Open Access Article
Open Access ArticleTumor suppression via diverting intracellular sialylation with multifunctional nanoparticles†
Yunlong
Chen
 ,
Yuanjiao
Yang
,
Qingqing
Tan
,
Huipu
Liu
and
Huangxian
Ju
,
Yuanjiao
Yang
,
Qingqing
Tan
,
Huipu
Liu
and
Huangxian
Ju
 *
*
State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China. E-mail: hxju@nju.edu.cn
First published on 26th January 2022
Abstract
Sialylation plays an important role in tumor-related physiological processes. Therefore, intervention of sialylation has great potential to explore new paths for tumor therapy. In view of the immune modulation of sialic acid (SA) on tumors, this work designs a multifunctional mesoporous silica nanoparticle (MFMSN) to divert intracellular sialylation for tumor suppression. The galactose groups covered on MFMSN act as sialylation substrates to bind intracellular SAs competitively, which inhibits the SA expression on the tumor cell surface. The diverted intracellular sialylation can be visualized on living cells and in vivo by specifically binding the sialylated galactose with a phenylboronic acid labeled ssDNA probe released from the pore of MFMSN to induce DNA strand displacement, which recovers the fluorescence of the dsDNA probe covered on MFMSN surface. The diverting of sialylation efficiently suppresses tumor growth in mice, demonstrating the great potential of the designed strategy for revealing SA-related biological processes and clinical cancer therapy.
Introduction
Sialic acid (SA) is a nine-carbon sugar molecule, which is often located at the terminal position of the glycan chain attached to glycoproteins or glycolipids on cell surfaces.1–4 The SA on the cancer cell surface mediates a series of physiological and pathological processes, including migration and metastasis,5–9 and plays a critical role in immune modulation through the interaction of SA with the immune-inhibitory SA-binding immunoglobulin-like lectin (Siglec) family.10–14 Down-regulation of SA can reduce immune evasion, which promotes the immune therapy of cancer cells.15–18 Generally, substrate-bound SA is produced by sialyltransferase (ST) mediated sialylation.19,20 According to the difference of carbohydrate linkages, there are four ST families in the human body: ST3Gal (α-2,3-ST), ST6Gal (α-2,6-ST), ST6GalNAc and ST8Sia (α-2,8-ST).21 The activities of these STs are directly related to SA expression on the cell surface, which further affects the physiological and pathological states of cancer cells. Thus, intervention of the sialylation may provide new paths to regulate SA expression of the cell surface for cancer therapy.Tumor suppression has been achieved through blockage or degradation of existing SAs to change their function on cancer cells. For example, a nano molecularly imprinted polymer has been designed to block the N-glycan on human epidermal growth factor receptor-2 (HER2), which suppresses the dimerization of HER2 with other HER family members to inhibit tumor growth.22 Sialidase linked to the HER2 antibody has been used to degrade SA on the HER2, which reduces the binding of the immune-inhibitory Siglec checkpoint receptor and thus enhances the activation of immune cells for tumor suppression.23,24 These works demonstrate the efficiency of cell surface SA regulation in tumor suppression, but may be limited by the continual generation of newly synthesized SAs.
Intracellular intervention of sialylation can endogenously change the SA expression of the cell surface. A SA mimic has been synthesized to block SA expression of tumor cells by direct inhibition of ST activity in vivo, which suppresses tumor growth by enhancing tumor-infiltrating natural killer cells and T-cells.25 However, the direct inhibition of ST activity in cancer cells with small molecules may suffer from drug resistance, and it is difficult to monitor the intracellular sialylation and intervention processes due to the lack of corresponding visual tools.
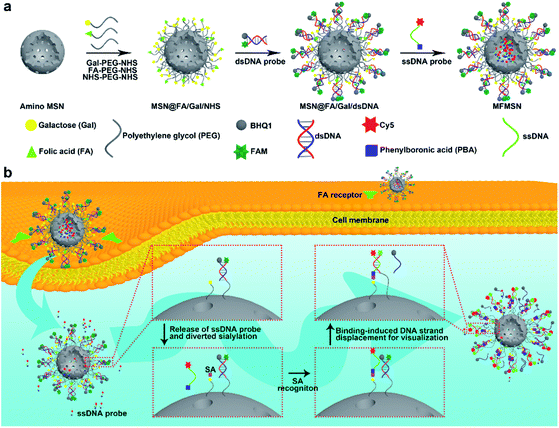
Inspired by the tumor suppression through a consumed lysosomal N-acetylglucosamine salvage pathway,26 this work developed a prime example of a theranostic strategy through diverting intracellular sialylation for tumor suppression. With this strategy, a multifunctional mesoporous silica nanoparticle (MFMSN) was designed for diverting of intracellular sialylation as well as its visualization to reduce SA expression on the cell surface. The MFMSN was constructed on an amino MSN, whose surface was firstly modified with three kinds of functional polyethylene glycols (PEGs) terminated with galactose (Gal), folic acid (FA) and succinimidyl carboxymethyl ester (NHS), respectively (Fig. 1a). Gal with the occupied site 2 mainly acted as the substrate of ST3Gal and ST6Gal mediated intracellular sialylation,21 FA targeted tumor cells for delivery of MFMSN,27 and NHS was used for further conjugation of the double stranded DNA (dsDNA) probe modified with a pair of carboxyfluorescein (FAM) and black hole quencher 1 (BHQ1) groups. Meanwhile, a single stranded DNA (ssDNA) probe terminated with Cy5 and phenylboronic acid (PBA) was loaded in the pores of the MSN for visual monitoring of the intracellular diverting of sialylation. After the MFMSNs were delivered into the cells, the Gal groups on the MFMSNs' surface were sialylated by ST3Gal or ST6Gal, and the sialylated Gal groups were then bound with the ssDNA probes released from the MFMSN through specific PBA-SA recognition28–30 to initiate the binding-induced DNA strand displacement,31 which recovered the fluorescence of FAM on the dsDNA probes (Fig. 1b). The in vivo visualization of the diverted sialylation on tumor bearing mice was achieved with an updated MFMSN′, which was loaded with a Cy5-free ssDNA probe in the pores and replaced the FAM-BHQ1 pair with the Cy5-BHQ2 pair on the dsDNA probe to avoid self-fluorescence of the mice. When visual monitoring was not required, the diverting of sialylation could be performed with MSN@FA/Gal, which was conveniently prepared by linking only FA-PEG-NHS and Gal-PEG-NHS on amino MSN. This work provides an ingenious tool to dynamically visualize the diverted sialylation in living cells and in vivo for efficient tumor suppression.
Results and discussion
Multifunctional MSNs
For the preparation of MFMSNs, MSNs with a diameter around 80 nm were firstly treated with piranha solution, followed by silylation with (3-aminopropyl) triethoxysilane to obtain amino MSNs for covalently binding Gal-PEG 35K-NHS, FA-PEG 10K-NHS, and NHS-PEG 20K-NHS to conjugate dsDNA probes. The dsDNA probe contained the FAM group at the 5′ end and an amino group at the 3′ end of DNA1 and BHQ1 at the 3′ end of DNA2, in which the fluorescence of FAM could be completely quenched by BHQ1. Considering that the diverting of intracellular sialylation on Gal and the binding-induced DNA strand displacement were the key steps of the designed strategy, Gal and dsDNA probes were conjugated on longer PEG chains. The two binding steps of the PEGs and dsDNA probes to MSNs were demonstrated by the changes of both the Zeta potential and hydration diameter measured with dynamic light scattering analysis (Fig. S1a and b†). The binding of three kinds functional PEGs resulted in a negative Zeta potential and increased hydration diameter of the nanoparticles. The Zeta potential became more negative after the dsDNA probe was bound to the surface of MSNs@FA/Gal/NHS, which also further increased the hydration diameter. Besides, TEM images and BET analysis demonstrated that the surface modification did not change the porous structure (Fig. S1c†) of the MSNs but did obviously increase the BET surface area (Fig. S1d†) due to the surface modification with PEG polymers and DNA molecules. Compared to MSNs without FA modification (MSNs@Gal/NHS), MSNs@FA/Gal/NHS exhibited characteristic peaks of FA around 1590–1700 cm−1 and 2850–2940 cm−1 in FTIR spectra (Fig. S1e†), which demonstrated the successful modification of FA.After the excess dsDNA probe was removed from the surface and the pores of MSNs by three washings with PBS (Fig. S2a†), the ssDNA probe labeled with the PBA group at the 5′ end and Cy5 at the 3′ end was loaded into the pores of MSNs to obtain the MFMSNs. The loading led to an obvious decrease of Cy5 fluorescence intensity of the ssDNA probe in the supernatant (Fig. S2b†). From the average molar mass of MSNs derived from the density of amorphous SiO2,32 the pore volume of the MSNs (1.4 mL g−1) provided by the manufacturer and Fig. S2,† the average loading amounts of dsDNA and ssDNA probes could be respectively calculated to be 167 and 4491 per MFMSN, which provides an abundant proportion of ssDNA to dsDNA for the DNA strand displacement. After MFMSNs were dispersed in PBS, the Cy5 fluorescence intensity in the supernatant continually increased up to 6 h (Fig. S3†), indicating the long-lasting release of the ssDNA probe from the pores of MFMSNs. Besides, after incubation with only high concentrations of ssDNA probes, MFMSNs did not exhibit any FAM fluorescence (Fig. S4†), indicating that the freely released ssDNA probe was not bound to the dsDNA probes to produce a false positive signal.
In vitro response of MFMSNs
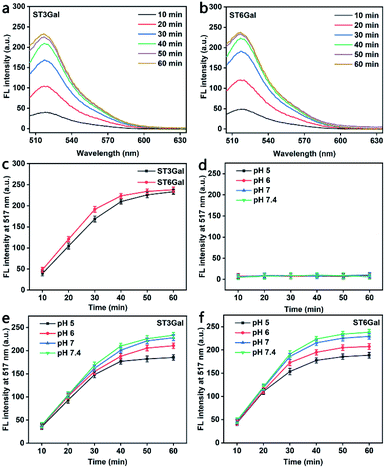
Generally, glycosyltransferase is sensitive to the availability of the reactive hydroxyl group and the stereoscopic structure of its substrates.33 The PEG-linked Gal used in this work contains two reactive hydroxyl groups at both positions 3 and 6 of the sugar ring (Fig. S5†), which can act as the acceptor sites of ST3Gal and ST6Gal, respectively. To demonstrate the responses of MFMSNs to these STs, MFMSNs were incubated with cytidine-5′-monophospho-N-acetylneuraminic acid sodium salt (CMP-Sia) and ST3Gal expressed in E. coli BL21 from Pasteurella multocida or ST6Gal expressed in E. coli BL21 from Photobacterium damsela in PBS (pH 7.4) at 37 °C for 10–60 min, respectively, which showed obviously increasing FAM fluorescence within an incubation time of 50 min due to the binding-induced DNA strand displacement (Fig. 2a–c), indicating the relatively quick sialylation process from CMP-Sia to sialylated Gal in vitro.Considering the weak acidic environment in the tumor cells and issues,34 where the PBA can keep the specific recognition to SA,29 the effect of the pH of the incubation solution on the binding-induced DNA strand displacement on the MFMSN surface was also examined. Compared to the completely quenched FAM fluorescence (Fig. 2d), the FAM fluorescence increased with the increasing incubation time of MFMSNs in PBS containing CMP-Sia and ST3Gal or ST6Gal at pH 5, 6, 7 and 7.4 (Fig. 2e and f). Thus, the MFMSN retain the response ability to ST3Gal or ST6Gal under a tumor microenvironment. The slight decrease of FAM fluorescence with the decreasing pH was attributed to the degradation of CMP-Sia in the acidic environment.35 Besides, the PBA-free and Gal-free MFMSNs did not exhibit any FAM fluorescence with either ST3Gal or ST6Gal at these pH values (Fig. S6†), indicating the PBA-SA recognition mediated signal response.
Visualization of intracellular diverted sialylation
As a proof-of-concept, human breast cancer MCF-7 cells were chosen as the cell model. After incubating MCF-7 cells with different concentrations of MFMSNs (1–6 nM) at 37 °C for 5 h, the confocal laser scanning microscopic (CLSM) images showed obvious FAM and Cy5 fluorescence (Fig. S7a†). Their intensity increased with the increasing concentration of MFMSNs and reached a plateau at the concentration of 5 nM, respectively (Fig. S7b and c†). As the mechanism described in Fig. 1b, the FAM fluorescence occurred at the dsDNA probe and could thus be completely covered by the Cy5 fluorescence from the ssDNA probes after DNA strand displacement, while the Cy5 fluorescence showed a wider distribution due to the release of excessive ssDNA probes from the pores of MFMSNs. Thus, the MFMSNs could respond to diverted sialylation within living cells, achieving visualization of the intracellular diverted sialylation on MFMSNs.The MFMSN responses inside the cells also showed time dependence (Fig. S8a†). After MCF-7 cells were incubated with 5 nM of MFMSNs at 37 °C for different times, the fluorescence intensity of both FAM and Cy5 increased with the increasing incubation time and reached a plateau at 4 h (Fig. S8b and c†), which implied the saturated binding-induced DNA strand displacement within the lasting release of the ssDNA probe (Fig. S3†). Thus, a concentration of 5 nM and an incubation time of 4 h of MFMSNs were chosen as the optimal conditions for visualization of the diverted sialylation in living cells.
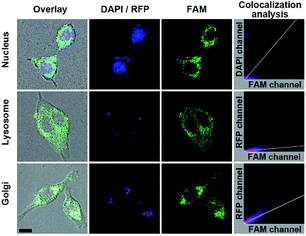
The FA mediated targeting delivery was verified by incubating MFMSNs with the normal human mammary epithelial MCF-10A cells, which exhibited negligible FAM and tiny Cy5 fluorescence comparing to that incubated with MCF-7 cells (Fig. S9†) due to low expression of FR receptor on MCF-10A.36,37 This phenomenon also indicated that FA conjugated on the shortest PEG chain did not affect its recognition with FA receptors. Interestingly, the MCF-7 cells incubated with FA-free MFMSN did not exhibit FAM fluorescence but instead exhibited obvious Cy5 fluorescence on the cell surface, indicating that the FA-free MFMSN could not be delivered into the cell but the released ssDNA probes could bind to the cell surface SAs. Besides, the MCF-7 cells incubated with PBA-free MFMSN did not exhibit FAM fluorescence but obvious Cy5 fluorescence occurred inside the cells, indicating that the PBA-free MFMSN could be delivered into the cells to release DNA3 but could not turn on FAM fluorescence. These results further demonstrated the FA mediated target delivery and PBA-SA recognition mediated signal response. The location of MFMSNs inside the cells was investigated with colocalization imaging of the nucleus, lysosomes and Golgi (Fig. 3), which were dyed with DAPI, lysosomes-RFP and Golgi-RFP, respectively. After the dyed cells were incubated with 5 nM of MFMSNs for 4 h, the fluorescence of FAM did not overlap with the fluorescence of the DAPI dyed nucleus (Pearson's correlation coefficient: −0.26), indicating that MFMSNs could not enter into the nucleus. Although a little fluorescence of FAM could be observed in the fluorescence area of the lysosomes-RFP dyed lysosomes (Pearson's correlation coefficient: 0.35), the FAM fluorescence mainly occurred beyond lysosomes-RFP, but overlapped with that of Golgi-RFP (Pearson's correlation coefficient: 0.77). Thus, the amount of MFMSNs in lysosomes was negligible, while most of the MFMSNs were located around the Golgi, which was the major place for sialylation.2
Regulation of intracellular sialylation
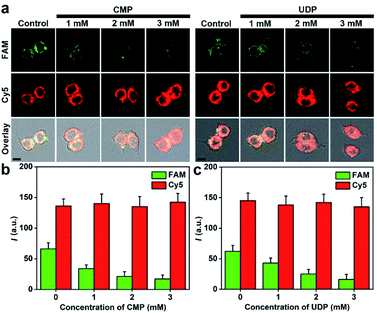
Sialylation in living cells can be inhibited with different inhibitors of ST, such as cytidine 5′-monophosphate disodium salt (CMP)38 and uridine 5′-diphosphate disodium salt hydrate (UDP).39 After the cells were treated with different concentrations of CMP or UDP and then incubated with 5 nM of MFMSNs for 4 h, an obvious decrease of FAM fluorescence was observed along with the increasing inhibitor concentrations, but the Cy5 fluorescence did not change (Fig. 4). These results indicated that the intracellular ST activity was inhibited, and the presence of the inhibitors did not affect the internalization of MFMSNs into the cells for releasing ssDNA probes. By using Cy5 fluorescence as the internal standard to eliminate the discrepancy in the amount of internalized MFMSN, the FAM fluorescence on MFMSNs could be directly used for qualitative assessment of ST activity inside living cells.In vivo visualization of diverted sialylation on tumor bearing mice
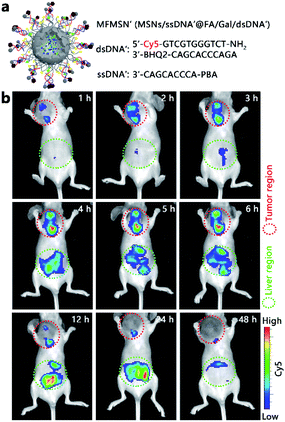
For the preliminary in vivo application of the designed strategy, the MCF-7 tumor xenograft mice models were established by subcutaneous inoculation of MCF-7 cells in the selected position of the mice. All animal experiments were performed according to the Laboratory Animal Management of Jiangsu Province published by the Department of Science and Technology of Jiangsu Province. To avoid the high background fluorescence of the mouse at the FAM-excitation region and increase the tissue depth of the in vivo fluorescence imaging, the fluorescence tag in MFMSNs was updated by replacing the FAM-BHQ1 pair with Cy5-BHQ2 in the dsDNA probe and by using the Cy5-free ssDNA probe to obtain the MFMSN' (Fig. 5a). As the Cy5-free ssDNA probe would continually release from MFMSN′, intratumoral injection was the only choice. The mice were intratumorally injected with 50 μL of MFMSNs' (5 nM) and imaged at different times after injection (Fig. 5b). Obviously, Cy5 fluorescence was observed at the tumor region, which increased in the first 6 h and then weakened from 12 to 48 h, indicating that the MFMSN′ could visualize the diverted sialylation at the tumor region in the first 6 h. Although the Cy5 fluorescence in the liver region appeared after 3 h of injection and continually increased from 4 to 24 h, it disappeared after 48 h of injection, indicating that most MFMSNs' were excreted by the body after 48 h. Thus, the designed MFMSN′ exhibited excellent capability for in vivo visualization of the diverted sialylation and physiological metabolizability.Cell surface SA expression upon diverted sialylation
Since the diverting of sialylation has been demonstrated through visualization both in living cells and in vivo, the consequence of the diverting was further explored. As the cell surface SAs were generated from intracellular sialylation,19,20 the variation of cell surface SA expression after diverting of sialylation was firstly examined. To reduce the interference factors, the MFMSN was simplified by removing its imaging module, which could be performed by modifying amino MSNs with only FA and Gal to obtain MSN@FA/Gal. After MCF-7 cells were incubated with different concentrations of MSN@FA/Gal for 4 h and recognized with FITC labeled Sambucus nigra agglutinin, a lectin specific to SA, for 1 h, the FITC fluorescence in both the CLSM images and flow cytometric analysis showed an obvious decrease with the increasing MSN@FA/Gal concentration (Fig. S10†), indicating the decreased cell surface SA expression. Thus, the Gal initiated diverting of intracellular sialylation could efficiently inhibit cell surface SA expression. As SAs on the cell surface are critical for tumor related processes,5–18 the designed diverting strategy could provide a new path for tumor suppression.Suppression of tumor growth
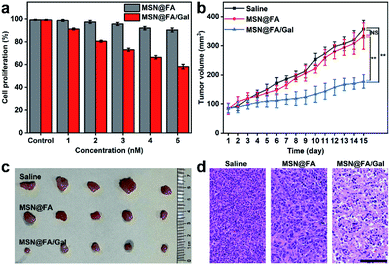
The effects of the designed diverting strategy on cell proliferation and tumor growth were examined with MSN@FA/Gal, using FA modified MSN (MSN@FA) as a negative control. With the increasing MSN@FA/Gal con-centration, the treated cells exhibited an obvious decrease of cell proliferation, while those on MSN@FA treated cells was not obvious (Fig. 6a). Therefore, the decrease of cell proliferation could be attributed to the Gal initiated diverting of sialylation and its further inhibition of cell surface SA expression, demonstrating the anti-tumor mechanism. Similarly, after MCF-7 tumor xenograft mice were intratumorally injected with 50 μL of MSNs@FA/Gal every 2 days, the tumor growth was obviously suppressed comparing to those injected with saline or MSN@FA (Fig. 6b). After 15 days, these groups showed a significant difference in tumor volume (Fig. 6c), but no discernible difference in their body weight (Fig. S11†). The histological analysis indicated a massive cell remission in tumor tissue (Fig. 6d), and negligible pathological abnormalities in the heart, liver, spleen, lung and kidney after MSNs@FA/Gal treatment (Fig. S12†), which verified the suppression effect of the designed diverting strategy on tumor growth and its tiny side-effects.Conclusions
In conclusion, a strategy for diverting intra-cellular sialylation has been developed for tumor suppression by using a newly designed MFMSN. The sialylation diverted by MFMSN both inside the cells and in vivo can be quickly achieved around the Golgi and is visualized through a binding-induced DNA strand displacement to recover the fluorescence of the dye co-labeled with BHQ on dsDNA. The diverting of sialylation inhibits the SA expression on cell surface and the cell proliferation, and exhibits efficient tumor suppression on tumor bearing mice with tiny side-effects. Moreover, the MFMSN is flexible and can be customized to perform different functions. By use of other glycosylation substrates and recognition compounds, the MFMSN can be expanded for intracellular visualization of different glycosylation pathways. The glycosylation diverting provides an efficient supplement for improving the therapeutic index of cancer in clinical applications.Data availability
All relevant data is presented in the paper and ESI.† Raw data is available upon request by email to the corresponding author.Author contributions
Y. L. C. and H. X. J. initiated the projects. Y. L. C. conceived the experiments. Y. L. C., Y. J. Y., Q. Q. T. and H. P. L. performed the experiments. Y. L. C. and H. X. J. analyzed the data and wrote the manuscript.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21974063, 21635005, 21827812, and 21890741), National Key Research and Development Program of China (2018YFC1004704), and Fundamental Research Funds for the Central Universities (14380200). This study was performed in strict accordance with the Laboratory Animal Management of Jiangsu Province for the care and use of laboratory animals (License No. SYXK 2017-0015) and was approved by the Department of Science and Technology of Jiangsu Province (Nanjing, China).Notes and references
- A. Varki, R. L. Schnaar and R. Schauer, Essentials of glycobiology, Cold Spring Harbor Laboratory Press, New York, 2015 Search PubMed.
- M. M. Fuster and J. D. Esko, Nat. Rev. Cancer, 2005, 5, 526–542 CrossRef CAS PubMed.
- C. Büll, M. A. Stoel, M. H. den Brok and G. J. Adema, Cancer Res., 2014, 74, 3199–3204 CrossRef PubMed.
- W. R. Alley Jr and M. V. Novotny, J. Proteome Res., 2010, 9, 3062–3072 CrossRef PubMed.
- A. Matsumoto, H. Cabral, N. Sato, K. Kataoka and Y. Miyahara, Angew. Chem., Int. Ed., 2010, 49, 5494–5497 CrossRef CAS PubMed.
- X. Chen and A. Varki, ACS Chem. Biol., 2010, 5, 163–176 CrossRef CAS PubMed.
- A. Varki, Trends Mol. Med., 2008, 14, 351–360 CrossRef CAS PubMed.
- D. H. Dube and C. R. Bertozzi, Nat. Rev. Drug Discovery, 2005, 4, 477–488 CrossRef CAS PubMed.
- Y. F. Xu, A. Sette, J. Sidney, S. J. Gendler and A. Franco, Immunol. Cell Biol., 2005, 83, 440–448 CrossRef CAS PubMed.
- M. S. Macauley, P. R. Crocker and J. C. Paulson, Nat. Rev. Immunol., 2014, 14, 653–666 CrossRef CAS PubMed.
- P. R. Crocker, J. C. Paulson and A. Varki, Nat. Rev. Immunol., 2007, 7, 255–266 CrossRef CAS PubMed.
- M. L. Perdicchio, A. M. Cornelissen, I. Streng-Ouwehand, S. Engels, M. I. Verstege, L. Boon, D. Geerts, Y. van Kooyk and W. W. J. Unger, Oncotarget, 2016, 7, 8771–8782 CrossRef PubMed.
- J. Lübbers, E. Rodríguez and Y. van Kooyk, Front. Immunol., 2018, 9, 2807 CrossRef PubMed.
- J. C. Paulson, M. S. Macauley and N. Kawasaki, Ann. N. Y. Acad. Sci., 2012, 1253, 37–48 CrossRef CAS PubMed.
- M. A. Stanczak, S. S. Siddiqui, M. P. Trefny, D. S. Thommen, K. F. Boligan, S. von Gunten, A. Tzankov, L. Tietze, D. Lardinois, V. Heinzelmann-Schwarz, M. von Bergwelt-Baildon, W. Zhang, H.-J. Lenz, Y. Han, C. I. Amos, M. Syedbasha, A. Egli, F. Stenner, D. E. Speiser, A. Varki, A. Zippelius and H. Läubli, J. Clin. Invest., 2018, 128, 4912–4923 CrossRef PubMed.
- J. E. Hudak, S. M. Canham and C. R. Bertozzi, Nat. Chem. Biol., 2014, 10, 69–75 CrossRef CAS PubMed.
- H. Läubli, O. M. T. Pearce, F. Schwarz, S. S. Siddiqui, L. Deng, M. A. Stanczak, L. Deng, A. Verhagen, P. Secrest, C. Lusk, A. G. Schwartz, N. M. Varki, J. D. Bui and A. Varki, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 14211–14216 CrossRef PubMed.
- C. Jandus, K. F. Boligan, O. Chijioke, H. Liu, M. Dahlhaus, T. Démoulins, C. Schneider, M. Wehrli, R. E. Hunger, G. M. Baerlocher, H.-U. Simon, P. Romero, C. Münz and S. von Gunten, J. Clin. Invest., 2014, 124, 1810–1820 CrossRef CAS PubMed.
- P. D. Bos, X. H.-F. Zhang, C. Nadal, W. Shu, R. R. Gomis, D. X. Nguyen, A. J. Minn, M. J. van de Vijver, W. L. Gerald, J. A. Foekens and J. Massagué, Nature, 2009, 459, 1005–1009 CrossRef CAS PubMed.
- A. Harduin-Lepers, M.-A. Krzewinski-Recchi, F. Colomb, F. Foulquier, S. Groux-Degroote and P. Delannoy, Front. Biosci., 2012, 4E, 499–515 CrossRef.
- F. Dall'Olio and M. Chiricolo, Glycoconjugate J., 2001, 18, 841–850 CrossRef PubMed.
- Y. Dong, W. Li, Z. Gu, R. Xing, Y. Ma, Q. Zhang and Z. Liu, Angew. Chem., Int. Ed., 2019, 58, 10621–10625 CrossRef CAS PubMed.
- H. Xiao, E. C. Woods, P. Vukojicic and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 10304–10309 CrossRef CAS PubMed.
- M. A. Gray, M. A. Stanczak, N. R. Mantuano, H. Xiao, J. F. A. Pijnenborg, S. A. Malaker, C. L. Miller, P. A. Weidenbacher, J. T. Tanzo, G. Ahn, E. C. Woods, H. Läubli and C. R. Bertozzi, Nat. Chem. Biol., 2020, 16, 1376–1384 CrossRef CAS PubMed.
- C. Büll, T. J. Boltje, N. Balneger, S. M. Weischer, M. Wassink, J. J. van Gemst, V. R. Bloemendal, L. Boon, J. van der Vlag, T. Heise, M. H. den Brok and G. J. Adema, Cancer Res., 2018, 78, 3574–3588 Search PubMed.
- R. Koide and S.-I. Nishimura, Angew. Chem., Int. Ed., 2019, 58, 14513–14518 CrossRef CAS PubMed.
- J. Tian, L. Ding, H.-J. Xu, Z. Shen, H. X. Ju, L. Jia, L. Bao and J.-S. Yu, J. Am. Chem. Soc., 2013, 135, 18850–18858 CrossRef CAS PubMed.
- A. Matsumoto, H. Cabral, N. Sato, K. Kataoka and Y. Miyahara, Angew. Chem., Int. Ed., 2010, 49, 5494–5497 CrossRef CAS PubMed.
- H. Otsuka, E. Uchimura, H. Koshino, T. Okano and K. Kataoka, J. Am. Chem. Soc., 2003, 125, 3493–3502 CrossRef CAS PubMed.
- A. Matsumoto, N. Sato, K. Kataoka and Y. Miyahara, J. Am. Chem. Soc., 2009, 131, 12022–12023 CrossRef CAS PubMed.
- F. Li, H. Zhang, C. Lai, X.-F. Li and X. C. Le, Angew. Chem., Int. Ed., 2012, 51, 9317–9320 CrossRef CAS PubMed.
- R. K. Iler, The chemistry of silica, John Wiley & Sons, Inc., New York, 1979 Search PubMed.
- D. K. Cooper, E. Koren and R. Oriol, Immunol. Rev., 1994, 141, 31–58 CrossRef CAS PubMed.
- I. F. Tannock and D. Rotin, Cancer Res., 1989, 49, 4373–4384 CAS.
- R. Schauer, M. Wember and C. F. do Amaral, Hoppe Seylers Z. Physiol. Chem., 1972, 353, 883–886 CrossRef CAS PubMed.
- R. J. Price, K. A. Lillycrop and G. C. Burdge, J. Nutr. Sci., 2016, 5, e17 CrossRef PubMed.
- J. Soleymani, M. Hasanzadeh, M. H. Somi, N. Shadjou and A. Jouyban, Biosens. Bioelectron., 2018, 115, 61–69 CrossRef CAS PubMed.
- X. Wang, L.-H. Zhang and X.-S. Ye, Med. Res. Rev., 2003, 23, 32–47 CrossRef CAS PubMed.
- S. Hinderlich, W. Weidemann, T. Yardeni, R. Horstkorte and M. Huizing, Topics in Current Chemistry, Springer, Berlin Heidelberg, 2013 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section, supporting figures and table. See DOI: 10.1039/d1sc05598d |
| This journal is © The Royal Society of Chemistry 2022 |