Open Access Article
Open Access ArticleCu(II)-based DNA labeling identifies the structural link between transcriptional activation and termination in a metalloregulator†
Joshua
Casto
 a,
Alysia
Mandato
a,
Lukas
Hofmann
b,
Idan
Yakobov
b,
Shreya
Ghosh
a,
Alysia
Mandato
a,
Lukas
Hofmann
b,
Idan
Yakobov
b,
Shreya
Ghosh
 a,
Sharon
Ruthstein
a,
Sharon
Ruthstein
 *b and
Sunil
Saxena
*b and
Sunil
Saxena
 *a
*a
aDepartment of Chemistry, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, USA. E-mail: sksaxena@pitt.edu
bDepartment of Chemistry, Faculty of Exact Sciences, The Institution of Nanotechnology and Advanced Materials, Bar-Ilan University, Ramat-Gan 5290002, Israel. E-mail: Sharon.Ruthstein@biu.ac.il
First published on 17th January 2022
Abstract
Understanding the structural and mechanistic details of protein-DNA interactions that lead to cellular defence against toxic metal ions in pathogenic bacteria can lead to new ways of combating their virulence. Herein, we examine the Copper Efflux Regulator (CueR) protein, a transcription factor which interacts with DNA to generate proteins that ameliorate excess free Cu(I). We exploit site directed Cu(II) labeling to measure the conformational changes in DNA as a function of protein and Cu(I) concentration. Unexpectedly, the EPR data indicate that the protein can bend the DNA at high protein concentrations even in the Cu(I)-free state. On the other hand, the bent state of the DNA is accessed at a low protein concentration in the presence of Cu(I). Such bending enables the coordination of the DNA with RNA polymerase. Taken together, the results lead to a structural understanding of how transcription is activated in response to Cu(I) stress and how Cu(I)-free CueR can replace Cu(I)-bound CueR in the protein-DNA complex to terminate transcription. This work also highlights the utility of EPR to measure structural data under conditions that are difficult to access in order to shed light on protein function.
Introduction
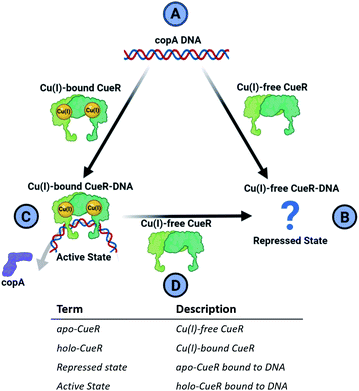
Metalloregulator proteins are transcription factors that perform a crucial role in the cellular defense of pathogenic bacteria by maintaining metal homeostasis.1 For example, while copper ions are essential for several important enzymes due to favorable redox properties, excess free copper ions induce cytotoxicity by participating in Fenton and Haber–Weiss-like reactions that produce biologically lethal hydroxyl radicals and carbonate ions.2,3 Cells have, therefore, evolved elaborate protective mechanisms to eliminate excess free copper. The copper efflux metalloregulator (CueR) in E. coli binds Cu(I) in the cell with an affinity of 1021 M and specific DNA sequences to activate the transcription of cueO or copA genes.4 The protein cueO oxidizes Cu(I) to less cytotoxic Cu(II), whereas copA is a transmembrane pump that exports free Cu(I) out of the cytosol. Single molecule fluorescence resonance energy transfer (smFRET) measurements4 indicate that the CueR transcription regulation mechanism can be thought of as a cycle depicted in Fig. 1.In this mechanism, the binding of Cu(I)-free CueR (i.e. apo CueR) to DNA putatively leads to a repressed state wherein transcription cannot occur (Fig. 1B). On the other hand, the binding of Cu(I)-bound CueR is critical to promoting the interaction with RNA polymerase (RNAp) to initiate transcription (Fig. 1C).5 Once Cu(I) concentrations fall below cytotoxic thresholds in the cell, transcription needs to be deactivated. Due to the 1021 M affinity4 of CueR to Cu(I), termination via dissociation of Cu(I) from CueR is unlikely. Alternatively, smFRET results suggest a substitution mechanism where Cu(I)-free CueR replaces Cu(I)-bound CueR on the DNA to end transcription (Fig. 1D).6
Crucially, therefore, the competition between Cu(I)-free CueR vs. Cu(I)-bound CueR for DNA is key to both activation and termination of transcription (Fig. 1C and D). However, despite much work there is still a lack of structural evidence that depicts how Cu(I)-free CueR participates in both repression and termination. Crystal7 and cryo-EM structures8,9 both show that Cu(I)-bound CueR bends the DNA. Alternatively, the crystal structure of the Cu(I)-free CueR–DNA complex7 indicates that the DNA is undistorted. These data pose an intriguing conundrum. How does Cu(I)-free CueR replace Cu(I)-bound CueR on a kinked duplex if it only coordinates to an undistorted duplex (Fig. 1B and D)?
Results and discussion
Herein we use pulsed Electron Paramagnetic Resonance (EPR) techniques to directly measure the conformational changes of the DNA at key steps of the regulation mechanism to elucidate the structural relationship between transcription activation, repression, and termination.Fig. 2A shows the DNA used for EPR measurements. The protein-binding sites of the copA DNA sequence are highlighted in grey. Each DNA strand contains a 2,2-dipicolylamine (DPA) phosphoramidite moiety that chelates Cu(II). The DPA residues, therefore, allow for attachment of two Cu(II) labels separated by 12 base pairs. The DPA-Cu(II) motif is a straightforward spin labeling technique that orients the DPA into the helix and reports directly on DNA backbone distances.10,11 Additionally, we have shown earlier that orientational selectivity12,13 effects for the DPA-Cu(II) motif are negligible even at Q-band, which enables the measurement of distances even from data at a single magnetic field.11 The labeling of DNA with Cu(II) is described in ESI.†11,14 Electrophoretic mobility shift assays (EMSA) show that CueR binds to the DPA-DNA duplex (Fig. S1†). Note that the native metal binding site in CueR coordinates only monovalent ions.15
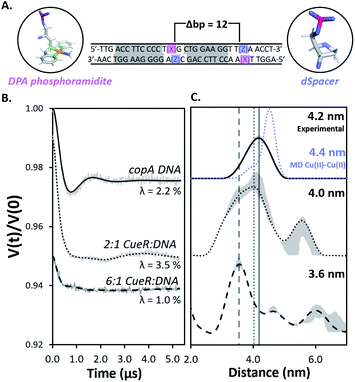 | ||
Fig. 2 (A) The 31 bp copA DNA sequence. DPA (pink) and dSpacer (blue) moieties were substituted into nucleotide positions that do not interact with CueR. The Cu(II) coordinates to DPA leading to two Cu(II) positioned 12 bp apart. The protein binding sites are highlighted in grey. (B) Background subtracted DEER time traces for the copA DNA and the duplex bound to CueR at different concentration ratios of protein to duplex. Modulation depths (λ) are shown. (C) The validated Cu(II)–Cu(II) distance distributions in copA DNA, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CueR 1 CueR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA, and 6 DNA, and 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CueR 1 CueR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA samples. The analysis was performed using DeerAnalysis. The Cu(II)–Cu(II) distance distribution from MD for copA DNA is presented as the blue dots. The MD and EPR distance distributions for copA DNA are normalized with respect to their probabilities. Upon increasing the ratio of protein to duplex from 2 DNA samples. The analysis was performed using DeerAnalysis. The Cu(II)–Cu(II) distance distribution from MD for copA DNA is presented as the blue dots. The MD and EPR distance distributions for copA DNA are normalized with respect to their probabilities. Upon increasing the ratio of protein to duplex from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 6 1 to 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the most probable distance decreases from 4.2 nm to 3.6 nm. The grey lines represent the most probable distance of each sample to provide a visual aid for comparison. The grey regions show the validated distribution from DEERAnalysis (details are provided in the ESI†). ComparativeDEERAnalysis consensus distance distributions are provided in the ESI (Fig. S7†). 1, the most probable distance decreases from 4.2 nm to 3.6 nm. The grey lines represent the most probable distance of each sample to provide a visual aid for comparison. The grey regions show the validated distribution from DEERAnalysis (details are provided in the ESI†). ComparativeDEERAnalysis consensus distance distributions are provided in the ESI (Fig. S7†). | ||
All EPR data were acquired in N-ethylmorpholine (NEM) buffer in order to ensure that free Cu(II) was EPR silent.16,17 Continuous wave (CW)-EPR experiments were performed to characterize sample Cu(II) coordination (cf. ESI Fig. S2†). Each CW spectrum consisted of a dominant component with hyperfine splitting characteristic of DPA-Cu(II) coordination.18 UV/vis measurements show no reduction of excess Cu(II) to Cu(I) (cf. Fig. S3†). Additional details of the sample preparation and experimental parameters are described further in the ESI.†
Next, double electron electron resonance19,20 (DEER) was performed to measure the distance distribution between the two Cu(II)–Cu(II) sites (cf.Fig. 1A). The primary DEER traces, basis EPR data, and biological repeats for all samples are provided in ESI (Fig. S4 and S5†). DEER data with optimal dipolar evolution times were achieved by using deuterated solvents and glycerol to increase the relaxation times of Cu(II) spin labels21 (Fig. S6†). First, DEER was performed on free DNA and then on the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CueR
1 CueR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA sample (Fig. 2B). The resulting distance distribution obtained from the background subtracted DEER time trace of the free duplex analyzed with DEERAnalysis22 returns a most probable distance centered at 4.2 nm (Fig. 2B). Such a distance is anticipated for a 12 base pair separation of a typical B-DNA and agrees well with a 2.25 μs molecular dynamics (MD) simulation of five averaged 450 ns replicates.
DNA sample (Fig. 2B). The resulting distance distribution obtained from the background subtracted DEER time trace of the free duplex analyzed with DEERAnalysis22 returns a most probable distance centered at 4.2 nm (Fig. 2B). Such a distance is anticipated for a 12 base pair separation of a typical B-DNA and agrees well with a 2.25 μs molecular dynamics (MD) simulation of five averaged 450 ns replicates.
The DEER time trace of the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 protein to DNA sample is distinctly different compared to the free DNA, which clearly indicates a different conformational ensemble for the DNA. An analysis of the distance distribution by DeerAnalysis reveals a bimodal distribution (Fig. 2C center panel). ComparativeDEERAnalysis consensus distance distributions utilizing DEERNet23 and automated Tikhonov regularization fitting show similar bimodal distributions as well (Fig. S7†). We attribute the larger distance at 5.7 nm to high order binding, where CueR oligomers and multiple DNA come together. Such high order binding is commonly seen in nucleoprotein complexes that require thermodynamically unfavorable distortion of the DNA.24,25 Therefore, two different DNA strands in the higher order nucleoprotein complex would give rise to a range of distances outside what is feasible for a 12 bp separation. When comparing the DEER traces from copA DNA and 2
1 protein to DNA sample is distinctly different compared to the free DNA, which clearly indicates a different conformational ensemble for the DNA. An analysis of the distance distribution by DeerAnalysis reveals a bimodal distribution (Fig. 2C center panel). ComparativeDEERAnalysis consensus distance distributions utilizing DEERNet23 and automated Tikhonov regularization fitting show similar bimodal distributions as well (Fig. S7†). We attribute the larger distance at 5.7 nm to high order binding, where CueR oligomers and multiple DNA come together. Such high order binding is commonly seen in nucleoprotein complexes that require thermodynamically unfavorable distortion of the DNA.24,25 Therefore, two different DNA strands in the higher order nucleoprotein complex would give rise to a range of distances outside what is feasible for a 12 bp separation. When comparing the DEER traces from copA DNA and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CueR–DNA time traces, the modulation depth increases from 2.2% to 3.5% (Fig. 2A). This increase in modulation depth is consistent with the presence of higher order complexes.26–28 Additionally, the EMSA data corroborates the presence of species with quadruplexes (Fig. S1†). In subsequent discussion, we focus on the more biophysically relevant distribution centered around 4.2 nm.
1 CueR–DNA time traces, the modulation depth increases from 2.2% to 3.5% (Fig. 2A). This increase in modulation depth is consistent with the presence of higher order complexes.26–28 Additionally, the EMSA data corroborates the presence of species with quadruplexes (Fig. S1†). In subsequent discussion, we focus on the more biophysically relevant distribution centered around 4.2 nm.
The distribution corresponding to the ca. 4.2 nm distance appears to broaden in the presence of a two-fold excess of CueR (Fig. 2C, top vs. middle panel). We hypothesize that this broadening may be due to the presence of bent and undistorted DNA in solution. The presence of the undistorted DNA is either from free DNA or from CueR–DNA complex with undistorted DNA. To test this hypothesis, we measured the DEER signal for a 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of protein to DNA. In this case a peak at 3.6 nm becomes dominant. The shortening of the distance is likely due to the DNA kinking upon protein binding. Prior work has shown that the Cu(II)–Cu(II) distance in DNA can be directly related to the backbone C′–C′ distance.14 A more careful model analysis that accounts for the positioning of the Cu(II) atoms with respect to the DNA backbone, yields a 5′ end – center – 3′ end angle of 55° for the duplex with a 3.6 nm Cu(II)–Cu(II) distance (Fig. S8†). Such a bending of the duplex makes sense as this DNA sequence has a 19 bp separation between the regions which interact with RNAp. Bending of the DNA brings sites needed for RNAp coordination on the same face of the DNA, which promotes transcription.9,29,30
1 ratio of protein to DNA. In this case a peak at 3.6 nm becomes dominant. The shortening of the distance is likely due to the DNA kinking upon protein binding. Prior work has shown that the Cu(II)–Cu(II) distance in DNA can be directly related to the backbone C′–C′ distance.14 A more careful model analysis that accounts for the positioning of the Cu(II) atoms with respect to the DNA backbone, yields a 5′ end – center – 3′ end angle of 55° for the duplex with a 3.6 nm Cu(II)–Cu(II) distance (Fig. S8†). Such a bending of the duplex makes sense as this DNA sequence has a 19 bp separation between the regions which interact with RNAp. Bending of the DNA brings sites needed for RNAp coordination on the same face of the DNA, which promotes transcription.9,29,30
The data with a CueR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA ratio of 6
DNA ratio of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 supports the hypothesis that at a ratio of at least 2
1 supports the hypothesis that at a ratio of at least 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 there is coexistence of undistorted DNA and a kinked DNA complex. However, as more CueR is added into the system, the kinked DNA complex becomes dominant. This concentration dependence is anticipated due to a lower affinity of Cu(I)-free CueR to DNA.5
1 there is coexistence of undistorted DNA and a kinked DNA complex. However, as more CueR is added into the system, the kinked DNA complex becomes dominant. This concentration dependence is anticipated due to a lower affinity of Cu(I)-free CueR to DNA.5
The formation of a kinked duplex in the presence of Cu(I)-free CueR is in apparent contradiction to the available crystal structure of the Cu(I)-free CueR–DNA complex (PDB: 4WLS) which shows an undistorted DNA (cf. Fig. S9† for details).7 More importantly, the crystal structure was solved using an approximate concentration ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 CueR dimer to DNA. This observation is unsurprising based on the DEER data, since even at 2
2 CueR dimer to DNA. This observation is unsurprising based on the DEER data, since even at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 protein to DNA ratio the dominant conformation corresponds to the undistorted DNA. More importantly, a key insight from our solution data is that higher concentrations of CueR promotes the formation of the kinked DNA complex, even in the absence of Cu(I). The observation that Cu(I)-free CueR can induce duplex kinking provides a clear structural understanding for how Cu(I)-free CueR can substitute and remove Cu(I)-bound CueR on kinked DNA to end transcription (see below).
1 protein to DNA ratio the dominant conformation corresponds to the undistorted DNA. More importantly, a key insight from our solution data is that higher concentrations of CueR promotes the formation of the kinked DNA complex, even in the absence of Cu(I). The observation that Cu(I)-free CueR can induce duplex kinking provides a clear structural understanding for how Cu(I)-free CueR can substitute and remove Cu(I)-bound CueR on kinked DNA to end transcription (see below).
Next, we examined the DNA structure in the Cu(I)-bound CueR–DNA complexes. Given that recent results have suggested that there are more than two Cu(I) binding sites in CueR,31 we measured DEER time traces for 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 6
1 and 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dimer to duplex at three different Cu(I) equivalents (Fig. 3). Notably the time traces for all six samples in Fig. 3A are similar, suggesting some uniformity among the spin pair ensembles. Importantly, the presence of Cu(I) promotes the formation of a 3.7 nm distance (within experimental error of the previously observed 3.6 nm) readily even at a 2
1 dimer to duplex at three different Cu(I) equivalents (Fig. 3). Notably the time traces for all six samples in Fig. 3A are similar, suggesting some uniformity among the spin pair ensembles. Importantly, the presence of Cu(I) promotes the formation of a 3.7 nm distance (within experimental error of the previously observed 3.6 nm) readily even at a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of protein to DNA (cf. compare Fig. 3B to Fig. 2C). Negligible differences in the distances are observed as Cu(I) concentrations are increased in both the 2
1 ratio of protein to DNA (cf. compare Fig. 3B to Fig. 2C). Negligible differences in the distances are observed as Cu(I) concentrations are increased in both the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 6
1 and 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes as anticipated from the time traces in Fig. 3A. The presence of a kinked DNA in the Cu(I)-bound CueR–DNA complex is consistent with the crystal structure of the complex measured using Ag(I) as a surrogate for Cu(I) (PDB: 4WLW).7 A detailed analysis is provided in the ESI (Fig. S9†).
1 complexes as anticipated from the time traces in Fig. 3A. The presence of a kinked DNA in the Cu(I)-bound CueR–DNA complex is consistent with the crystal structure of the complex measured using Ag(I) as a surrogate for Cu(I) (PDB: 4WLW).7 A detailed analysis is provided in the ESI (Fig. S9†).
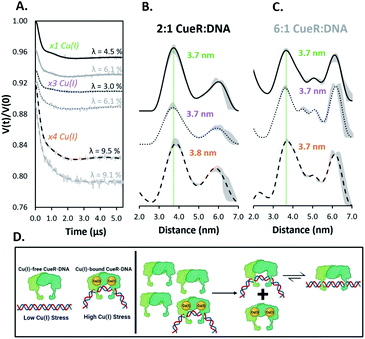 | ||
Fig. 3 (A) Background subtracted DEER time traces for the copA DNA at varying CueR dimer to duplex ratios (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 black, 6 1 black, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 grey) and monomeric equivalents of Cu(I). Modulation depths (λ) are shown. (B and C) Distance distributions of CueR complex ratios with equivalents of Cu(I) per dimer as described. Distributions were obtained from their respective time traces using DeerAnalysis. ComparativeDEERAnalysis consensus distance distributions are provided in the ESI (Fig. S7†). The green dashed line is a visual aid to compare the most probable distances. The grey regions show the validated distribution from DEERAnalysis (details are provided in the ESI†). (D) Pictorial summary of the presented work. Under high Cu(I) stress, the Cu(I)-bound CueR–DNA complex with a bent DNA is formed to promote transcription. After Cu(I) homeostasis is restored, a surge of Cu(I)-free CueR can readily substitute Cu(I)-bound CueR on DNA to end transcription, since Cu(I)-free CueR can bind in the bent state. Subsequently, Cu(I)-free CueR can interconvert to a thermodynamically favorable linear duplex state or unbind, in order to end transcription. 1 grey) and monomeric equivalents of Cu(I). Modulation depths (λ) are shown. (B and C) Distance distributions of CueR complex ratios with equivalents of Cu(I) per dimer as described. Distributions were obtained from their respective time traces using DeerAnalysis. ComparativeDEERAnalysis consensus distance distributions are provided in the ESI (Fig. S7†). The green dashed line is a visual aid to compare the most probable distances. The grey regions show the validated distribution from DEERAnalysis (details are provided in the ESI†). (D) Pictorial summary of the presented work. Under high Cu(I) stress, the Cu(I)-bound CueR–DNA complex with a bent DNA is formed to promote transcription. After Cu(I) homeostasis is restored, a surge of Cu(I)-free CueR can readily substitute Cu(I)-bound CueR on DNA to end transcription, since Cu(I)-free CueR can bind in the bent state. Subsequently, Cu(I)-free CueR can interconvert to a thermodynamically favorable linear duplex state or unbind, in order to end transcription. | ||
Additionally, modulation depths of ca. 9% were observed with ×4 equivalents of Cu(I) per CueR monomer (Fig. 3A). A consistent increase of modulation depth for these samples further supports the claim that additional spin pairs form via high order binding. The greater modulation depths for the Cu(I)-bound CueR–DNA complexes compared to the Cu(I)-free protein complexes suggest a greater amount of high order binding occurring in the presence of Cu(I).
Importantly, the data suggest that the presence of Cu(I) plays a key role in DNA distortion. A distorted DNA is observed even at a lower ratio of protein to DNA compared to the Cu(I)-free CueR–DNA case (cf.Fig. 2). Thus, Cu(I) acts as the key to drive the dominant formation of the kinked DNA complex that will allow RNAp coordination for transcription (Fig. 1C and D).
Conclusions
In summary, these EPR results, used in conjunction with insight gained from prior biochemical,4 smFRET,6 and structural measurements,7 provide a deeper understanding of transcription activation and termination in E. coli. We have shown that Cu(I)-free CueR can distort DNA in a similar fashion as Cu(I)-bound CueR (cf.Fig. 2vs.Fig. 3), but a large concentration of Cu(I)-free CueR is needed to bend the DNA. Under normal conditions a large concentration of CueR is unlikely in E. coli cells, given that CueR is expressed through copper sensing mechanisms that activate in response to the presence of Cu(I).32,33 Thus, the copA expression is not activated (cf.Fig. 3D, left panel). On the other hand, the presence of even a small amount of Cu(I) generates a Cu(I)-bound CueR–DNA complex in which the DNA is bent, leading to the activation of transcription and the remediation response (Fig. 3D, left panel). When Cu(I) homeostasis is restored, there now exists an excess of Cu(I)-free CueR created by copper sensing. The substitution of Cu(I)-bound CueR by a single Cu(I)-free CueR becomes thermodynamically and kinetically straightforward as minimal structural perturbation of DNA is needed (Fig. 3D, right panel). This complex can then transition to a state with an undistorted DNA or the Cu(I)-free CueR can dissociate from the duplex to end transcription. Indeed, the presence of a Cu(I)-free CueR-undistorted DNA complex and unbinding has been inferred from smFRET results.6 These data, taken together with recent results using Cu(II) labeling of the CueR,34 illustrate the potential of site-directed Cu(II) labeling. More broadly, this work highlights the utility of such DNA labeling approaches35–37 to enable the measurement of conformational ensembles under conditions that are not easily accessible (e.g. concentration dependent measurements in this case) in order to generate a more holistic picture of protein function. In the future, incorporating RNAp into our measurements would allow for a direct comparison to cryo-EM structures8,9 and permit the capturing of structural details for the DNA transcription bubble in solution.Data availability
The data supporting this work have been uploaded as part of the ESI:†Protein purification and expression, sample preparations, EMSA assay methods, MD simulation details, CW-EPR spectra and simulations, raw DEER data, biological replicate DEER data, primary DEER data, comparative method analysis for DEER time trace fitting, DNA bending model, EPR and crystal structure data comparisons.
Raw and processed datasets are available for preview and download at DOI: 10.17632/zrxjrc7fgn.1.
Author contributions
J. C. expressed, purified, and prepared the protein used with DNA samples. J. C. also carried out the primary investigation and analysis of EPR data. A. M. conducted the MD simulations and created the bent duplex model. L. H. and I. Y. provided initial protein resources, EMSA data, the protein plasmid used for expression, and aided with project development. S. G. was responsible for initial project development. S. R. and S. S. supervised all aspects of the research. J. C., S. R., and S. S. wrote the manuscript. All authors reviewed the manuscript.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
We thank Riti Sen and Dr Xing Yee Gan at the University of Pittsburgh for their time and assistance with respect to preparation and storage of air sensitive stock solutions used in this work. We also thank Dr Lada Gevorkyan-Airapetov for her assistance with the EMSA experiments. All MD simulations were carried out at the University of Pittsburgh Center for Research Computing. Cartoon figures were created with http://Biorender.com. S. S. and S. R. acknowledge the support from the National Science Foundation-Binational Science Foundation (NSF-BSF, NSF no. MCB-2006154; BSF. 2019723).References
- D. P. Giedroc and A. I. Arunkumar, Dalton Trans., 2007, 29, 3107–3120 RSC.
- J. M. Argüello, D. Raimunda and T. Padilla-Benavides, Front. Cell. Infect. Microbiol., 2013, 3, 1–14 Search PubMed.
- A. M. Fleming and C. J. Burrows, Chem. Soc. Rev., 2020, 49, 6524–6528 RSC.
- F. W. Outten, C. E. Outten, J. Hale and T. V. O'Halloran, J. Biol. Chem., 2000, 275(40), 31024–31029 CrossRef CAS PubMed.
- D. J. Martell, C. P. Joshi, A. Gaballa, A. G. Santiago, T. Y. Chen, W. Jung, J. D. Helmann and P. Chen, PNAS, 2015, 112(44), 13467–13472 CrossRef CAS PubMed.
- C. P. Joshi, D. Panda, D. J. Martell, N. M. Andoy, T.-Y. Chen, A. Gaballa, J. D. Helmann and P. Chen, PNAS, 2012, 109(38), 15121–15126 CrossRef CAS PubMed.
- S. J. Philips, M. Canalizo-Hernandez, I. Yildirim, G. C. Schatz, A. Mondragón and T. V. O'Halloran, Science, 2015, 349(6250), 877–881 CrossRef CAS PubMed.
- C. Fang, S. J. Philips, X. Wu, K. Chen, J. Shi, L. Shen, J. Xu, Y. Feng, T. V. O'Halloran and Y. Zhang, Nat. Chem. Biol., 2021, 17, 57–64 CrossRef CAS PubMed.
- W. Shi, B. Zhang, Y. Jiang, C. Liu, W. Zhou, M. Chen, Y. Yang, Y. Hu and B. Liu, iScience, 2021, 24(5), 102449 CrossRef CAS PubMed.
- M. J. Lawless, J. L. Sarver and S. Saxena, Angew. Chem., Int. Ed., 2017, 56(8), 2115–2117 CrossRef CAS PubMed.
- A. Gamble Jarvi, X. Bogetti, K. Singewald, S. Ghosh and S. Saxena, Acc. Chem. Res., 2021, 54(6), 1481–1491 CrossRef CAS PubMed.
- Z. Yang, M. Ji and S. Saxena, Appl. Magn. Reson., 2010, 39, 487–500 CrossRef CAS.
- J. E. Lovett, A. M. Bowen, C. R. T. Timmel, M. W. Jones, J. R. Dilworth, D. Caprotti, S. G. Bell, L. L. Wong and J. Harmer, Phys. Chem. Chem. Phys., 2009, 11, 6840–6848 RSC.
- S. Ghosh, J. Casto, X. Bogetti, C. Arora, J. Wang and S. Saxena, Phys. Chem. Chem. Phys., 2020, 22(46), 26707–26719 RSC.
- A. Changela, K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran and A. Mondragon, Science, 2003, 301(5638), 1383–1387 CrossRef CAS PubMed.
- C. D. Syme, R. C. Nadal, S. E. J. Rigby and J. H. Viles, J. Biol. Chem., 2004, 279(18), 18169–18177 CrossRef CAS PubMed.
- B. K. Shin and S. Saxena, J. Phys. Chem. A, 2011, 115(34), 9590–9602 CrossRef CAS PubMed.
- S. Ghosh, M. J. Lawless, H. J. Brubaker, K. Singewald, M. R. Kurpiewski, L. Jen-Jacobson and S. Saxena, Nucleic Acids Res., 2020, 48(9), 1–11 CrossRef PubMed.
- A. D. Milov, A. G. Maryasov and Y. D. Tsvetkov, Pulsed Electron, Appl. Magn. Reson., 1998, 15, 107–143 CrossRef CAS.
- M. Pannier, S. Veit, A. Godt, G. Jeschke and H. W. Spiess, J. Magn. Reson., 2000, 142(2), 331–340 CrossRef CAS PubMed.
- J. Casto, A. Mandato and S. Saxena, J. Phys. Chem. Lett., 2021, 12(19), 4681–4685 CrossRef CAS PubMed.
- G. Jeschke, V. Chechik, P. Ionita, A. Godt, H. Zimmermann, J. Banham, C. R. Timmel, D. Hilger and H. Jung, Appl. Magn. Reson., 2006, 30, 473–498 CrossRef CAS.
- S. G. Worswick, J. A. Spencer, G. Jeschke and I. Kuprov, Sci. Adv., 2018, 4(8), 1–17 Search PubMed.
- W. Jung, K. Sengupta, B. M. Wendel, J. D. Helmann and P. Chen, Nucleic Acids Res., 2020, 48(5), 2199–2208 CrossRef CAS PubMed.
- R. Grosschedl, Curr. Opin. Cell Biol., 1995, 7(3), 362–370 CrossRef CAS PubMed.
- B. E. Bode, D. Margraf, J. Plackmeyer, D. Dürner, T. F. Prisner and O. Schiemann, J. Am. Chem. Soc., 2007, 129(21), 6736–6745 CrossRef CAS PubMed.
- A. Gamble Jarvi, A. Sargun, X. Bogetti, J. Wang, C. Achim and S. Saxena, J. Phys. Chem. B, 2020, 124(35), 7544–7556 CrossRef CAS PubMed.
- M. J. Lawless, S. Ghosh, T. F. Cunningham, A. Shimshi and S. Saxena, Phys. Chem. Chem. Phys., 2017, 19(31), 20959–20967 RSC.
- A. Z. Ansari, J. E. Bradner and T. V. O'Halloran, Nat, 1995, 374, 370–375 CrossRef PubMed.
- A. Z. Ansari, M. L. Chael and T. V. O'Halloran, Nat, 1992, 355, 87–89 CrossRef CAS PubMed.
- R. K. Balogh, B. Gyurcsik, E. Hunyadi-Gulyas, J. Schell, P. W. Thulstrup, L. Hemmingsen and A. Jancso, Chemistry, 2019, 25(66), 15030–15035 CrossRef CAS PubMed.
- K. Yamamoto and A. Ishihama, Mol. Microbiol., 2005, 56(1), 215–227 CrossRef CAS PubMed.
- K. S. Chaturvedi and J. P. Henderson, Frontiers in Cellular and Infection Microbiology, 2014, 1–12 Search PubMed.
- H. Sameach, S. Ghosh, L. Gevorkyan-Airapetov, S. Saxena and S. Ruthstein, Angew. Chem., Int. Ed., 2019, 58(10), 3053–3056 CrossRef CAS PubMed.
- C. M. Grytz, A. Marko, P. Cekan, S. T. Sigurdsson and T. F. Prisner, Phys. Chem. Chem. Phys., 2016, 18(4), 2993–3002 RSC.
- G. W. Reginsson, S. A. Shelke, C. Rouillon, M. F. White, S. T. Sigurdsson and O. Schiemann, Nucleic Acids Res., 2013, 41(1), 1–10 CrossRef PubMed.
- G. Y. Shevelev, O. A. Krumkacheva, A. A. Lomzov, A. A. Kuzhelev, O. Y. Rogozhnikova, D. V. Trukhin, T. I. Troitskaya, V. M. Tormyshev, M. V. Fedin and D. V. Pyshnyi, J. Am. Chem. Soc., 2014, 136(28), 9874–9877 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc06563g |
| This journal is © The Royal Society of Chemistry 2022 |