Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Metal binding and interdomain thermodynamics of mammalian metallothionein-3: enthalpically favoured Cu+ supplants entropically favoured Zn2+ to form Cu4+ clusters under physiological conditions†
Matthew R.
Mehlenbacher
a,
Rahma
Elsiesy
b,
Rabina
Lakha
b,
Rhiza Lyne E.
Villones
c,
Marina
Orman
b,
Christina L.
Vizcarra
*b,
Gabriele
Meloni
 *c,
Dean E.
Wilcox
*c,
Dean E.
Wilcox
 *a and
Rachel N.
Austin
*a and
Rachel N.
Austin
 *b
*b
aDepartment of Chemistry, Dartmouth College, Hanover, NH 03755, USA
bDepartment of Chemistry, Barnard College of Columbia University, New York, NY 10027, USA. E-mail: raustin@barnard.edu
cDepartment of Chemistry and Biochemistry, University of Texas at Dallas, Richardson, TX 75080, USA
First published on 4th April 2022
Abstract
Metallothioneins (MTs) are a ubiquitous class of small metal-binding proteins involved in metal homeostasis and detoxification. While known for their high affinity for d10 metal ions, there is a surprising dearth of thermodynamic data on metals binding to MTs. In this study, Zn2+ and Cu+ binding to mammalian metallothionein-3 (MT-3) were quantified at pH 7.4 by isothermal titration calorimetry (ITC). Zn2+ binding was measured by chelation titrations of Zn7MT-3, while Cu+ binding was measured by Zn2+ displacement from Zn7MT-3 with competition from glutathione (GSH). Titrations in multiple buffers enabled a detailed analysis that yielded condition-independent values for the association constant (K) and the change in enthalpy (ΔH) and entropy (ΔS) for these metal ions binding to MT-3. Zn2+ was also chelated from the individual α and β domains of MT-3 to quantify the thermodynamics of inter-domain interactions in metal binding. Comparative titrations of Zn7MT-2 with Cu+ revealed that both MT isoforms have similar Cu+ affinities and binding thermodynamics, indicating that ΔH and ΔS are determined primarily by the conserved Cys residues. Inductively coupled plasma mass spectrometry (ICP-MS) analysis and low temperature luminescence measurements of Cu-replete samples showed that both proteins form two Cu4+–thiolate clusters when Cu+ displaces Zn2+ under physiological conditions. Comparison of the Zn2+ and Cu+ binding thermodynamics reveal that enthalpically-favoured Cu+, which forms Cu4+–thiolate clusters, displaces the entropically-favoured Zn2+. These results provide a detailed thermodynamic analysis of d10 metal binding to these thiolate-rich proteins and quantitative support for, as well as molecular insight into, the role that MT-3 plays in the neuronal chemistry of copper.
Introduction
Metallothioneins (MTs) are small cysteine-rich proteins found in all domains of life, with mammals expressing four isoforms, MT-1, MT-2, MT-3 and MT-4.1,2 Mammalian MTs contain 61–68 amino acids, 20 of which are highly conserved cysteines that bind up to seven divalent metal ions in two metal–thiolate clusters in two distinct domains of the protein (Fig. 1).3,4 The structurally characterized Zn- and Cd-bound MTs feature a M3Cys9 cluster in the N-terminal β-domain and a M4Cys11 cluster in the C-terminal α-domain.5,6 The primary roles of MTs are to sequester toxic metal ions7 and to buffer the concentration of Zn2+ and potentially Cu+ ions.4,8–12 They can also reductively quench reactive oxygen species (ROS) and reactive nitrogen species (RNS),7,13–17 and they have been linked to protective roles in neurodegenerative diseases and other CNS pathologies.7,18–25 Nevertheless, a complete understanding of the biological roles of MTs, especially isoform-specific functions, remains elusive.4,26–34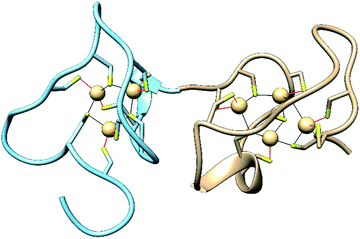 | ||
| Fig. 1 Structures of the N-terminal β-domain (blue, PDB: 1MHU), containing 3 Cd2+ ions bound to 9 cysteine residues, and the C-terminal α-domain (tan, PDB: 2MHU), containing 4 Cd2+ ions bound to 11 cysteine residues, determined by NMR measurements,6 of the human MT-2 protein. | ||
Metal homeostasis in all living organisms is tightly controlled by biological molecules that bind metal ions with affinities spanning many orders of magnitude.35–40 The rich palette of potential ligands, coupled with the impact of concentration gradients on metal-binding thermodynamics, makes well-orchestrated metal trafficking pathways possible.37,41–43 Cu+ is the most tightly controlled essential metal ion, with one of the lowest availabilities of free ions. The biological window of free copper concentrations in eukaryotic cells has been estimated to be between 10−21 and 10−18 M.44,45 A large thermodynamic gradient moves Cu+ into copper-requiring biomolecules.37,41,43,46,47
Metallothioneins are implicated in metal homeostasis, which makes it important to understand the metal-binding thermodynamics of the different isoforms.48 MT-3 is a particularly enigmatic member of the MT family.7,18,49–54 First identified in brain extracts as a factor that inhibited neuronal sprouting, it was initially named “neuronal growth inhibitor factor (GIF)”15,55,56 and only later reclassified as a metallothionein.55 The MT-3 isoform is distinctive in several ways.57 It is expressed primarily in neuronal tissues where it is localized to specific regions of the Central Nervous System (CNS),50,58 its sequence has two unique features (a conserved threonine insert at position 5 followed by a pair of proline residues in the β-domain (residues 7 and 9) and an acidic insert in the α-domain), and it inhibits neuronal growth.27,29,32,49,54,57,59–67 In contrast to MT-1 and MT-2, the expression of MT-3 is not upregulated by Zn2+ nor other divalent toxic metals (e.g. Cd2+), suggesting that its role in metal homeostasis is different from that of the more ubiquitously expressed MTs.68–70 MT-3 is, however, upregulated by hypoxia.52,71–73
Metallothionein-3 may modulate copper toxicity.74–76 Its sequence and structural properties give it the most pronounced Cu–thionein, in contrast to Zn–thionein, character of the MTs,30,74,77 and it is isolated from mammalian brains in a surprisingly air-stable Cu4+Zn3–42+MT-3 form.77–79 MT-3 is capable of exchanging Zn2+ for copper that is bound to several neurologically important peptides, including amyloid β and α-synuclein, thereby eliminating harmful redox chemistry associated with Cu2+ in these coordination environments.61,80,81 The driving force for this “metal swap” is presumed to be the high affinity of MT-3 for Cu+, coupled with its ability to reduce Cu2+ to Cu+via intramolecular disulfide bond formation.82 MT-3 may also scavenge Cu+ in the reducing environment of the cell and help to maintain the low levels of free copper in neuronal cells.
In this study we have quantified the thermodynamics of Cu+ binding to MT-3 and MT-2, as well as the thermodynamics of Zn2+ binding to MT-3 and its isolated domains at physiological pH. We also provide evidence that Cu+ forms Cu4+–thiolate clusters that are structurally similar to those found in the β-domain of Cu4+Zn3–42+MT-3 isolated from neuronal tissue.78,82,83 The thermodynamics of the competition between these two metal ions for MT-3 and domain–domain interaction in metal binding by MT, determined here, provide new insight on the molecular basis for the biological roles of these proteins.
Materials and methods
Reagents
Buffer salts, including MES, Bis–Tris, Tris, TAPSO and phosphate, were purchased in the highest purity available from Sigma-Aldrich, as were metal salts, dithiothreitol (DTT), 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB), 2,2-dithiodipyridine, imidazole, β-mercaptoethanol (βME), phenylmethylsulfonyl fluoride (PMSF), reduced L-glutathione (GSH), acetonitrile (MeCN), tris(2-carboxyethyl)phosphine (TCEP), ethylenediaminetetraacetic acid (EDTA), DNAse, and diethylenetriaminepentaacetic acid (DTPA), and used without further purification. Ampicillin and isopropylthio-galactoside (IPTG) were purchased from GoldBio. HisPur cobalt resin, Ni NTA resin, Lysogeny Broth (LB) and Terrific Broth (TB) were purchased from Thermo Fisher. The HiLoad 26/600 Superdex 75 pg column, HiTrap 5 mL desalting column and HisTrap FF 5 mL affinity column were all obtained from GE Healthcare, now Cytiva. Amicon supplied the 3 kDa concentrator tubes. Recombinant His6-tagged TEV protease was expressed at 18 °C in BL21-DE3* cells and purified via cobalt-affinity chromatography as described below for MT-3. The only difference was an additional wash with 5 mM ATP in TALON wash buffer (50 mM Na2HPO4, 300 mM NaCl, pH 7.0, 1 mM βME). The eluted protein was dialyzed against 50 mM Tris, 250 mM NaCl, 0.5 mM TCEP, pH 8.0 for 3 h, followed by overnight dialysis into 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 glycerol-to-buffer. The sample was centrifuged at 60
1 glycerol-to-buffer. The sample was centrifuged at 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min, flash frozen, and stored at −80 °C.
000 × g for 10 min, flash frozen, and stored at −80 °C.
Buffer solutions for ITC measurements were prepared with Nanopure (18 MΩ) water in acid-washed glassware and subsequently treated overnight with Chelex 100® cation exchange resin (Sigma Aldrich) to ensure the absence of metal contaminants. Buffer solutions were then filtered and degassed under vacuum with stirring for at least 1 h, or until no further bubbles formed, and finally moved into an anaerobic Coy glovebox with a 95% N2 and 5% H2 environment. Chelator and metal stock solutions were prepared with oxygen-free buffer solutions and stored in the glovebox. Solutions of GSH were made fresh for each measurement. The concentration of metal stock solutions was confirmed with ITC by titrating the metal solution into a known concentration of EDTA and verifying with the binding stoichiometry, as well as confirming the known binding enthalpy. Similarly, the concentration of chelator stock solutions was confirmed with ITC by titrating a metal solution of known concentration into the chelator solution and verifying with the binding stoichiometry, as well as confirming the known binding enthalpy.
Metallothionein-3 (MT-3) expression, purification and characterization
An Äkta Pure 25 FPLC was used for protein purification and a Coy Chamber was used for anaerobic protein manipulations. Lower speed centrifugations were performed on a Beckman Coulter Allegra X-30 R and higher speed centrifugations above 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm were performed on a Thermo Scientific WV+ Ultra 100. Mass spectrum measurements were obtained on a Bruker ultrafleXtreme MALDI-TOF/TOF at the Columbia University mass spectrometry facility in the Chemistry Department.
000 rpm were performed on a Thermo Scientific WV+ Ultra 100. Mass spectrum measurements were obtained on a Bruker ultrafleXtreme MALDI-TOF/TOF at the Columbia University mass spectrometry facility in the Chemistry Department.
Mouse MT-3 (musMT3) plasmids were constructed from synthetic gene fragments that were codon-optimized for expression in E. coli (Integrated DNA Technologies) and a modified version of the pET15b vector. MT-3 genes were inserted in frame with an N-terminal His6-Green Fluorescence Protein (GFP) tag and a TEV protease site using FastCloning.84 Complete gene sequences are provided in Fig. S1.† BL21-DE3 cells were transformed with the His6-GFP-tev-musMT-3 plasmid. A single colony was inoculated and grown overnight at 37 °C in LB with ampicillin (100 μg mL−1). In the morning, 2.8 L flasks, each containing 500 mL of TB, were inoculated with 10 mL of the overnight culture. TB cultures were then incubated at 37 °C and 250 rpm until the optical density at 600 nm (OD600) reached 0.7, whereupon 1 mM IPTG was added to each flask. Cultures were incubated for an additional hour at 25 °C and 250 rpm, and then 0.5 mM ZnCl2 was added to each flask, which was shaken for an additional 4–5 h. The MT-3-GFP cultures were centrifuged at 7500 × g for 15 min at 4 °C. Cell pellets were collected and resuspended into 4 mL of 50 mM sodium phosphate, 300 mM NaCl, pH 8.0, 1 mM βME per 1 g of cell pellet and either flash frozen in liquid nitrogen prior to storage at −80 °C or lysed immediately for protein isolation. Prior to cell lysis, 0.1 mM PMSF and 4 μg μL−1 DNAse were added to the cell suspension. Cells were either French pressed in a single pass with a Glen Mill press operating at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kPa or sonicated at 20 W for 5 s on and 10 s rest for a total of 3 min of sonication. Lysed cells were centrifuged at 40
000 kPa or sonicated at 20 W for 5 s on and 10 s rest for a total of 3 min of sonication. Lysed cells were centrifuged at 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min at 4 °C and the supernatant was collected.
000 × g for 30 min at 4 °C and the supernatant was collected.
The crude cell lysate was added to HisPur cobalt resin and nutated for 1 h at 4 °C. The resin was washed with extraction buffer (50 mM Na2HPO4, 300 mM NaCl, 1 mM βME, pH 8.0), followed by wash buffer (50 mM Na2HPO4, 300 mM NaCl, pH 7.0, 1 mM βME). His6-GFP-tev-MT-3 was eluted from the resin with wash buffer containing 200 mM imidazole. A PD10 column, or a 3 kDa spin-concentrator, or dialysis for 3 h, was then used to exchange the buffer of the elution fractions into PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3, 1 mM DTT). The GFP molar extinction coefficient at 488 nm, ε488 = 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1, was used to determine the concentration of His6-GFP-tev-MT-3. Fractions were analyzed by SDS-PAGE.
000 M−1 cm−1, was used to determine the concentration of His6-GFP-tev-MT-3. Fractions were analyzed by SDS-PAGE.
To remove the His6-GFP tag, His6-TEV protease was added in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 protease
100 protease![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GFP-tev-MT-3 molar ratio. The sample tube was flushed with argon and incubated at 4 °C overnight. The sample was either loaded onto a HisTrap 5 mL FF column equilibrated with PBS buffer or rocked with pre-washed Ni-NTA resin with PBS buffer for 1 h. The flowthrough, containing cleaved MT-3, was collected from the column, concentrated in a 3 kDa spin-concentrator at 3900 × g at 4 °C, and further purified on a HiLoad 26/600 Superdex 75 pg size exclusion chromatography (SEC) column equilibrated with MES buffer (100 mM MES, 150 mM NaCl, 1 mM DTT, pH 6.4). Eluted fractions from the gel filtration column were analyzed by SDS-PAGE. Fractions with the desired protein were combined and concentrated using a 3 kDa spin-concentrator. MT-3 was characterized by SDS-PAGE using the monobromobimane-labeling method optimized by Meloni et al.62,85 and by MALDI-TOF mass spectrometry.
GFP-tev-MT-3 molar ratio. The sample tube was flushed with argon and incubated at 4 °C overnight. The sample was either loaded onto a HisTrap 5 mL FF column equilibrated with PBS buffer or rocked with pre-washed Ni-NTA resin with PBS buffer for 1 h. The flowthrough, containing cleaved MT-3, was collected from the column, concentrated in a 3 kDa spin-concentrator at 3900 × g at 4 °C, and further purified on a HiLoad 26/600 Superdex 75 pg size exclusion chromatography (SEC) column equilibrated with MES buffer (100 mM MES, 150 mM NaCl, 1 mM DTT, pH 6.4). Eluted fractions from the gel filtration column were analyzed by SDS-PAGE. Fractions with the desired protein were combined and concentrated using a 3 kDa spin-concentrator. MT-3 was characterized by SDS-PAGE using the monobromobimane-labeling method optimized by Meloni et al.62,85 and by MALDI-TOF mass spectrometry.
Metal ions were removed from the protein by lowering the pH to 3 with 1 M HCl in the anaerobic chamber. The resulting apo-protein was then desalted with a HiTrap desalting column that was equilibrated with 100 mM MES, 150 mM NaCl, pH 6.4, in order to separate free metal ions and exchange the protein into a buffer without DTT. The protein concentration was determined by measuring the absorption at 220 nm in 0.1 M HCl, using ε220 = 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1.86 The free thiol concentration was determined spectrophotometrically, after the reaction of MT-3 with DTNB in 25 mM Tris, 50 mM NaCl, 1 mM EDTA, pH 8.0, using ε412 = 14
000 M−1 cm−1.86 The free thiol concentration was determined spectrophotometrically, after the reaction of MT-3 with DTNB in 25 mM Tris, 50 mM NaCl, 1 mM EDTA, pH 8.0, using ε412 = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 M−1 cm−1 for free thiols.66,87 With the MT-3 protein concentration determined from the thiol content, the appropriate stoichiometry of Zn2+ from solutions of ZnCl2 was added to generate Zn7MT-3.
150 M−1 cm−1 for free thiols.66,87 With the MT-3 protein concentration determined from the thiol content, the appropriate stoichiometry of Zn2+ from solutions of ZnCl2 was added to generate Zn7MT-3.
The individual α- and β-domains of MT-3 were purified in an identical manner. All constructs are listed in the ESI (Fig. S1†).
Metallothionein-2 (MT-2) expression, purification and characterization
The recombinant human MT-2 expression plasmid was generated by synthetic DNA synthesis upon codon optimization (Genscript) and cloned in a pET-3d plasmid (Novagen) between the NcoI/BamHI restriction sites. MT-2 was expressed in E. coli BL21(DE3)pLys cells and purified following the method of Faller et al.88 In this method, MT-2 is expressed and purified in its Cd-bound form upon addition of 0.4 mM CdSO4 30 min after IPTG induction. For the anion-exchange chromatography step, MT-2 was eluted with a linear gradient of 0 to 200 mM NaCl in 25 mM Tris, pH 8.6, at a flow-rate of 8 mL min−1 using a HiPrep DEAE FF 16/10 column connected to an Äkta Pure chromatographic system (GE Healthcare, now Cytiva). The Cd-containing fractions, as determined by ICP-MS quantification, were pooled. The apo-protein was then generated and reconstituted to the Zn7MT-2 form using the method of Vašák,89 by addition of HCl, followed by the addition of ZnCl2 and adjustment of the pH to 8.0 with 1 M Tris base. Zn7MT-2 was further purified by size exclusion chromatography to ensure the complete removal of low molecular weight protein contaminants. The MT-2 protein concentration was determined spectrophotometrically in 0.1 M HCl, which releases the Zn2+ to generate apo MT-2, using a Cary 300 UV-vis spectrophotometer (Agilent) and ε220 = 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1. The Zn2+ concentration was determined with quantification by ICP-MS (Agilent 7900), using protein samples digested in 45% HNO3 (v/v) overnight and subsequently diluted to 1% HNO3. The Cys concentration was determined spectrophotometrically by sulfhydryl group quantification, after sample reaction with 2,2-dithiodipyridine in 0.2 M sodium acetate, 1 mM EDTA, pH 4.0, using ε343 = 7600 M−1 cm−1 for free thiols.87 Zinc-to-protein ratios of 7.0 ± 0.5 and Cys-to-protein ratios of 20 ± 3 were obtained. The protein purity was confirmed by SDS-PAGE using MT samples subjected to cysteine modification by monobromobimane, following the method of Meloni et al.62
200 M−1 cm−1. The Zn2+ concentration was determined with quantification by ICP-MS (Agilent 7900), using protein samples digested in 45% HNO3 (v/v) overnight and subsequently diluted to 1% HNO3. The Cys concentration was determined spectrophotometrically by sulfhydryl group quantification, after sample reaction with 2,2-dithiodipyridine in 0.2 M sodium acetate, 1 mM EDTA, pH 4.0, using ε343 = 7600 M−1 cm−1 for free thiols.87 Zinc-to-protein ratios of 7.0 ± 0.5 and Cys-to-protein ratios of 20 ± 3 were obtained. The protein purity was confirmed by SDS-PAGE using MT samples subjected to cysteine modification by monobromobimane, following the method of Meloni et al.62
ITC data collection and analysis
All ITC measurements were obtained with a Malvern (MicroCal) VP-ITC housed in a custom plexiglass glovebox under a N2 atmosphere at 25 ± 0.2 °C. The samples were stirred at a constant rate in the range 307 to 437 rpm and titrant injection volumes were 4 to 12 μL, with a spacing of 240 to 600 seconds between each injection. Heat associated with the last few injections quantifies the heat of dilution, which was subtracted from the heat of each injection. The ITC data are presented as the baseline adjusted heat flow vs. time in the upper panel and the integrated, concentration-normalized, molar heat per injection vs. molar ratio of titrant-to-titrand in the lower panel. Data were analysed by either a one-site or two-sites binding model with Origin 7.0 software. The tabulated experimental values represent the average and standard deviation from at least three independent measurements, unless otherwise noted. The change in entropy is reported as −TΔS at 25 °C, which has the same units and sign convention as ΔH, for ready comparison of thermodynamic values.Protein samples were prepared in buffer solutions that were identical to those containing the metal ion or chelate titrant. For Zn2+ chelation measurements, the chelate concentration was 120 to 140 times greater than the concentration of the Zn2+-bound protein sample (∼5 μM). For the Cu+ binding measurements, GSH was present at the same concentration in both titration solutions, with a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of GSH to Cu+ (∼0.5 mM) in the syringe and a 2000
1 ratio of GSH to Cu+ (∼0.5 mM) in the syringe and a 2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of GSH to protein (∼2.5 μM) in the cell.
1 ratio of GSH to protein (∼2.5 μM) in the cell.
For each type of ITC measurement, the experimental change in enthalpy (ΔHITC) in different buffers was used to quantify the number of protons that bind to, or are released from, the buffer upon addition of the titrant to the sample in the cell.90 This value was then used to determine the contribution of buffer protonation to ΔHITC, so it could be subtracted. It was also used to quantify the number of protons displaced from the protein upon metal binding at the experimental pH. An analysis based on Hess's law was used to determine the metal-binding enthalpy from the ΔHITC value. The metal-binding equilibrium constant was determined from the experimental binding constant (KITC) for direct metal titrations as described previously48,91 and for chelation titrations as described in the ESI.†
Metal content and 77 K luminescence characterization of Cun+MT-2 and Cun+MT-3
The Cun+MT species formed upon reaction of Cu+ with Zn7MT-2 or Zn7MT-3 in the presence of excess GSH were prepared to mimic the conditions utilized in the ITC measurements and correspond to the complete Cu+ titration of the MT samples. Zn7MT-2 and Zn7MT-3 stock solutions and the GSH-containing buffer (100 mM MOPS, pH 7.4, 150 mM NaCl, 20 mM GSH) were made oxygen-free by three vacuum/nitrogen cycles on a Schlenk line, while a stock solution of [Cu(MeCN)4]PF6 (100 mM in MeCN) was prepared inside a constant-flow nitrogen-purged anaerobic glovebox. A 10 mM Cu+ stock solution was subsequently prepared in the glovebox with the anaerobic GSH-containing buffer and used for reactions with the Zn7MT samples.20 equivalents/mole of Cu+ were mixed with Zn7MT-2 (25 mM Tris, 50 mM NaCl, pH 8.0) and Zn7MT-3 (100 mM Bis–Tris, 150 mM NaCl, pH 7.4) samples (500 μL, 10 μM) and incubated for 2.5 h at 25 °C inside the nitrogen-purged glovebox. The mixtures were subsequently purified by injection onto a Superdex 75 10/300 column connected to an Äkta Pure Chromatographic System (GE Healthcare) and elution with 100 mM MOPS, pH 7.4, 150 mM NaCl, 20 mM GSH. The fractions corresponding to MT-2 or MT-3 were identified by subjecting aliquots from each peak of the SEC chromatogram to monobromobimane modification62 and SDS-PAGE to confirm the presence and purity of MT-2 or MT-3 (Fig. S2†).
The Cu- and Zn-to-MT stoichiometries of the purified MT samples were determined by metal and protein quantification. Cu and Zn were quantified by ICP-MS (Agilent 7900) after digesting the MT samples in 45% HNO3 (v/v) overnight at room temperature and subsequently diluting each sample to a final 1% HNO3 (v/v) with ultrapure H2O. Prior to determining the protein concentration, samples were acidified with 1 M HCl to pH < 1 to ensure metal release from the protein. The acidified samples were subsequently washed by three consecutive dilution–concentration cycles (3 × 500 μL) with 100 mM HCl in a 3 kDa cutoff Amicon Ultra centrifugal filter. Protein concentrations were determined spectrophotometrically on a Cary 300 UV-vis spectrophotometer (Agilent) by recording the absorbance at 220 nm and using ε220 = 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 and 48
000 M−1 cm−1 and 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1 for MT-3 and MT-2, respectively.
200 M−1 cm−1 for MT-3 and MT-2, respectively.
The formation and nature of the Cu+–thiolate clusters in the Cun+MT samples were characterized by low temperature (77 K) luminescence emission spectra of the purified MT-2 and MT-3 fractions using a FluoroMax-4 spectrofluorometer (Horiba Scientific). 400 μL samples were flash frozen using liquid N2 inside a 4 mm inner diameter quartz tube, placed in a quartz dewar, and the emission spectra recorded from 380 to 750 nm (λexc: 320 nm, slit width: 5 nm) with a 10 μs initial delay and a 300 μs sample window. The final spectra for each sample were recorded three times upon rotating the quartz tube inside the dewar and averaged to minimize freezing inhomogeneities. Spectra were subsequently collected for three independent sample replicates and the corresponding standard deviation calculated. Decay lifetimes of the emissive bands at 425 nm and 575 nm were determined on the same samples using a 75 μs initial delay and 300 μs sample window. Delay increments of 10 μs and 20 μs and maximum delays of 500 μs and 1000 μs were used for the 425 nm and 575 nm bands, respectively. Lifetimes at 425 nm and 575 nm for both MT-2 and MT-3 were calculated by fitting the data on three independent replicates with a single decay exponential function.
Results, analysis, and discussion
Measurements of metal ions binding to metallothionein (MT) are challenging, due in large part to the susceptibility of the many conserved cysteine residues to oxidation. However, the Zn2+-bound form of the protein is less prone to oxidation and amenable to storage and reproducible characterization. Therefore, the thermodynamics of Zn2+ and Cu+ binding to MT-3 were determined with two types of anaerobic ITC measurements, both involving samples of Zn7MT-3. For Zn2+, the metal ion was extracted from the protein by titration with a chelating ligand whose affinity for Zn2+ is higher than that of the protein. This type of ITC measurement has been used previously to quantify the binding of metal ions to proteins,91 including MT-3.48 The thermodynamics of Zn2+ binding to MT-3 were then determined from an analysis of the chelation experimental data that includes competition between the chelating ligand and MT-3 for the Zn2+ and the concept of microscopic reversibility. Since MT has a higher affinity for Cu+ than it does for Zn2+, titration of the former into samples of Zn7MT-3 results in displacement of the latter.4,36 However, Cu+ is unstable to disproportionation in aqueous solution, which can be suppressed by the addition of a ligand that is introduced for this purpose.92 The thermodynamics of Cu+ binding to MT-3 were determined with an analysis that accounts for competition between the Cu+-stabilizing ligand and MT-3 for Cu+, as well as Zn2+ in the initial Zn7MT-3 sample. The resulting Cu+-bound protein was characterized further by ICP-MS to determine the Cu-to-protein stoichiometry and by low temperature luminescence to provide information on the structure of copper clusters in the protein that results under these conditions.Zn2+ binding to MT-3
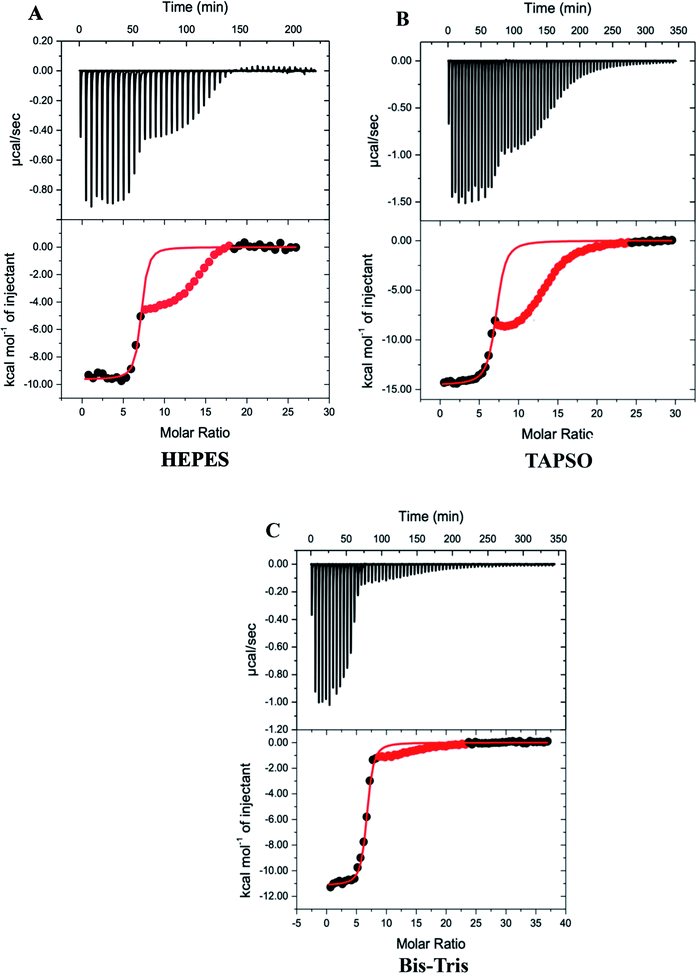
Our previous ITC measurements of the thermodynamics of Zn2+ binding to MT-3 were part of a study of Pb2+ binding to the protein, which was conducted at pH 6.0 to ensure the Pb2+ remained soluble.48 In that study, the challenges of working with metal-free (apo) MT-3 were avoided by titrating the chelating ligand ethylenediaminetetraacetic acid (EDTA) into Zn7MT-3 and analysing the experimental binding isotherms with microscopic reversibility. The data revealed three sequential chelation events with different enthalpies, which were analysed to quantify the thermodynamics of triphasic Zn2+ binding to apo MT-3 to form Zn7MT-3 at pH 6.0. For the current study, similar measurements with EDTA at pH 7.4 gave different results that were not readily interpreted, revealing complex and pH-dependent equilibria for chelation titrations with EDTA. Therefore, other chelating ligands were evaluated for ITC measurements at pH 7.4, and diethylenetriaminepentaacetic acid (DTPA), which binds Zn2+ with an affinity that is two orders of magnitude higher than that of EDTA, was found to give consistent results.The DTPA titrations of Zn7MT-3, however, are unusual in that they have both a net exothermic chelation of Zn2+ with an inflection at the stoichiometry of the Zn2+ in the Zn7MT-3 sample, followed by another exothermic event upon further addition of DTPA, suggesting the formation of a higher-order DTPA–Zn2+ complex (Fig. 2). While initially puzzling, control titrations of DTPA into Zn2+ in the same buffers at pH 7.4 revealed a similar pattern, with initial formation of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DTPA–Zn2+ complex and subsequent formation of a 3
1 DTPA–Zn2+ complex and subsequent formation of a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 DTPA–Zn2+ complex, whose formation enthalpy depends on the buffer that is present (Fig. S3†). This latter observation provides an important correlation to the MT-3 data, as the magnitude of the formation enthalpy for this subsequent DTPA–Zn2+ species in different buffers is Bis–Tris < HEPES < TAPSO < Tris for both the control titrations and the protein titrations. Therefore, the isotherms for DTPA titrations of Zn7MT-3 also contain the subsequent formation of a species when excess DTPA is added to the DTPA–Zn2+ formed by chelation of Zn2+ from the protein.
2 DTPA–Zn2+ complex, whose formation enthalpy depends on the buffer that is present (Fig. S3†). This latter observation provides an important correlation to the MT-3 data, as the magnitude of the formation enthalpy for this subsequent DTPA–Zn2+ species in different buffers is Bis–Tris < HEPES < TAPSO < Tris for both the control titrations and the protein titrations. Therefore, the isotherms for DTPA titrations of Zn7MT-3 also contain the subsequent formation of a species when excess DTPA is added to the DTPA–Zn2+ formed by chelation of Zn2+ from the protein.
Since the first event in the experimental titrations is the chelation of Zn2+ from Zn7MT-3, data after the inflection were masked (red points) and only data associated with the initial inflection were fitted to a one-site binding model (Fig. 2). In contrast to ITC data at pH 6.0, where EDTA chelation isotherms contain three inflections and were fitted with a three-site binding model,48 the DTPA chelation of each Zn2+ at pH 7.4 has the same net change in enthalpy as each metal ion is removed from the protein at this pH under these conditions. The average best-fit experimental values for titrations in three different buffers are found in Table 1.
| Protein | Buffer | n ITC | K ITC | ΔH°ITC (kcal mol−1) |
|---|---|---|---|---|
| Zn7MT-3 | HEPES | 6.90 ± 0.05 | 1.27 (±0.09) × 107 | −9.5 ± 0.2 |
| Bis–Tris | 6.5 ± 0.1 | 1.1 (±0.9) × 107 | −11.0 ± 0.2 | |
| TAPSO | 7.08 ± 0.06 | 4.5 (±0.6) × 106 | −14.1 ± 0.7 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Zn4αMT-3 | Bis–Tris | 4.4 ± 0.3 | 9 (±5) × 107 | −10.1 ± 0.6 |
| TAPSO | 4.2 ± 0.2 | 6.5 (±0.3) × 106 | −14.1 ± 0.1 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Zn3βMT-3 | Bis–Tris | 2.6 ± 0.1 | 2.8 (±0.6) × 107 | −10.9 ± 0.5 |
| TAPSO | 2.8 ± 0.2 | 4 (±1) × 106 | −14.1 ± 0.4 | |
The experimental ITC values obtained from chelation titrations require additional analysis to determine the buffer- and chelate-independent thermodynamics of Zn2+ binding to MT-3 at pH 7.4. This post hoc analysis takes into account the contributions from coupled and competing equilibria. For the enthalpy, these include deprotonation of the DTPA chelating ligand (ΔHLH1 and ΔHLH2), protonation of the buffer (ΔHBH), the desired dissociation of Zn2+ from MT-3 (ΔHMP), protonation of MT-3 upon release of Zn2+ (ΔHPH) and Zn2+ binding to DTPA (ΔHML). This analysis requires the net protonation, or deprotonation, of the buffer, which is determined from the dependence of the experimental enthalpy (ΔHITC) on the buffer protonation enthalpy (ΔHBH) for data in different buffers (Fig. S4†). This value, which is determined from the slope of these plots, allows the enthalpic contribution from buffer protonation to be subtracted and, through a proton inventory (Scheme S1†), the protonation of MT-3 upon release of Zn2+ to be quantified (9.5 ± 0.3 H+). As summarized in Scheme S1,† the weighted sum of the enthalpies for each of these events equals the experimentally measured enthalpy (ΔHITC). The condition-independent Zn2+-binding enthalpy determined here is the average value per Zn2+ for the formation of Zn7MT-3 at pH 7.4: 7Zn2+ + MT-3 → Zn7MT-3 + 9.5H.
To determine the buffer- and chelate-independent binding constant for Zn2+ binding to MT-3, coupled and competing equilibria must be taken into account in a post hoc analysis similar to that for the binding enthalpy, as outlined in the ESI.† The results of these analyses of the net experimental binding enthalpy (exothermic) and binding constant to determine the condition-independent binding enthalpy (endothermic) and binding constant provide the thermodynamic values for Zn2+ binding to MT-3 at pH 7.4 found in Table 2.
| Protein | n (Zn2+) | K | ΔG° (kcal mol−1 Zn2+) | ΔH° (kcal mol−1 Zn2+) | −TΔS° (kcal mol−1 Zn2+) |
|---|---|---|---|---|---|
| MT-3 | 6.8 ± 0.3 | 4 (±2) × 1011 | −15.7 ± 0.3 | 13.4 ± 0.2 | −29 ± 0.5 |
| αMT-3 | 4.3 ± 0.3 | 4 (±3.5) × 1011 | −15 ± 1 | 4.26 ± 0.05 | −20 ± 1 |
| βMT-3 | 2.7 ± 0.2 | 9 (±8.5) × 1011 | −16.0 ± 0.8 | 5.76 ± 0.06 | −21.8 ± 0.8 |
Data from our earlier study of Zn2+-binding to MT-3 at pH 6.0 revealed that Zn2+ binds to MT-3 sequentially in three populations with different enthalpies, an initial ∼4 that bind most tightly with terminal tetra-thiolate coordination, a subsequent ∼2 that bind less tightly with bridging thiolate coordination and finally ∼1 that binds even less tightly.48 For DTPA chelation measurements at pH 7.4, we measure only a single set of thermodynamic values for all seven Zn2+ ions bound to MT-3. This result is clearly different from our chelation ITC measurements at lower pH, as well as results from chromophore chelation measurements and fitting in an earlier study of MT-2 by Krężel and Maret.93 While the DTPA chelation data could suggest a cooperative chelation process at pH 7.4, this would be inconsistent with other experimental results (e.g. zinc titrations followed by ESI-MS analysis) and computational simulations of MTs binding and releasing Zn2+.93–95 Of relevance is the Zn2+ affinity of the chelate used. The chromophore chelates have affinities that are lower than that of MT, while DTPA has an affinity that is 6–7 orders of magnitude greater than that of MT, as required for ITC measurements, and two orders of magnitude greater than that of EDTA. The much higher Zn2+ affinity of DTPA and the properties of Zn7MT-3 at pH 7.4 result in conditions where the ITC chelation measurements are only able to discern an average Zn2+ affinity of MT-3. This value, which is obtained from the post hoc analysis of the ITC chelation data (K = 4 (±2) × 1011 at pH 7.4), is comparable to two reported values (K = 6.2 × 1010 at pH 8.0 and K = 2.4 × 1011 for the β-domain)96,97 but somewhat higher than another (K = 7.7 × 109).57
The binding of Zn2+ to MT-3 at pH 7.4 is enthalpically disfavoured (ΔH = +13 kcal mol−1) and entropically driven (ΔS = 97 cal mol−1 K−1; −TΔS = −29 kcal mol−1 at 25 °C). This is similar to results for Zn2+ binding to peptides with tetra-thiolate ligation, including a Gly-rich peptide prepared and studied by Gibney and co-workers98 (ΔH = +5.6 kcal mol−1; ΔS = 77 cal mol−1 K−1; −TΔS = −23 kcal mol−1 at 25 °C) and a peptide corresponding to the C-terminal Zn-binding sequence of the glucocorticoid receptor (GR-2) studied by Rich et al.99 (ΔH = +10 ± 2 kcal mol−1; ΔS = 70 cal mol−1 K−1; −TΔS = −20 ± 2 kcal mol−1 at 25 °C). The net endothermic binding is attributed to a large contribution from the enthalpic penalty to deprotonate the Cys thiols. The net increase in entropy is attributed to desolvation of the Zn2+ ions and protein and loss of the Cys protons, in spite of conformational restrictions imposed on the protein by Zn2+ coordination.
Further analysis of the Zn2+-binding thermodynamics of MT-3, however, reveals an important, and previously unknown, property of this protein. The endothermic contribution from deprotonation of the Cys thiols can be subtracted from the net Zn2+ binding enthalpy to estimate the Zn2+–thiolate bond enthalpy. When this penalty is subtracted from data for the tetra-Cys peptide GR-2,99 the residual enthalpy is −25 kcal mol−1 Zn2+. When a similar subtraction is made for Cys thiols that are deprotonated upon Zn2+ binding to MT-3, the corresponding value is +1.5 kcal mol−1 Zn2+ (ΔHMP in Scheme S1†). The enthalpy of a μ2-bridging Zn2+–thiolate bond, which is the case for some of the Zn2+ in MT, is expected to be smaller than that of a terminal Zn2+–thiolate bond and thus the average bond enthalpy for Zn2+ ions bound to MT-3 should be less than that of tetra-thiolate coordination. However, the enthalpy of Zn2+–thiolate bonds must be favourable (negative) for stable binding. Therefore, this unusual result for MT-3 reveals there must be another endothermic contribution to Zn2+ binding to MT-3 (vide infra).
The high affinity of MT-3 for Zn2+ is due to a very favourable change in entropy upon binding the metal ion, which is even more favourable than that of Zn2+ binding to tetra-Cys peptides. The two cases would have similar entropic contributions per Zn2+ from desolvation of the metal ion and an inventory of species reveals a comparable net increase in the translational entropy (eqn (1) and (2)),
| MT-3 + 6.8 Zn2+ → Zn6.8MT-3 + 9.5 H+, Δspecies = 10.5 − 7.8 = +2.7 | (1) |
| GR-2 + Zn2+ → ZnGR-2 + 4H+, Δspecies = 5 − 2 = +3 | (2) |
Zn2+ binding to the individual domains of MT-3
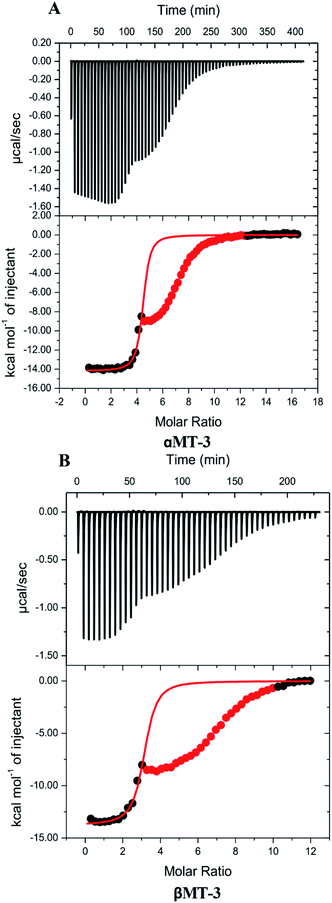
Quantitative insight on inter-domain interactions of MT-3 can be obtained from measurements of metal ions binding to the individual domains for comparison to results on MT-3. Fig. 3 shows ITC data for DTPA titrations of Zn4αMT-3 and Zn3βMT-3 at pH 7.4. Similar to the whole protein, isotherms of the individual domains have an initial exothermic event with a stoichiometry that matches the Zn2+ bound to the domain in the sample, followed by the exothermic binding of excess DTPA to the DTPA–Zn2+ from the Zn2+ chelation. As with Zn7MT-3, only the data associated with the first event were fitted with a one-site binding model. Average best-fit experimental ITC values for DTPA chelation of Zn2+ from Zn4αMT-3 and Zn3βMT-3 in two different buffers are found in Tables S1 and S2,† respectively.Our ITC data for Zn2+ binding to the individual α and β domains of MT-3 allow us to evaluate the thermodynamics of inter-domain interaction.95,102,103 The results in Table 3 show that the free energy of Zn7MT-3 formation is identical to the sum of the free energies of formation of Zn4αMT-3 and Zn3βMT-3. However, there are significant enthalpic and entropic differences between MT-3 and the sum of the values for the isolated domains, with an unfavourable difference in the formation enthalpy (ΔΔH = +57 kcal mol−1 protein) and a favourable difference in the formation entropy (−TΔΔS = −52 kcal mol−1 protein at 25 °C) when the two domains are linked in the protein. This reveals that domain–domain interactions provide an entropic benefit but an enthalpic penalty to Zn2+ binding to form the Zn4 cluster in the α-domain and the Zn3 cluster in the β-domain. These two effects, however, are similar in magnitude, so they cancel, in another example of enthalpy–entropy compensation (EEC) in biological coordination chemistry.98,99
| Sample | n (Zn2+) | n (H+) | ΔG° (kcal mol−1) | ΔH° (kcal mol−1) | −TΔS° (kcal mol−1) |
|---|---|---|---|---|---|
| MT-3 | 6.8 ± 0.3 | 9.5 ± 0.3 | −106.8 | 91.1 | −197.2 |
| α domain | 4.3 ± 0.3 | 7.0 ± 0.6 | −64.5 | 18.3 | −86.0 |
| β domain | 2.7 ± 0.2 | 4.1 ± 0.4 | −43.2 | 15.6 | −58.9 |
| α value + β value | 7.0 ± 0.4 | 11.1 ± 0.7 | −107.7 | 33.9 | −144.9 |
| Difference: MT-3 − (α + β) | — | — | +0.9 | +57.2 | −52.3 |
In the analysis above of the net enthalpy of Zn2+ binding to holo MT-3, subtraction of the endothermic Cys deprotonation gave an enthalpy, which is expected to be that of Zn2+ binding to the Cys thiolates, that was unexpectedly endothermic (+1.5 kcal mol−1 Zn2+). A similar analysis that subtracts the Cys thiol deprotonation enthalpy from the net Zn2+-binding enthalpy of the isolated α and β domains gives the favourable values of −9.6 and −7.1 kcal mol−1 Zn2+, respectively. This supports the notion that the endothermic (unfavourable) value for the whole MT-3 contains an additional endothermic contribution, which appears to be the endothermic inter-domain interaction (+57 kcal mol−1 protein). In fact, when this value is put on a per Zn2+ basis (+8.4 kcal mol−1 Zn2+) and subtracted from the Zn2+–thiolate binding enthalpy, a value of −6.9 kcal mol−1 Zn2+ is now found for MT-3. Thus, while the thermodynamics of interdomain interaction do not impact the affinity of MT-3 for Zn2+, they do modulate the relative contributions of the binding enthalpy (disfavouring) and binding entropy (favouring) to the affinity of this two-domain protein for metal ions.
Cu+ binding to MT-3 and MT-2
Quantitative measurements of Cu+ coordination chemistry in aqueous solution are challenging due to its ready oxidation to Cu2+ and favourable disproportionation to Cu2+ and Cu0. While the former can be prevented with anaerobic conditions, the latter needs to be suppressed by a ligand that favours Cu+ over Cu2+, such as 1,1,4,7,10,10-hexamethyltriethylenetetramine (Me6Trien), bicinchoninic acid (BCA), bathocuproine disulfonate (BCS) or glutathione (GSH).92 Since the Cu+-stabilizing ligand competes with the protein for Cu+, it was necessary to choose a ligand with an affinity for Cu+ that is less than that of the protein, yet one whose competition with MT-3 falls within the accuracy range of the titration calorimeter (∼3 < log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kcompetition < ∼8). After initial measurements in MeCN (see ESI†), GSH proved to be ideal and Cu+ binding to MT-3 was measured with Cu+ titrations of Zn7MT-3 in a solution containing excess GSH, the only physiologically relevant ligand from among those noted above. To minimize the heat of dilution in the ITC measurements, the concentration of GSH was matched in the titrant and titrand solutions. Therefore, the 10-fold excess of GSH to Cu+ in the syringe required a ∼2000-fold excess of GSH to Zn7MT-3 in the cell. Based on the affinity of MT-3 for Zn2+ determined here (log
Kcompetition < ∼8). After initial measurements in MeCN (see ESI†), GSH proved to be ideal and Cu+ binding to MT-3 was measured with Cu+ titrations of Zn7MT-3 in a solution containing excess GSH, the only physiologically relevant ligand from among those noted above. To minimize the heat of dilution in the ITC measurements, the concentration of GSH was matched in the titrant and titrand solutions. Therefore, the 10-fold excess of GSH to Cu+ in the syringe required a ∼2000-fold excess of GSH to Zn7MT-3 in the cell. Based on the affinity of MT-3 for Zn2+ determined here (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 11.6) and the stability of Zn2+(GSH)2 (log
K = 11.6) and the stability of Zn2+(GSH)2 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β2 ∼ 12),104 we estimate pZn2+free = 8.2 for the former equilibrium and pZn2+free = 10.6 for the latter equilibrium under our experimental conditions. These values indicated that the large excess of GSH would remove Zn2+ from Zn7MT-3 in situ. The resulting apo MT-3 appears to be stable in the excess GSH for the duration of the anaerobic ITC measurement, as there was no evidence of protein precipitation in the samples or the ITC isotherms (excessive heats, baseline noise, etc.) and the results were reproducible.
β2 ∼ 12),104 we estimate pZn2+free = 8.2 for the former equilibrium and pZn2+free = 10.6 for the latter equilibrium under our experimental conditions. These values indicated that the large excess of GSH would remove Zn2+ from Zn7MT-3 in situ. The resulting apo MT-3 appears to be stable in the excess GSH for the duration of the anaerobic ITC measurement, as there was no evidence of protein precipitation in the samples or the ITC isotherms (excessive heats, baseline noise, etc.) and the results were reproducible.
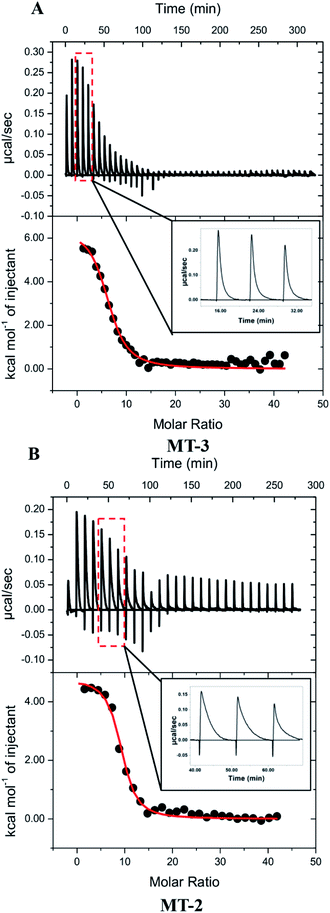
Representative ITC data for Cu+ titrated into Zn7MT-3 with both titrant and titrand in a buffered 5 mM GSH solution are shown in Fig. 4A, where the binding isotherm reflects the competition between the protein and GSH for Cu+. The isotherms were fit to a one-site binding model, indicating similar thermodynamics for all Cu+ binding to MT-3 under these conditions, with average experimental ITC values from data in three buffers found in Table 4. Further analysis of these net endothermic data that accounts for coupled and competing equilibria requires the quantification of buffer protonation upon Cu+ binding. The experimental binding enthalpy in solutions with different buffers was used in this analysis (Fig. S5A†), which shows that 0.5 ± 0.2 protons dissociate from the buffer upon Cu+ binding to the protein at pH 7.4. Since the two GSH that are initially bound to each Cu+ would bind 1.6 ± 0.2 protons after the Cu+ has been released at this pH, a proton inventory (Scheme S2†) shows that 9.1 ± 0.2 protons are displaced from MT-3 when 6 ± 1 Cu+ ions bind to the protein at pH 7.4 in these ITC measurements. If Zn2+ had remained bound to MT-3 in the presence of the large excess GSH, then Cu+ would have displaced the Zn2+ without the release of protons. This confirms that Cu+ is, in fact, binding to in situ generated apo MT-3 under these conditions.
| Protein | Buffer | n ITC | K ITC | ΔH°ITC (kcal mol−1) |
|---|---|---|---|---|
| MT-3 | MOPS | 6.3 ± 0.7 | 5 (±1) × 107 | 4.5 ± 0.7 |
| Bis–Tris | 6 ± 1 | 4 (±2) × 105 | 6.6 ± 1.5 | |
| TAPSO | 4.7 ± 0.2 | 2.1 (±0.2) × 106 | 6.9 ± 0.5 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| MT-2 | MOPS | 8.5 ± 1.5 | 6 (±1) × 105 | 3.6 ± 0.2 |
| TAPSO | 8.1 ± 0.9 | 1.5 (±0.4) × 106 | 5.2 ± 0.4 | |
The condition-independent thermodynamic values for Cu+ binding to MT-3 were obtained from a post hoc analysis, which includes the competition between GSH and MT-3 for Cu+ and the protonation of GSH when Cu+ binds to the protein (Scheme S2†). These condition-independent values for Cu+ binding to MT-3 at pH 7.4 are found in Table 5. The average Cu+ affinity of MT-3 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 19.6) is comparable to that of several Cu+-binding proteins with thiolate coordination105–107 and very similar to log
K = 19.6) is comparable to that of several Cu+-binding proteins with thiolate coordination105–107 and very similar to log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 19.3 reported by Calvo et al.77 for MT-3 determined with a different method. The binding of Cu+ is both enthalpically favoured (exothermic) and entropically favoured (Table 5). Further analysis of the Cu+ binding enthalpy that subtracts the endothermic penalty for Cys deprotonation reveals an average enthalpy for binding to Cys thiolates of −23 kcal mol−1 Cu+, which gives an estimate of −7 kcal mol−1 for the average Cu+–thiolate bond enthalpy. The average Cu+ binding entropy is also favourable, although not as much so as the Zn2+ binding entropy. This is due, in part, to the smaller desolvation contribution from a monopositive, instead of a dipositive, metal ion binding to the protein. The contribution from translational entropy is comparable to that for Zn2+ binding (eqn (1)vs.(3)).
K = 19.3 reported by Calvo et al.77 for MT-3 determined with a different method. The binding of Cu+ is both enthalpically favoured (exothermic) and entropically favoured (Table 5). Further analysis of the Cu+ binding enthalpy that subtracts the endothermic penalty for Cys deprotonation reveals an average enthalpy for binding to Cys thiolates of −23 kcal mol−1 Cu+, which gives an estimate of −7 kcal mol−1 for the average Cu+–thiolate bond enthalpy. The average Cu+ binding entropy is also favourable, although not as much so as the Zn2+ binding entropy. This is due, in part, to the smaller desolvation contribution from a monopositive, instead of a dipositive, metal ion binding to the protein. The contribution from translational entropy is comparable to that for Zn2+ binding (eqn (1)vs.(3)).
| MT-3 + 6 Cu+ → Cu6MT-3 + 9.1 H+ Δspecies = 10.1 − 7 = +3.1 | (3) |
| Protein | K | ΔG° (kcal mol−1 Cu+) | ΔH° (kcal mol−1 Cu+) | −TΔS° (kcal mol−1 Cu+) |
|---|---|---|---|---|
| MT-3 | 4 (±4) × 1019 | −26.9 ± 0.5 | −10 ± 1 | −17 ± 1.5 |
| MT-2 | 8 (±5) ± 1019 | −27.1 ± 0.4 | −12 ± 1 | −15 ± 1 |
Calorimetric data for Cu+ titrations into Zn7MT-2 were obtained with conditions identical to those used with Zn7MT-3 (Fig. 4B and Table 4). An analysis of buffer protonation for the Cu+ titrations (Fig. S5B†) and a proton inventory showed that 12.4 ± 0.2 H+ are displaced from MT-2 when 8 ± 1 Cu+ ions bind to the protein at pH 7.4 in these ITC measurements. Therefore, as was the case with MT-3, the large excess of GSH extracts Zn2+ from Zn7MT-2 and these data quantify Cu+ binding to in situ generated apo MT-2. The condition-independent thermodynamics of Cu+ binding to MT-2 at pH 7.4 were then determined from a post hoc analysis of the binding enthalpy and binding constant, similar to that used for MT-3 (Table 5).
To the extent that ITC measurements of Cu+ binding can distinguish thermodynamic differences, Cu+ coordination by MT-3 and MT-2 are not significantly different in the presence of excess GSH, affirming that metal coordination is predominantly dictated by the conserved Cys residues in all mammalian MT isoforms. Nevertheless, a clear difference between the binding of Cu+ to MT-3 and MT-2 is seen in the experimental ITC data. In contrast to the monophasic injections with MT-3, the MT-2 measurements show biphasic injections, which are attributed to a rapid exothermic contact binding followed by a slower endothermic (protein) reorganization to the final equilibrium (Fig. 4, insets). This difference between MT-2 and MT-3 reflects different kinetics of Cu+ binding under these conditions and is consistent with other reports of unique metal-binding kinetics77 and metalloprotein dynamics96 for MT-3.
The reported Cu+-binding stoichiometries of MTs range from 4 to 20, with the maximum corresponding to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Cu+
1 Cu+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cys ratio, depending on the conditions and the type of measurement.108–112 The presence of excess GSH in our measurements limits the stoichiometry of bound Cu+ to those whose coordination to the protein is more stable than that of the Cu+ (GSH)2 complex (log
Cys ratio, depending on the conditions and the type of measurement.108–112 The presence of excess GSH in our measurements limits the stoichiometry of bound Cu+ to those whose coordination to the protein is more stable than that of the Cu+ (GSH)2 complex (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β2 = 15.3),104 a point we believe is relevant to in vivo conditions.113,114 While the average Cu+ affinity and binding thermodynamics of MT-3 and MT-2 are comparable, within experimental error (Table 5), the number of Cu+ that bind to MT-3 (6 ± 1) and MT-2 (8 ± 1) that were determined by ITC measurements are different, as are the number of protons that are displaced upon formation of Cu6±1MT-3 (9.1 ± 0.2) and Cu8±1MT-2 (12.4 ± 0.2).
β2 = 15.3),104 a point we believe is relevant to in vivo conditions.113,114 While the average Cu+ affinity and binding thermodynamics of MT-3 and MT-2 are comparable, within experimental error (Table 5), the number of Cu+ that bind to MT-3 (6 ± 1) and MT-2 (8 ± 1) that were determined by ITC measurements are different, as are the number of protons that are displaced upon formation of Cu6±1MT-3 (9.1 ± 0.2) and Cu8±1MT-2 (12.4 ± 0.2).
Binding stoichiometries determined from ITC measurements are affected by conditions (rapid stirring) and several sources of error (concentrations, volumes) and can have low accuracy and precision. Therefore, to accurately determine the stoichiometry of the resulting Cu+-bound MT-3 and MT-2, ICP-MS was employed. For both MT-3 and MT-2, quantification of the copper and zinc content by ICP-MS and analysis of the protein concentration allowed the Cu- and Zn-to-protein ratios in the final reaction products to be determined. These results show that the Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MT ratios were 8
MT ratios were 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mol mol−1 for both isoforms (8.5 ± 0.2 mol mol−1 for MT-3; 7.8 ± 0.2 mol mol−1 for MT-2), confirming the presence of Cu8+MT and suggesting the presence of two Cu4+ clusters, one in each domain. The somewhat lower Cu+ stoichiometry of MT-3 that was determined with ITC measurements is likely due to a small amount of protein oxidation. This would lower both the number of Cu+ that bind and the H+ that are displaced but not significantly affect the Cu+ affinity and binding thermodynamics, which are determined on a per Cu+ basis. Finally, zinc was not detected in the reaction products (<0.1 mol mol−1 for both MT-3 and MT-2), in agreement with Cu+ binding to apo MTs under the conditions used for ITC measurements.
1 mol mol−1 for both isoforms (8.5 ± 0.2 mol mol−1 for MT-3; 7.8 ± 0.2 mol mol−1 for MT-2), confirming the presence of Cu8+MT and suggesting the presence of two Cu4+ clusters, one in each domain. The somewhat lower Cu+ stoichiometry of MT-3 that was determined with ITC measurements is likely due to a small amount of protein oxidation. This would lower both the number of Cu+ that bind and the H+ that are displaced but not significantly affect the Cu+ affinity and binding thermodynamics, which are determined on a per Cu+ basis. Finally, zinc was not detected in the reaction products (<0.1 mol mol−1 for both MT-3 and MT-2), in agreement with Cu+ binding to apo MTs under the conditions used for ITC measurements.
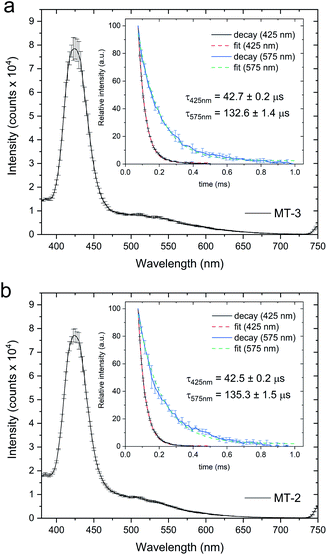
Cu4+–thiolate clusters are readily identified by intense and characteristic low temperature luminescence spectra and emission lifetimes. For Cu4+-cluster(s) in MTs, this includes a sharp emission band at 425 nm with a triplet cluster-centered (CC) origin and a broad emission band at 560–575 nm with a triplet charge-transfer (CT) origin. The presence of the triplet CC band at 425 nm, with a characteristic lifetime, is indicative of short Cu+–Cu+ distances (<2.8 Å), d10–d10 overlap and metal–metal bonding, and is absent in clusters with higher nuclearity (i.e., Cu6+–thiolate clusters in MTs).82,115–117 Low temperature (77 K) luminescence measurements for both MT-3 and MT-2 samples prepared by Cu+ addition to Zn7MT in excess GSH show emission features at 425 and 575 nm (Fig. 5), with the emission peak at 425 nm diagnostic of Cu4+–thiolate cluster formation.82,83 The lifetimes of these emission bands in both protein samples (Fig. 5, inserts) are also consistent with Cu4+ clusters. Thus, Cu+ titrations of Zn7MT-3 and Zn7MT-2 in the presence of excess GSH result in the formation of β(Cu+)4α(Cu+)4MT-3 and β(Cu+)4α(Cu+)4MT-2.
Conclusions
Biological implications from Zn2+ and Cu+ binding thermodynamics
A goal of in vitro measurements of protein coordination chemistry is to provide molecular insight on the in vivo binding of metals. A putative biological role for MT-3 is scavenging copper, which could be Cu+ in neuronal cells or Cu2+ from intracellular proteins under conditions of excess copper or from extracellular neuronal tissue.61,80,81,118,119 The thermodynamics of Cu2+ scavenging are complicated by reduction of the Cu2+ by Cys residues of MT-3, or other thiol-containing species,70 prior to Cu+ binding to the protein, which then contains oxidized Cys disulfide bonds. Calorimetric measurements for this proposed binding are difficult to interpret as they involve both redox chemistry and metal ion binding. Efforts to determine the formation thermodynamics of the native Cu4Zn3–4MT-3 through selective chelation of Zn2+ with EDTA and Cu+ with BCS in ITC measurements have so far proven to be challenging. Nevertheless, by individually quantifying the thermodynamics of Cu+ and Zn2+ binding to MT-3, we can determine the competition between these two metal ions for the protein. Table 6 compares the Cu+ and Zn2+ binding thermodynamics at pH 7.4, thereby quantifying Cu+ displacement of Zn2+ as the difference between their MT-3 binding thermodynamics. This reveals that the entropically favoured binding of Zn2+ (−TΔΔS = +12 ± 1 kcal mol−1) is overwhelmed by the enthalpically favoured binding of Cu+ (ΔΔH = −23 ± 1 kcal mol−1) due to the larger enthalpy of Cu+–thiolate bonds. These results support and quantify the proposed in vivo role for MT-3 in modulating the bioavailability of Cu+ ions.| Metal ion | n | K | ΔG° (kcal mol−1) | ΔH° (kcal mol−1) | −TΔS° (kcal mol−1) |
|---|---|---|---|---|---|
| Zn2+ | 6.8 ± 0.3 | 4 (±2) × 1011 | −15.7 ± 0.3 | 13.4 ± 0.2 | −29 ± 0.5 |
| Cu+ | 6 ± 1 | 4 (±4) × 1019 | −27.1 ± 0.4 | −10 ± 1 | −17 ± 1 |
| Difference: Cu+ − Zn2+ | — | — | −11.4 ± 0.4 | −23 ± 1 | +12 ± 1 |
The presence of an unusually air stable Cu4+–thiolate cluster in the β-domain of MT-3 is thought to be important in its ability to detoxify Cu2+in vivo.82 The Cu+ clusters formed in the samples of this study differ in important ways from those formed in the reaction of MT-3 and Cu2+, as the binding of Cu+ does not require the concomitant oxidation of MT-3. However, the resulting Cu+4–thiolate cluster, which is thought to form an intriguing adamantane-like shape,120 must be similar, whether formed with Cu2+in vivo or by binding Cu+in vitro, given the similarities in their luminescence spectra.82 The approximately ten-fold difference in Cu+ affinity between MT-3 and MT-2 in scavenging reactions, where Cu2+ is reduced and bound by the protein, as measured by Calvo et al.,77 likely reflects the formation of intramolecular disulfide bonds and is the subject of ongoing studies.
Data availability
The data supporting this article has either been included in the main body of the article or uploaded as part of the ESI.†Author contributions
MRM, RE, RL, RLEV, MO performed the experiments and helped analyse the data. CLV, GM, DEW, and RNA oversaw the experimental work and supervised the data analysis and drafted the manuscript. All authors reviewed and commented on multiple drafts.Conflicts of interest
There are no conflicts to declare.Acknowledgements
CLV and RNA thank former students Mehrose Ahmed, Yeliz Sezgin, Pia Chakravarty, Liz Irvin, Christina Juste, Laura Arbelaez, and Alison Milano for a great deal of hard work that helped to advance this project at various times and Dr Brandon Fowler and Dr Fereshteh Zandkarimi for help with MS measurements. Dr Jenifer Calvo is thanked for expressing and purifying MT-2. NSF grants CHE 1710176 (CLV and RNA) and CHE 1904705 (DEW) are gratefully acknowledged for supporting this work. GM is supported by the Robert A. Welch Foundation (AT-2073-20210327), the NIH (R35GM128704) and the NSF (CHE 2045984).Notes and references
- N. Romero-Isart and M. Vašák, Advances in the structure and chemistry of metallothioneins, J. Inorg. Biochem., 2002, 88, 388–396 CrossRef CAS PubMed.
- M. Capdevila, R. Bofill, O. Palacios and S. Atrian, State-of-the-art of metallothioneins at the beginning of the 21st century, Coord. Chem. Rev., 2012, 256, 46–62 CrossRef CAS.
- H.-J. Hartmann and U. Weser, Metallothioneins, in Biological Inorganic Chemistry, ed. I. Bertini, H. B. Gray, E. I. Stiefel, and J. S. Valentine, University Science Books, Sausalito, 2007, pp. 156–174 Search PubMed.
- A. Krężel and W. Maret, The functions of metamorphic metallothioneins in zinc and copper metabolism, Int. J. Mol. Sci., 2017, 18, 1237–1257 CrossRef PubMed.
- K. Zangger, G. Öz, J. D. Otvos and I. M. Armitage, Three-dimensional solution structure of mouse [Cd7]-metallothionein-l by homonuclear and heteronuclear NMR spectroscopy, Protein Sci., 1999, 8, 2630–2638 CrossRef CAS PubMed.
- B. A. Messerle, A. Schäffer, M. Vašák, J. H. R. Kägi and K. Wüthrich, Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy, J. Mol. Biol., 1990, 214, 765–779 CrossRef CAS PubMed.
- S. Bolognin, B. Cozzi, P. Zambenedetti and P. Zatta, Metallothioneins and the central nervous system: From a deregulation in neurodegenerative diseases to the development of new therapeutic approaches, J. Alzheimer’s Dis., 2014, 41, 29–42 CAS.
- E. J. Kelly and R. D. Palmiter, A murine model of Menkes disease reveals a physiological function of metallothionein, Nat. Genet., 1996, 13, 219–222 CrossRef CAS PubMed.
- C. M. Ackerman and C. J. Chang, Copper signaling in the brain and beyond, J. Biol. Chem., 2018, 293, 4628–4635 CrossRef CAS PubMed.
- N. O. Nartey, J. V. Frei and M. G. Cherian, Hepatic copper and metallothionein distribution in Wilson’s disease (hepatolenticular degeneration), Lab. Invest., 1987, 57, 397–401 CAS.
- W. E. Evering, S. Haywood, I. Bremner, A. M. Wood and J. Trafford, The protective role of metallothionein in copper-overload: II. Transport and excretion of immunoreactive MT-1 in blood, bile and urine of copper-loaded rats, Chem.-Biol. Interact., 1991, 78, 297–305 CrossRef CAS PubMed.
- Y. Luo, Y. Xu, Q. Bao, Z. Ding, C. Zhu, Z. X. Huang and X. Tan, The molecular mechanism for human metallothionein-3 to protect against the neuronal cytotoxicity of Aβ(1-42) with Cu ions, J. Biol. Inorg Chem., 2013, 18, 39–47 CrossRef CAS PubMed.
- Y. J. Kang, Metallothionein redox cycle and function, Exp. Biol. Med., 2006, 231, 1459–1467 CrossRef CAS PubMed.
- W. Maret and B. L. Vallee, Thiolate ligands in metallothionein confer redox activity on zinc clusters, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 3478–3482 CrossRef CAS PubMed.
- Y. Uchida, F. Gomi, T. Masumiza and Y. Miura, Growth inhibitory factor prevents neurite extension and the death of cortical neurons caused by high oxygen exposure through hydroxyl radical scavenging, J. Biol. Chem., 2002, 277, 32353–32359 CrossRef CAS PubMed.
- B. Ruttkay-Nedecky, L. Nejdjl, J. Gumulec, O. Zitka, M. Masarik, T. Eckschlager, M. Stiborova, V. Adam and R. Kizek, The role of metallothionein in oxidative stress, Int. J. Mol. Sci., 2013, 14, 6044–6066 CrossRef CAS PubMed.
- Y. Chen, Y. Irie, W. M. Keung and W. Maret, S-Nitrosothiols react preferentially with zinc thiolate clusters of metallothionein III through transnitrosation, Biochemistry, 2002, 41, 8360–8367 CrossRef CAS PubMed.
- D. L. Pountney, T. C. Dickson, J. H. Power, J. C. Vickers, A. J. West and W. P. Gai, Association of metallothionein-III with oligodendroglial cytoplasmic inclusions in multiple system atrophy, Neurotoxic. Res., 2011, 19, 115–122 CrossRef CAS PubMed.
- R. S. Chung, C. Howells, E. D. Eaton, L. Shabala, K. Zovo, P. Palumaa, R. Sillard, A. Woodhouse, W. R. Bennet, S. Ray, J. C. Vickers and A. K. West, The native copper- and zinc-binding protein metallothionein blocks copper-mediated Aβ aggregation and toxicity in rat cortical neurons, PLoS One, 2010, 5, e12030 CrossRef PubMed.
- R. S. Chung, M. Penkowa, J. Dittmann, C. E. King, C. Bartlett, J. W. Asmussen, J. Hidalgo, J. Carrasco, Y. K. J. Leung, A. K. Walker, S. J. Fung, S. A. Dunlop, M. Fitzgerald, L. D. Beazley, M. I. Chuah, J. C. Vickers and A. K. West, Redefining the role of metallothionein within the injured brain: extracellular metallothioneins play an important role in the astrocyte-neuron response to injury, J. Biol. Chem., 2008, 283, 15349–15358 CrossRef CAS PubMed.
- A. K. West, J. Hidalgo, D. Eddins, E. D. Levin and M. Aschner, Metallothionein in the central nervous system: Roles in protection, regeneration and cognition, Neurotoxicology, 2008, 29, 489–503 CrossRef CAS PubMed.
- C. R. A. Santos, A. Martimho, T. Quintela and I. Gonçalves, Neuroprotective and neurogenerative properties of metallothioneins, IUBMB Life, 2012, 64, 126–135 CrossRef CAS PubMed.
- E. Atrián-Blasco, A. Santoro, D. L. Pountney, G. Meloni, C. Hureau and P. Faller, Chemistry of mammalian metallothioneins and their interaction with amyloidogenic peptides and proteins, Chem. Soc. Rev., 2017, 46, 7683–7693 RSC.
- Y. Irie and W. M. Keung, Metallothionein-III antagonizes the neurotoxic and neurotrophic effects of amyloid β peptides, Biochem. Biophys. Res. Commun., 2001, 282, 416–420 CrossRef CAS PubMed.
- F. Ma, H. Wang, B. Chen, F. Wang and H. Xu, Metallothionein 3 attenuated the apoptosis of neurons in the CA1 region of the hippocampus in the senescence-accelerated mouse/PRONE8 (SAMP8), Arq Neuropsiquiart, 2011, 69, 105–111 CrossRef PubMed.
- R. S. Chung, J. Hidalgo and A. K. West, New insight into the molecular pathways of metallothionein-mediated neuroprotection and regeneration, J. Neurochem., 2008, 104, 14–20 CAS.
- J.-Y. Koh and S.-J. Lee, Metallothionein-3 as a multifunctional player in the control of cellular processes and diseases, Mol. Brain, 2020, 13, 116 CrossRef CAS PubMed.
- K. S. Vignesh and G. S. Deepe Jr, Metallothioneins: Emerging Modulators in Immunity and Infection, Int. J. Mol. Sci., 2017, 18, 2197 CrossRef PubMed.
- C. Howells, A. K. West and R. S. Chung, Neuronal growth-inhibitory factor (metallothionein-3): evaluation of the biological function of growth-inhibitory factor in the injured and neurodegenerative brain, FEBS J., 2010, 277, 2931–2939 CrossRef CAS PubMed.
- E. Artells, O. Palacios and S. Atrian, In vivo-folded metal-metallothionein 3 complexes reveal the Cu–thionein rather than Zn–thionein character of this brain-specific mammalian metallothionein, FEBS J., 2014, 281, 1659–1678 CrossRef CAS PubMed.
- S. Krizkova, M. Ryvolova, J. Hrabeta, V. Adam, M. Stilborova, T. Eckschlager and R. Kizek, Metallothioneins and zinc in cancer diagnosis and therapy, Drug Metab. Rev., 2012, 44, 287–301 CrossRef CAS PubMed.
- S.-J. Lee, B.-R. Seo and J.-Y. Koh, Metallothionein-3 modulates the amyloid β endocytosis of astrocytes through its effects on actin polymerization, Mol. Brain, 2015, 8, 84–95 CrossRef PubMed.
- D. Chowdhury, H. Alrefai, J. A. Landero Figueroa, K. Candor, A. Porollo, R. Fecher, S. Divanovic, G. S. Deepe Jr and K. Subramanian Vignesh, Metallothionein 3 Controls the Phenotype and Metabolic Programming of Alternatively Activated Macrophages, Cell Rep., 2019, 27, 3873–3886 CrossRef CAS PubMed.
- R. D. Palmiter, Constitutive expression of metallothionein-III (MT-III), but not MT-I, inhibits growth when cells become zinc deficient, Toxicol. Appl. Pharmacol., 1995, 135, 139–146 CrossRef CAS PubMed.
- J. H. Kaplan and E. B. Maryon, How mammalian cells acquire copper: An essential but potentially toxic metal, Biophys. J., 2016, 110, 7–13 CrossRef CAS PubMed.
- L. Banci, I. Bertini, S. Ciofi-Baffoni, T. Kozyreva, K. Zovo and P. Palumaa, Affinity gradients drive copper to cellular destinations, Nature, 2010, 465, 645–648 CrossRef CAS PubMed.
- K. J. Waldron, J. C. Rutherford, D. Ford and N. J. Robinson, Metalloproteins and metal sensing, Nature, 2009, 460, 823–830 CrossRef CAS PubMed.
- J. Wątły, M. Łuczkowski, M. Padjasek and A. Krężel, Phytochelatins as a dynamic system for Cd(II) buffering from the micro- to femtomolar range, Inorg. Chem., 2021, 60, 4657–4675 CrossRef PubMed.
- M. L. Turski and D. J. Thiele, New roles for copper metabolism in cell proliferation, signaling, and disease, J. Biol. Chem., 2009, 284, 717–721 CrossRef CAS PubMed.
- W. Maret, Metals on the move: zinc ions in cellular regulation and in the coordination dynamics of zinc proteins, BioMetals, 2011, 24, 411–418 CrossRef CAS PubMed.
- N. J. Robinson and D. R. Winge, Copper metallochaperones, Annu. Rev. Biochem., 2010, 79, 537–562 CrossRef CAS PubMed.
- C. E. Outten and T. V. O’Halloran, Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis, Science, 2001, 292, 2488–2492 CrossRef CAS PubMed.
- D. Osman, M. A. Martini, A. W. Foster, J. Chen, A. J. P. Scott, R. J. Morton, J. W. Steed, E. Lurie-Luke, T. G. Huggins, A. D. Lawrence, E. Deery, M. J. Warren, P. T. Chivers and N. J. Robinson, Bacterial sensors define intracellular free energies for correct enzyme metalation, Nat. Chem. Biol., 2019, 15, 241–249 CrossRef CAS PubMed.
- T. D. Rae, P. J. Schmidt, R. A. Pufahl, V. C. Culotta and T. V. O’Halloran, Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase, Science, 1999, 284, 805–808 CrossRef CAS PubMed.
- M. T. Morgan, D. Bourassa, S. Harankhedkar, A. M. McCallum, S. A. Zlatic, J. S. Calvo, G. Meloni, V. Faundez and C. J. Fahrni, Ratiometric two-photon microscopy reveals attomolar copper buffering in normal and Menkes mutant cells, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 12167–12172 CrossRef CAS PubMed.
- K. P. Carter, A. M. Young and A. E. Palmer, Fluorescent sensors for measuring metal ions in living systems, Chem. Rev., 2014, 114, 4564–4601 CrossRef CAS PubMed.
- M. E. Helsel and K. J. Franz, Pharmacological activity of metal binding agents that alter copper bioavailability, Dalton Trans., 2015, 44, 8760–8770 RSC.
- M. C. Carpenter, A. Shami Shah, S. DeSilva, A. Gleaton, A. Su, B. Goundie, M. L. Croteau, M. J. Stevenson, D. E. Wilcox and R. N. Austin, Thermodynamics of Pb(II) and Zn(II) binding to MT-3, a neurologically important metallothionein, Metallomics, 2016, 8, 605–617 CrossRef CAS PubMed.
- M. Vašák and G. Meloni, Metallothionein-3, zinc, and copper in the central nervous system, Met. Ions Life Sci., 2009, 5, 319–351 Search PubMed.
- C. A. Sogawa, M. Asanuma, N. Sogawa, I. Miyazaki, T. Nakanishi, H. Furuta and N. Ogawa, Localization, regulation, and function of metallothionein-III/growth inhibitory factor in the brain, Acta Med. Okayama, 2001, 55, 1–9 CAS.
- M. Knapp, G. Meloni, B. Roschitski and M. Vašák, Zn7Metallothionein-3 and the synaptic vesicle cycle: Interaction of metallothionein-3 with the small GTPase Rab3A, Biochemistry, 2005, 44, 3159–3165 CrossRef PubMed.
- K.-H. Tsui, C.-P. Hou, K.-S. Chang, Y.-H. Lin, T.-H. Feng, C.-C. Chen, Y.-S. Shin and H.-H. Juang, Metallothionein 3 is a hypoxia-upregulated oncogene enhancing cell invasion and tumorigenesis in human bladder carcinoma cells, Int. J. Mol. Sci., 2019, 20, 980–995 CrossRef CAS PubMed.
- J. C. Erickson, G. Hollopeter, S. A. Thomas, G. J. Froelick and R. D. Palmiter, Disruption of the metallothionein-III gene in mice: Analysis of brain zinc, behavior, and neuron vulnerability to metals, aging, and seizures, J. Neurosci., 1997, 17, 1271–1281 CrossRef CAS PubMed.
- Y. Manso, J. Carrasco, G. Comes, G. Meloni, P. A. Adlard, A. I. Bush, M. Vašák and J. Hidalgo, Characterization of the role of metallothionein-3 in an animal model of Alzheimer’s disease, Cell. Mol. Life Sci., 2012, 69, 3683–3700 CrossRef CAS PubMed.
- Y. Uchida, K. Takio, K. Titani, Y. Ihara and M. Tomonaga, The growth inhibitory factor that is deficient in the Alzheimer's disease brain is a 68 amino acid metallothionein-like protein, Neuron, 1991, 7, 337–347 CrossRef CAS PubMed.
- Y. Uchida and M. Tomonaga, Neurotrophic action of Alzheimer’s disease brain extract is due to the loss of inhibitory factors for survival and neurite formation of cerebral cortical neurons, Brain Res., 1989, 481, 190–193 CrossRef CAS.
- Z.-C. Ding, F.-Y. Ni and Z.-X. Huang, Neuronal growth-inhibitory factor (metallothionein-3): structure-function relationships, FEBS J., 2010, 277, 2912–2920 CrossRef CAS PubMed.
- B. A. Masters, C. J. Quaife, J. C. Erickson, E. J. Kelly, G. J. Froelick, B. P. Zambrowicz, R. L. Brinster and R. D. Palmiter, Metallothionein III is expressed in vesicles that sequester zinc in synaptic vesicles, J. Neurosci., 1994, 14, 5844–5857 CrossRef CAS PubMed.
- M. Vašák and G. Meloni, Mammalian metallothionein-3: new functional and structural insights, Int. J. Mol. Sci., 2017, 18, 1117 CrossRef PubMed.
- S.-J. Lee, M. H. Park, H. J. Kim and J.-Y. Koh, Metallothionein-3 regulates lysosomal function in cultured astrocytes under both normal and oxidative conditions, Glia, 2010, 58, 1186–1196 CrossRef PubMed.
- G. Meloni, V. Sonois, T. Delaine, L. Guilloreau, A. Gillet, J. Teissie, P. Faller and M. Vašak, Metal swap between Zn7-metallothionein-3 and amyloid β-Cu protects against amyloid-β toxicity, Nat. Chem. Biol., 2008, 4, 366–372 CrossRef CAS PubMed.
- G. Meloni, M. Knipp and M. Vašák, Detection of neuronal growth inhibitory factor (metallothionein-3) in polyacrylamide gels and by western blot analysis, J. Biochem. Biophys. Methods, 2005, 64, 76–81 CrossRef CAS PubMed.
- P. Palumaa, E. Eriste, O. Njunkova, L. Pokras, H. Jornvall and R. Sillard, Brain-specific metallothionein-3 has higher metal-binding capacity than ubiquitous metallothioneins and binds metals noncooperatively, Biochemistry, 2002, 41, 6158–6163 CrossRef CAS PubMed.
- P. Palumaa, I. Tammiste, K. Kruusel, L. Kangur, H. Jornvall and R. Sillard, Metal binding of metallothionein-3 versus metallothionein-2: lower affinity and higher plasticity, Biochem. Biophys. Acta., 2005, 1747, 205–211 CrossRef CAS PubMed.
- A. K. Sewell, L. T. Jensen, J. C. Erickson, R. D. Palmiter and D. R. Winge, Bioactivity of metallothionein-3 correlates with its novel β domain sequence rather than metal binding properties, Biochemistry, 1995, 34, 4740–4747 CrossRef CAS PubMed.
- S. Somji, S. H. Garrett, M. A. Sens and D. E. Sens, The unique N-terminal sequence of metallothionein-3 is required to regulate the choice between apoptotic or necrotic cell death of human proximal tubule cells exposed to Cd(II), Toxicol. Sci., 2006, 90, 369–376 CrossRef CAS PubMed.
- S. Toriumi, T. Saito, T. Hosokawa, T. Takashi, T. Numata and M. Kuraski, Metal binding ability of metallothionein-3 expressed in Escherichia coli, Basic Clin. Pharmacol. Toxicol., 2005, 96, 295–301 CrossRef CAS PubMed.
- J. Bousleiman, A. Pinsky, S. Ki, A. Su, I. Morozova, S. Kalachikov, A. Wiqas, R. Silver, M. Sever and R. N. Austin, Function of metallothionein-3 in neuronal cells: Do metal ions alter expression levels of MT3?, Int. J. Mol. Sci., 2017, 18, 1133 CrossRef PubMed.
- B. Sullivan, G. Robison, J. Osborn, M. Kay, P. Thompson, K. Davis, T. Zakharova, O. Antipova and Y. Pushkar, On the nature of the Cu-rich aggregates in brain astrocytes, Redox Biol., 2017, 11, 231–239 CrossRef CAS PubMed.
- A. Santoro, N. E. Wezynfeld, M. Vašák, W. Bal and P. Faller, Cysteine and glutathione trigger the Cu–Zn swap between Cu(II)-amyloid-β(4-16) and Zn7-metallothionein-3, Chem. Commun., 2017, 53, 11634–11637 RSC.
- B. Wang, I. S. Wood and P. Trayhurn, PCR arrays identify metallothionein-3 as a highly hypoxia-inducible gene in human adipocytes, Biochem. Biophys. Res. Commun., 2008, 368, 88–93 CrossRef CAS PubMed.
- S. Yanagitani, H. Miyazaki, Y. Nakahashi, K. Kuno, Y. Ueno, M. Matsushita, Y. Naitoh, S. Taketani and K. Inoue, Ischemia Induces metallothionein III expression in neurons of rat brains, Life Sci., 1999, 64, 707–715 CrossRef CAS.
- K. Tanji, Y. Irie, Y. Uchida, F. Mori, K. Satoh, Y. Mizushima and K. Wakabayashi, Expression of metallothionein-III induced by hypoxia attenuates hypoxia-induced cell death in vitro, Brain Res., 2003, 976, 125–129 CrossRef CAS PubMed.
- S. Atrian, Ò. Palacios and M. Capdevila, Metallothioneins and Copper, in Encyclopedia of Metalloproteins, ed. R. H. Kretsinger, V. N. Uversky and E. A. Permyakov, Springer New York, New York, NY, 2013, pp. 1379–1383 Search PubMed.
- S. L. Hands, R. Mason, M. U. Sajjad, F. Giorgini and A. Wyttenbach, Metallothioneins and copper metabolism are candidate therapeutic targets in Huntington’s disease, Biochem. Soc. Trans., 2010, 38, 552–558 CrossRef CAS PubMed.
- J. Calvo, H. Jung and G. Meloni, Copper metallothioneins, IUBMB Life, 2017, 69, 236–245 CrossRef CAS PubMed.
- J. S. Calvo, V. M. Lopez and G. Meloni, Non-coordinative metal selectivity bias in human metallothioneins metal–thiolate clusters, Metallomics, 2018, 10, 1777–1791 CrossRef CAS PubMed.
- D. L. Pountney, S. M. Fundel, P. Faller, N. E. Birchler, P. E. Hunziker and M. Vašák, Isolation, primary structures, and metal binding properties of neuronal growth inhibitory factor (GIF) from bovine and equine brain, FEBS Lett., 1994, 345, 193–197 CrossRef CAS PubMed.
- R. Bogumil, P. Faller, D. L. Pountney and M. Vašák, Evidence for Cu(I) clusters and Zn(II) clusters in neuronal growth-inhibitory factor isolated from bovine brain, Eur. J. Biochem., 1996, 238, 698–705 CrossRef CAS PubMed.
- G. Meloni, P. Faller and M. Vašák, Redox silencing of copper in metal-linked neurodegenerative disorders: Reaction of Zn7Metallotheionein-3 with Cu(II) Ions, J. Biol. Chem., 2007, 282, 16068–16078 CrossRef CAS PubMed.
- J. S. Calvo, N. V. Mulpuri, A. Dao, N. K. Qaxi and G. Meloni, Membrane insertion exacerbates the α-Synuclein-Cu(II) dopamine oxidase activity: Metallothionein-3 targets and silences all α-synuclein-Cu(II) complexes, Free Radicals Biol. Med., 2020, 158, 149–161 CrossRef CAS PubMed.
- J. S. Calvo, R. L. E. Villones, N. J. York, E. Stefaniak, G. E. Hamilton, A. L. Stelling, W. Bal, B. S. Pierce and G. Meloni, Evidence for a long-lived, Cu-coupled and oxygen-inert disulfide radical anion in the assembly of metallothionein-3 Cu(I)4–thiolate cluster, J. Am. Chem. Soc., 2022, 144, 709–722 CrossRef CAS PubMed.
- B. Roschitzki and M. Vašák, A distinct Cu4–thiolate cluster of human metallothionein-3 is located in the N-terminal domain, J. Biol. Inorg Chem., 2002, 7, 611–616 CrossRef CAS PubMed.
- C. Li, A. Wen, B. Shen, J. Lu, Y. Huang and Y. Chang, FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method, BMC Biotechnol., 2011, 11, 92 CrossRef CAS PubMed.
- E. M. Kosower and N. S. Kosower, Bromobimane probes for thiols, Methods Enzymol., 1995, 251, 133–146 CAS.
- G. Meloni, T. Polanski, O. Braun and M. Vašák, Effects of Zn(II), Ca(II), and Mg(II) on the structure of Zn7metallothionein-3: evidence for an additional zinc binding site, Biochemistry, 2009, 48, 5700–5707 CrossRef CAS PubMed.
- A. O. Pedersen and J. Jacobsen, Reactivity of the thiol group in human and bovine albumin at pH 3–9, as measured by exchange with 2,2′-dithiodipyridine, Eur. J. Biochem., 1980, 106, 291–295 CrossRef CAS PubMed.
- P. Faller, D. W. Hasler, O. Zerbe, S. Klauser, D. R. Winge and M. Vašák, Evidence for a dynamic structure of human neuronal growth inhibitory factor and for major rearrangements of its metal–thiolate clusters, Biochemistry, 1999, 38, 10158–10167 CrossRef CAS PubMed.
- M. Vašák, Metal removal and substitution in vertebrate and invertebrate metallothioneins, Methods Enzymol., 1991, 205, 452–458 Search PubMed.
- C. F. Quinn, M. C. Carpenter, M. L. Croteau and D. E. Wilcox, Isothermal Titration Calorimetry Measurements of Metal Ions Binding to proteins, Methods Enzymol., 2016, 567, 3–20 CAS.
- M. C. Carpenter and D. E. Wilcox, Thermodynamics of Formation of the Insulin Hexamer: Metal-stabilized Proton-Coupled Assembly of Quaternary Structure, Biochemistry, 2014, 53, 1296–1301 CrossRef CAS PubMed.
- D. K. Johnson, M. J. Stevenson, Z. A. Almadidy, S. E. Jenkins, D. E. Wilcox and N. E. Grossoehme, Stabilization of Cu(I) for binding and calorimetric measurements in aqueous solution, Dalton Trans., 2015, 44, 16494–16505 RSC.
- A. Krężel and W. Maret, Dual nanomolar and picomolar Zn(II) binding properties of metallothionein, J. Am. Chem. Soc., 2007, 129, 10911–10921 CrossRef PubMed.
- D. P. Jayawardena, I. U. Heinemann and M. J. Stillman, Zinc binds non-cooperatively to human liver metallothionein 2a at physiological pH, Biochem. Biophys. Res. Commun., 2017, 493, 650–653 CrossRef CAS PubMed.
- M. D. Peris-Díaz, R. Guran, C. Domene, V. de los Rios, O. Zitka, V. Adam and A. Krężel, An integrated mass spectrometry and molecular dynamics simulations approach reveals the spatial organization impact of metal-binding sites on the stability of metal-depleted metallothionein-2 species, J. Am. Chem. Soc., 2021, 143, 16486–16501 CrossRef PubMed.
- D. W. Hasler, L. T. Jensen, O. Zerbe, D. TR. Winge and M. Vašák, Effect of the two conserved prolines of human growth inhibitory factor (metallothionein-3) on its biological activity and structure fluctuations: comparison with a mutant protein, Biochemistry, 2000, 39, 14567–14575 CrossRef CAS PubMed.
- B. Roschitzki and M. Vašák, Redox labile site in a Zn4 cluster of Cu4,Zn4-metallothionein-3, Biochemistry, 2003, 42, 9822–9828 CrossRef CAS PubMed.
- A. R. Reddi, T. R. Guzman, R. M. Breece, D. L. Tierney and B. R. Gibney, Deducing the energetic cost of protein folding in zinc finger proteins using designed metallopeptides, J. Am. Chem. Soc., 2007, 129, 12815–12827 CrossRef CAS PubMed.
- A. M. Rich, E. Bombarda, A. D. Schenk, P. E. Lee, E. H. Cox, A. M. Spuches, L. D. Hudson, B. Keifer and D. E. Wilcox, Thermodynamics of Zn2+ binding to Cys2His2 and Cys2HisCys zinc fingers and a Cys4 transcription factor site, J. Am. Chem. Soc., 2012, 134, 10405–10418 CrossRef CAS PubMed.
- A. Tamura and P. L. Privalov, The entropy cost of protein association, J. Mol. Biol., 1997, 273, 1048–1060 CrossRef CAS PubMed.
- M. K. Gilson, J. A. Given, B. L. Bush and J. A. McCammon, The statistical-thermodynamic basis for computation of binding affinities: A critical review, Biophys. J., 1997, 72, 1047–1069 CrossRef CAS PubMed.
- L.-J. Jiang, M. Vašák, B. L. Vallee and W. Maret, Zinc transfer potentials of the α- and β-clusters of metallothionein are affected by domain interactions in the whole molecule, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 2503–2508 CrossRef CAS PubMed.
- A. Drozd, D. Wojewska, M. D. Peris-Díaz, P. Jakimowicz and A. Krężel, Crosstalk of the structural and zinc buffering properties of mammalian metallothionein-2, Metallomics, 2018, 10, 595–613 CrossRef CAS PubMed.
- M. J. Walsh and B. A. Ahner, Determination of stability constants of Cu(I), Cd(II), and Zn(II) complexes with thiols using fluorescent probes, J. Inorg. Biochem., 2013, 128, 112–123 CrossRef CAS PubMed.
- Z. Xiao, F. Loughlin, G. N. George, G. J. Howlett and A. G. Wedd, C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) Ions as a cuprous–thiolate polynuclear cluster: Sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking, J. Am. Chem. Soc., 2004, 126, 3081–3090 CrossRef CAS PubMed.
- A. Badarau and C. Dennison, Thermodynamics of copper and zinc distribution in the cyanobacterium Synechocystis PCC 6803, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 13007–13012 CrossRef CAS PubMed.
- Z. Xiao, J. Brose, S. Schimo, S. M. Ackland, S. La Fontaine and A. G. Wedd, Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: Detection probes and affinity standards, J. Biol. Chem., 2011, 286, 11047–11055 CrossRef CAS PubMed.
- A. Presta, A. R. Green, A. Zelazowski and M. J. Stillman, Copper binding to rabbit liver metallothionein. Formation of a continuum of copper(I)–thiolate stoichiometric species, Eur. J. Biochem., 1995, 227, 226–240 CrossRef CAS PubMed.
- M. Salgado, K. L. Bacher and M. J. Stillman, Probing structural changes in the α and β domains of copper- and silver-substituted metallothionein by emission spectroscopy and electrospray ionization mass spectrometry, J. Biol. Inorg Chem., 2007, 12, 294–312 CrossRef CAS PubMed.
- J. S. Scheller, G. W. Irvine, D. L. Wong, A. Hartwig and M. J. Stillman, Stepwise copper(I) binding to metallothionein: a mixed cooperative and non-cooperative mechanism for all 20 copper ions, Metallomics, 2017, 9, 447–462 CrossRef CAS PubMed.
- M. J. Stillman, D. Thomas, C. Trevithick, X. Guo and M. Siu, Circular dichroism, kinetic and mass spectrometric studies of copper(I) and mercury(II) binding to metallothionein, J. Inorg. Biochem., 2000, 79, 11–19 CrossRef CAS PubMed.
- A. Melenbacher, N. C. Korkola and M. J. Stillman, The pathways and domain specificity of Cu(I) binding to human metallothionein 1A, Metallomics, 2020, 12, 1951–1964 CrossRef CAS PubMed.
- Y.-M. Go and D. P. Jones, Redox compartmentalization in eukaryotic cells, Biochim. Biophys. Acta, 2008, 1780, 1273–1290 CrossRef CAS PubMed.
- A. Santoro, B. Vileno, Ò. Palacios, M. D. Peris-Diaz, G. Riegel, C. Gaidoon, A. Krężel and P. Faller, Reactivity of Cu(II)-, Zn(II)- and Fe(II)–thiosemicarbazone complexes with glutathione and metallothioenein: from stability to dissociation to transmetallation, Metallomics, 2019, 11, 994–1004 CrossRef CAS PubMed.
- D. L. Pountney, I. Schauwecker, J. Zarn and M. Vašák, Formation of mammalian Cu8–metallothionein in vitro: Evidence for the existence of two Cu(I)4–thiolate clusters, Biochemistry, 1994, 33, 9699–9705 CrossRef CAS PubMed.
- P. Faller and M. Vašák, Distinct metal–thiolate clusters in the N-terminal domain of neuronal growth inhibitory factor, Biochemistry, 1997, 36, 13341–13348 CrossRef CAS PubMed.
- D. W. Hasler, P. Faller and M. Vašák, Metal–thiolate clusters in the C-terminal domain of human neuronal growth inhibitory factor (GIF), Biochemistry, 1998, 37, 14966–14973 CrossRef CAS PubMed.
- Y. Okita, A. N. Rcom-H’cheo-Gauthier, M. Goulding, R. S. Chung, P. Faller and D. L. Pountney, Metallothionein, copper and α-synuclein in α-synucleinopathies, Front. Neurosci., 2017, 11, 114 CrossRef PubMed.
- J. Kardos, L. Héja, Á. Simon, I. Jablonkai, R. Kovács and K. Jemnitz, Copper signalling: causes and consequences, Cell. Commun. Signal., 2018, 16, 71 CrossRef CAS PubMed.
- E. H. Griffith, G. W. Hunt and E. L. Amma, The Adamantane Structure in Polynuclear Cu4S6 cores: The crystal and molecular structures of Cu4[SC(NH2)2]6(NO3)4·4H2O and Cu4[SC(NH2)2]9(NO3)4·4H2O, J. Chem. Soc., Chem. Commun., 1976, 432–433 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Cloning constructs and protein sequences, supporting ITC data and analysis, tables of experimental ITC data, schemes of Hess's law analysis of binding enthalpy, analysis for K from chelation ITC, and ITC results for titrations with Cu+ stabilized by MeCN are provided in the ESI. See https://doi.org/10.1039/d2sc00676f |
| This journal is © The Royal Society of Chemistry 2022 |