec, B. J. Ravoo and J. Müller, Chem. Commun., 2012, 48, 11844–11846 RSC.
- S. Hensel, N. Megger, K. Schweizer and J. Müller, Beilstein J. Org. Chem., 2014, 10, 2139–2144 CrossRef PubMed.
- N. Sandmann, D. Defayay, A. Hepp and J. Müller, J. Inorg. Biochem., 2019, 191, 85–93 CrossRef CAS PubMed.
- K. Schweizer, J. Kösters and J. Müller, J. Biol. Inorg Chem., 2015, 20, 895–903 CrossRef CAS PubMed.
- B. A. Otter, E. A. Falco and J. J. Fox, J. Org. Chem., 1969, 34, 2636–2642 CrossRef CAS PubMed.
- C. Cadena-Amaro and S. Pochet, Tetrahedron, 2005, 61, 5081–5087 CrossRef CAS.
- It has been reported that the pKa2 of 1-methyl-2-oxo-imidazole-4-carboxylate is greater than 13 (ref. 54). Therefore, the N3 atom of the ImOC nucleobase is considered to be protonated under neutral conditions.
- T. E. Spratt, Biochemistry, 2001, 40, 2647–2652 CrossRef CAS PubMed.
- M. D. McCain, A. S. Meyer, S. S. Schultz, A. Glekas and T. E. Spratt, Biochemistry, 2005, 44, 5647–5659 CrossRef CAS PubMed.
- E.-K. Kim and C. Switzer, Org. Lett., 2014, 16, 4059–4061 CrossRef CAS PubMed.
- S. Katz, Biochim. Biophys. Acta, 1963, 68, 240–253 CrossRef CAS.
- Y. Miyake, H. Togashi, M. Tashiro, H. Yamaguchi, S. Oda, M. Kudo, Y. Tanaka, Y. Kondo, R. Sawa, T. Fujimoto, T. Machinami and A. Ono, J. Am. Chem. Soc., 2006, 128, 2172–2173 CrossRef CAS PubMed.
- A. Ono, S. Cao, H. Togashi, M. Tashiro, T. Fujimoto, T. Machinami, S. Oda, Y. Miyake, I. Okamoto and Y. Tanaka, Chem. Commun., 2008, 44, 4825–4827 RSC.
- Y. Tanaka, S. Oda, H. Yamaguchi, Y. Kondo, C. Kojima and A. Ono, J. Am. Chem. Soc., 2007, 129, 244–245 CrossRef CAS PubMed.
- J. Kondo, T. Yamada, C. Hirose, I. Okamoto, Y. Tanaka and A. Ono, Angew. Chem., Int. Ed., 2014, 53, 2385–2388 CrossRef CAS PubMed.
- M. Flamme, C. Figazzolo, G. Gasser and M. Hollenstein, Metallomics, 2021, 13, mfab016 CrossRef PubMed.
- T. Kobayashi, Y. Takezawa, A. Sakamoto and M. Shionoya, Chem. Commun., 2016, 52, 3762–3765 RSC.
- C. Kaul, M. Müller, M. Wagner, S. Schneider and T. Carell, Nat. Chem., 2011, 3, 794–800 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2sc00926a |
| This journal is © The Royal Society of Chemistry 2022 |