Open Access Article
Open Access ArticleHydroFlipper membrane tension probes: imaging membrane hydration and mechanical compression simultaneously in living cells†
José
García-Calvo‡
,
Javier
López-Andarias‡
,
Jimmy
Maillard
,
Vincent
Mercier
,
Chloé
Roffay
,
Aurélien
Roux
 ,
Alexandre
Fürstenberg
,
Alexandre
Fürstenberg
 ,
Naomi
Sakai
,
Naomi
Sakai
 and
Stefan
Matile
and
Stefan
Matile
 *
*
School of Chemistry and Biochemistry, NCCR Chemical Biology, University of Geneva, Geneva, Switzerland. E-mail: stefan.matile@unige.ch; Web: http://www.unige.ch/sciences/chiorg/matile Tel: +41 22 379 6523
First published on 3rd February 2022
Abstract
HydroFlippers are introduced as the first fluorescent membrane tension probes that report simultaneously on membrane compression and hydration. The probe design is centered around a sensing cycle that couples the mechanical planarization of twisted push–pull fluorophores with the dynamic covalent hydration of their exocyclic acceptor. In FLIM images of living cells, tension-induced deplanarization is reported as a decrease in fluorescence lifetime of the dehydrated mechanophore. Membrane hydration is reported as the ratio of the photon counts associated to the hydrated and dehydrated mechanophores in reconvoluted lifetime frequency histograms. Trends for tension-induced decompression and hydration of cellular membranes of interest (MOIs) covering plasma membrane, lysosomes, mitochondria, ER, and Golgi are found not to be the same. Tension-induced changes in mechanical compression are rather independent of the nature of the MOI, while the responsiveness to changes in hydration are highly dependent on the intrinsic order of the MOI. These results confirm the mechanical planarization of push–pull probes in the ground state as most robust mechanism to routinely image membrane tension in living cells, while the availability of simultaneous information on membrane hydration will open new perspectives in mechanobiology.
The detection and study of membrane mechanics in living cells is a topic of current concern.1–14 To enable this research, appropriate chemistry tools, that is small-molecule fluorescent probes that allow imaging of membrane tension, are needed.15 With the direct imaging of physical forces being intrinsically impossible, design strategies toward such probes have to focus on the suprastructural changes caused by changes in membrane tension.15 These suprastructural changes are divers, often interconnected, and vary with the composition of the membrane.15–25 Beyond the fundamental lipid compression and decompression, they include changes in membrane curvature, from rippling, buckling and budding to tubules extending from the membrane and excess lipid being ejected. Of similar importance are changes in membrane organization, particularly tension-induced phase separation and mixing, i.e. assembly and disassembly of microdomains. Consequences of these suprastructural changes include microdomain strengthening and softening and changes in membrane hydration and viscosity.16–25
The currently most developed fluorescent flipper probes have been introduced26,27 to image membrane tension by responding to a combination of mechanical compression and microdomain assembly in equilibrium in the ground state.15 Extensive studies, including computational simulations,28 have shown that flipper probes align non-invasively along the lipid tails of one leaflet and report changes in membrane order and tension as changes in fluorescent lifetimes and shifts of excitation maxima.15 Among other candidates, solvatochromic probes respond off-equilibrium in the excited state to changes in membrane hydration and have very recently been considered for the imaging of membrane tension in living cells.29–36 So far not considered to image tension, ESIPT probes also report off equilibrium in the excited state on membrane hydration, but for different reasons.37,38 Mechanosensitive molecular rotors respond off equilibrium in the excited state to changes in microviscosity.17,30,32,39–53 The same principle holds for the planarization of bent, papillon or flapping fluorophores.54–57 The response of all possible probes to tension can further include less desired changes in positioning and partitioning between different domains, not to speak of more catastrophic probe aggregation, precipitation, disturbance of the surrounding membrane structure, and so on. Although the imaging of membrane tension is conceivable in principle with most of above approaches, the complex combination of parameters that has to be in place can thus far only be identified empirically, followed by much optimization.15
The force-induced suprastructural changes are accompanied by the alteration in several unrelated physical properties of membranes. It is, for instance, well documented that membrane hydration increases with membrane disorder, from solid-ordered (So) to liquid-disordered (Ld) phases.29,58 Increasing cholesterol content decreases membrane hydration in solid- and liquid-ordered membranes.59 However, studies in model membranes also indicate that membrane hydration and membrane fluidity do not necessarily correlate.59 The dissection of the individual parameters contributing to the response of fluorescent membrane tension probes would be important for probe design and understanding of their responses, but it remains a daunting challenge. In this study, we introduce fluorescent flipper probes that simultaneously report on mechanical membrane compression and membrane hydration at equilibrium in the ground state. Changes of both in response to changes in membrane tension and membrane composition are determined in various organelles in living cells.
The dual hydration and membrane tension probes are referred to as HydroFlippers to highlight the newly added responsiveness to membrane hydration. The mechanosensing of lipid compression in bilayer membranes by flipper probes has been explored extensively.15 Fluorescent flippers27 like 1 are designed as bioinspired60 planarizable push–pull probes26 (Fig. 1). They are constructed from two dithienothiophene fluorophores that are twisted out of co-planarity by repulsion of methyls and σ holes on sulfurs61,62 next to the twistable bond. The push–pull system is constructed first from formal sulfide and sulfone redox bridges in the two twisted dithienothiophenes. These endocyclic donors and acceptors are supported by exocyclic ones, here a trifluoroketone acceptor and a triazole donor.63 To assure stability, these endo- and exocyclic donors are turned off in the twisted ground state because of chalcogen bonding and repulsion, respectively.62
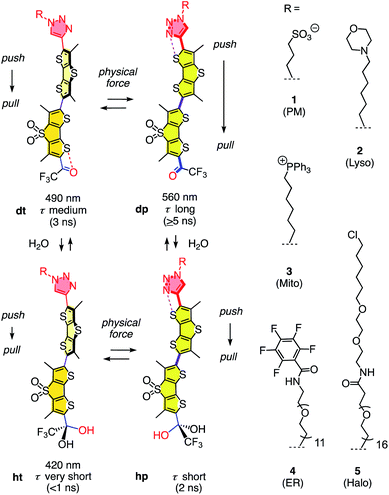 | ||
| Fig. 1 The dual sensing cycle of HydroFlippers 1–5, made to target the indicated MOIs in living cells and responding to membrane compression by planarization and to membrane hydration by dynamic covalent ketone hydration. With indication of excitation maxima (ref. 63) and fluorescence lifetimes (this study). | ||
Mechanical planarization of the flipper probe establishes conjugation along the push–pull systems, electrons flow from endocyclic donors to acceptors, which turns on the exocyclic donors and acceptors to finalize the push–pull system.62 This elaborate, chalcogen-bonding cascade switch has been described elsewhere in detail, including high-level computational simulations.62 The planar high-energy conformer 1dp excels with red shifted excitation and increased quantum yield and lifetime compared to the twisted conformer 1dt because the less twisted Franck-Condon state favors emission through planar intramolecular charge transfer (PICT) over non-radiative decay through twisted ICT, or conical intersections.15
Flipper probe 1 was considered for dual responsiveness to membrane tension and hydration because of the trifluoroketone acceptor.63 Dynamic covalent hydration of 1dt yields hydrate 1ht.64–76 Blue-shifted excitation and short lifetime of 1ht are not expected to improve much upon planarization because the hydrate is a poor acceptor and thus, the push–pull system in 1hp is weak. The dynamic covalent chemistry of the trifluoroketone acceptor has been characterized in detail in solution and in lipid bilayer membranes.63
To explore dual responsiveness to membrane tension in any membrane of interest (MOI) in living cells, HydroFlippers 2–5 were synthesized. While HydroFlipper 1 targets the plasma membrane (PM), HydroFlippers 2–4 were equipped with empirical targeting motifs.77 HydroFlipper 5 terminates with a chloroalkane to react with the self-labeling HaloTag protein, which can be expressed in essentially any MOI.78 Their substantial multistep synthesis was realized by adapting reported procedures (Schemes S1–S4†).
The MOIs labeling selectivity of HydroFlippers was determined in HeLa Kyoto (HK) cells by confocal laser scanning microscopy. Co-localization experiments of flippers 1–4 with the corresponding trackers gave Pearson correlation coefficients (PCCs) >0.80 for the targeting of mitochondria, lysosomes and the endoplasmic reticulum (ER, Fig. S4–S6†). HydroFlipper 5 was first tested with stable HGM cells, which express both HaloTag and GFP on mitochondria (referred to as 5M).78,79 The well-established chloroalkane penetration assay demonstrated the efficient labeling of HaloTag protein by 5 as previously reported HaloFlippers (Fig. S3†).78 By transient transfection, HydroFlippers 5 were also directed to lysosomes (5L), Golgi apparatus (GA, 5G)80 and peroxisomes (5P) with HaloTag and GFP expressed on their surface.78 PCCs >0.80 for co-localization of flipper and GFP emission confirmed that MOI labeling with genetically engineered cells was as efficient as with empirical trackers (Fig. S7–S11†).
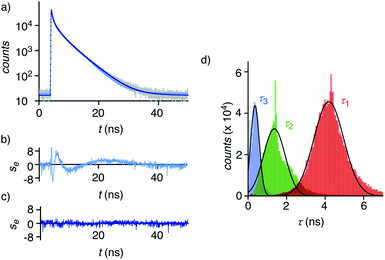
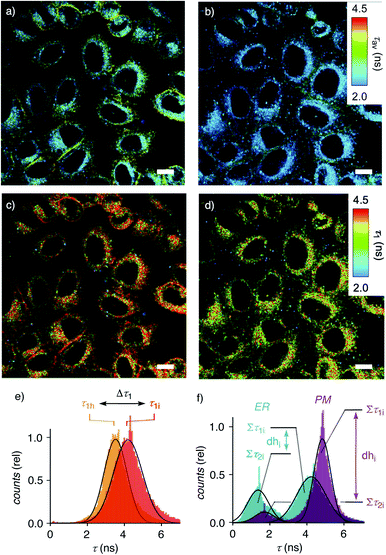
Dual imaging of membrane compression and hydration was envisioned by analysis of fluorescence lifetime imaging microscopy (FLIM) images using a triexponential model (Fig. 2).81 FLIM images of ER HydroFlipper 4 in iso-osmotic HK cells were selected to illustrate the concept (Fig. 3a). Contrary to classical flipper probes, the fluorescence decay curve of the total FLIM image (Fig. 2a, grey) showed a poor fit to a biexponential model (Fig. 2a, cyan, b). Consistent with their expected dual sensing mode, a triexponential fit was excellent (Fig. 2a, dark blue, c). Lifetimes τ1i = 4.3 ns (Table 1, entry 5) and τ2i = 1.5 ns (Table S4†) were obtained besides background. This three-component model was then applied to every pixel of FLIM images (Fig. 3c). The resulting reconvoluted FLIM histogram revealed three clearly separated populations for τ1 (red), τ2 (green), and background (τ3, blue, Fig. 2d). Maxima of these three clear peaks were at the lifetimes estimated by triexponential fit of the global decay curve, thus demonstrating the validity of the methodology at necessarily small photon counts. Irreproducible fitting would give randomly scattered data without separated peaks.
| Probeb | dhic | dhhd | Δdhe (%) | τ 1i (ns) | τ 1h (ns) | Δτ1h (%) | |
|---|---|---|---|---|---|---|---|
| a From triexponential fit of FLIM images in HK cells (errors, see ESI). b Flipper (target MOI). c dhi = area Στ1i/area Στ2i in FLIM histogram under iso-osmotic (i) conditions (e.g.Fig. 3f). d dhh = area Στ1h/area Στ2h in FLIM histogram under hyper-osmotic (h) conditions. e Flipper hydration change in response to membrane tension: Δdh = (1 – dhh/dhi) × 100%. f Fluorescence lifetime value of the slowest component from the fitted fluorescence decay under iso-osmotic (i) conditions (e.g.Fig. 2d). g Same as f, under hyper-osmotic (h) conditions. h Flipper planarization in response to membrane tension: Δτ1 = (1 – τ1h/τ1i) × 100%. i Measured after cholesterol (C) removal from cells with MβCD. j Compared to dhi of 1 (6.6) in untreated cells measured on the same day. k Compared to τih of 1 (5.0) in untreated cells measured on the same day. l As j using 4 and compared to dhi = 1.8. m As k using 4 compared to τih = 4.5. n Measured in transiently transfected HK cells with ST-HaloTag-HA expressed inside GA.80 o Measured in transiently transfected HK cells with HaloTag-Sec61B expressed inside ER.78 p Measured in SM/C GUVs. q Measured in DOPC GUVs. | |||||||
| 1 | 1 (PM) | 6.3 | 6.5 | -3 | 4.8 | 4.4 | 8 |
| 2 | 1 (-C)i | 6.1 | — | 8j | 4.8 | — | 3k |
| 3 | 2 (Lyso) | 2.9 | 2.8 | 4 | 4.4 | 4.0 | 10 |
| 4 | 3 (Mito) | 2.3 | 1.9 | 17 | 4.4 | 4.0 | 8 |
| 5 | 4 (ER) | 1.8 | 1.5 | 17 | 4.3 | 3.7 | 15 |
| 6 | 4 (–C)i | 1.1 | — | 39l | 4.1 | — | 10m |
| 7 | 5 G (GA)n | 2.5 | 2.3 | 8 | 4.2 | 3.8 | 10 |
| 8 | 5 E (ER)o | 1.7 | 1.2 | 29 | 3.8 | 3.7 | 5 |
| 9 | 1 (Lo)p | 11 | — | — | 5.2 | — | — |
| 10 | 1 (Ld)q | 1.2 | — | — | 3.4 | — | — |
Extensive lifetime data for monofunctional flipper probes supported that the intensities associated to τ1i (i for iso-osmotic, see below) originate from at least partially planarized flippers 4d in the ER (Fig. 2d, red, 3c, 1). The population of the τ2i component in the reconvoluted FLIM histogram was attributed to the presence of hydrated 4h in the ER (Fig. 2d, green, 1). This assignment was consistent with lifetime differences in solution between τ = 2.7 ns for the dehydrated and τ = 0.7 ns for the hydrated form of a hydrophobic flipper analog in dioxane-water mixtures (Fig. S2†), and model studies in GUVs (see below).63
The ratio between the τ1i (red) and τ2i (green) populations in the reconvoluted FLIM histogram was used to extract a quantitative measure for hydration of the MOI (Fig. 2d, 3f). A dehydration factor dh was defined by dividing the total integrated counts for τ1 (Στ1) by Στ2. For 4 in iso-osmotic ER, dhi = 1.8 ± 0.1 was obtained (Table 1, entry 5). For comparison, 1 in iso-osmotic PM gave a dhi = 6.3 ± 0.8 (Fig. 3f, Table 1, entry 1). This difference was consistent with poor hydration of the more ordered plasma membrane and high hydration of the less ordered ER membranes. Further in support of this interpretation, 1 in uniform liquid-ordered (Lo) sphingomyelin/cholesterol (SM/C) giant unilamellar vesicles (GUVs) gave significantly larger dhi = 11 ± 3 (τ1 = 5.2 ns, τ2 = 1.9 ns) than in Ld dioleoyl-phosphocholine (DOPC) GUVs (dhi = 1.2 ± 0.1, τ1 = 3.4 ns, τ2 = 1.3 ns, Fig. S12,†Table 1, entries 9, 10). Previous fluorescence and NMR studies in large unilamellar vesicles and in solution demonstrated flipper 1 to be mainly in dehydrated planar 1dp form in Lo membranes, whereas in hydrated twisted 1ht form in Ld membranes.63 Thus, these results implied that the dehydration factor dh obtained from reconvoluted triexponential FLIM images reports quantitatively on membrane hydration, that is the local water concentration around HydroFlippers in their MOI.
In uniform model membranes composed of only one lipid, flipper probes like 1 respond to increasing membrane tension with decreasing lifetimes.15,18 This response can be explained by flipper deplanarization upon lipid decompression. In the mixed membranes composed of different lipids, flipper probes reliably respond to increasing membrane tension with increasing lifetimes, and lifetime changes can be calibrated quantitatively to the applied physical force.18,77 This indicates that in these biologically relevant membranes, the response is dominated by factors other than lipid decompression. Tension-induced microdomain formation is confirmed to account for, or at least contribute to, increasing lifetimes with increasing tension, or membrane decompression.15,18 Not only microdomain disassembly but also changes in membrane curvature from rippling, budding and microdomain softening to tube formation and lipid ejection combine to afford decreasing lifetimes with membrane compression, or decreasing tension.17,18
Membrane tension was applied to the ER by extracellular hyper-osmotic stress. This causes membrane tension to decrease, i.e., membrane compression to increase.18,77 Consistent with tension-induced deplanarization from 4p to 4t (Fig. 1), lifetimes of 4 visibly decreased in response to decreasing membrane tension (Fig. 3b). The reconvoluted FLIM histogram clearly shows that compression caused the decrease of τ1 of 4 in the ER from τ1i = 4.3 ns to τ1h = 3.7 ns, whereas τ2i = 1.5 ns was less mechanosensitive (τ2h = 1.4 ns, Fig. 3e, 4a–c). These different mechanosensitivities were meaningful considering that in three-component histograms, τ1 originates from dehydrated HydroFlipper 4d that loses a strong push–pull dipole and thus shortens lifetime upon tension-induced deplanarization from 4dp to 4dt (Fig. 1). In contrast, hydrated HydroFlipper 4h accounting for τ2 lacks a strong dipole and thus features short lifetimes with poor sensitivity for tension-induced deplanarization from 4hp and 4ht. This result was consistent with the central importance of turn-on push–pull systems for flipper probes to function as mechanosensitive planarizable push–pull probes.81
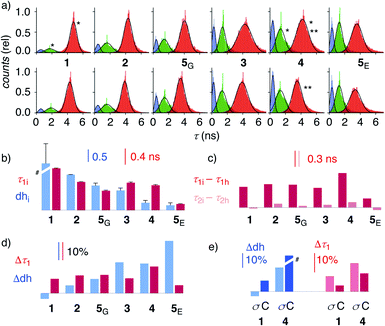 | ||
| Fig. 4 (a) Reconvoluted FLIM histograms for 1–5 obtained by fitting each pixel of the FLIM image to a three-exponential model under iso-osmotic (top) and hyper-osmotic (bottom) conditions in HK cells; *dhi analysis in Fig. 3f; **Δτ1 analysis in Fig. 3e. (b–e) Trend plots for membrane compression (τ1) and hydration (dh) for 1–5 in HK cells without (b, e) and in response to hyper-osmotic membrane tension (c–e). (b) τ1i (iso-osmotic compression) vs. dhi (iso-osmotic hydration). (c) τ1i–τ1hvs. τ2i–τ2h (compression response in ns). (d) Δτ1 (compression response, %) vs. Δdh (hydration response, %), (e) Δτ1 and Δdh upon compression (σ) and cholesterol depletion (C). #Discontinuous, see Table 1. | ||
In response to decreasing membrane tension, or increasing compression, with hyper-osmotic stress, the dehydration factor dhi = 1.8 ± 0.1 of 4 in the ER decreased to dhh = 1.5 ± 0.1 (Table 1, entry 5, S1, entry 6). Decreasing dehydration factors dh corresponded to increasing counts from hydrated flipper 4h upon decreasing membrane tension. Increasing flipper hydration from 4d to 4h was consistent with increasing flipper deplanarization from 4p to 4t with decreasing membrane tension, or increasing compression. Like for planarization, these results implied that the response of the dual probe to tension-induced membrane hydration in mixed membranes is dominated by the same combination of membrane deformation and microdomain softening and disassembly, and not by lipid compression.17,18
The uniform response of HydroFlipper planarization and hydration thus provided corroborative support that membrane deformation and reorganization dominate the fluorescence imaging of membrane tension under the condition that the probe partitions equally between different phases.63 However, the dual response HydroFlipper dissects the consequences of these tension-induced suprastructural changes. HydroFlipper planarization 4t/4p detected by τ1 reports on lipid compression in the local environment in the MOI. HydroFlipper hydration 4d/4h detected by the dehydration factor dh reports on local membrane hydration. Pertinent reports from model membranes in the literature indicate that the two do not have to be the same.59
To elaborate on these implications, FLIM images were recorded for all HydroFlippers 1–5 in their respective MOIs before and after the application of hyper-osmotic stress and then analyzed using the three-component model (Fig. 4a, Table 1). As already stated above, excellent correlations were found between lifetimes at the maxima in histograms (Fig. 4a) and estimated by global triexponential fit (Table 1), confirming validity of the results. Three clear peaks in the FLIM histograms of all tested membranes thus revealed remarkable robustness of the introduced method. However, results must be interpreted carefully. Namely, quantitative comparison of results from changes during experiments, including hyper-osmotic stress, is reliable, also for experiments made in series (e.g., Δτ11, 4 ± C, Table 1). In contrast, absolute values from parallel experiments with different cell cultures (e.g., τ1i of trackers 1–4vs. HaloFlipper 5 in GA (5G) vs.5 in ER (5E)) and identical cell cultures measured at different days (e.g., τ1i1, 4 ± C, Table 1), with different levels of transfection, etc., should not be overinterpreted. These absolute values (but not their relative changes) from different cell culture and experiments from different days can fluctuate within a small range for reasons beyond experimental control (e.g., cell cycle progression, transfection side effects, media). The values for τ2i vary also with changing membrane environments within a small range below 2 ns (Fig. 3f, 4a). However, these changes do not affect dhi, which compares areas rather than maxima in the histograms.
Trends for membrane hydration and compression reported by dhi and τ1i, respectively, should reflect the overall composition and thus nature of the different membranes. For PM 1, Lyso 2, GA 5G and ER 5E, coinciding trends were found for hydration (dhi, blue) and compression (τ1i, red, Fig. 4b). Hydration and deplanarization increased in parallel, consistent with increasingly disordered membranes. With Mito 3 and ER 4, increasing hydration (blue) was not reflected in increasing deplanarization (red, Fig. 4b).
For the comprehensive analysis of the changes caused by hyper-osmotic stress, the differences in lifetimes for τ1 and τ2 were clarified first. Whereas τ1i–τ1h values (red) around 0.3 ns were large and significant in all MOIs, τ2i–τ2h values (pink) were negligible (Fig. 4c). The mechano-insensitive τ2, corresponding to hydrate 4h, were thus not further considered as a valid measure of membrane compression.
To facilitate direct comparability, membrane compression Δτ1 and membrane dehydration Δdh in response to hyper-osmotic stress were converted in percentage of decrease (positive) or increase (negative) from the value under iso-osmotic conditions (Fig. 4d, Table 1). Throughout the different MOIs, decompression Δτ1 reported reliably an about 10% decrease upon hyper-osmotic stress (Fig. 4d, red). In clear contrast, dehydration Δdh varied from 3% increase to 29% decrease (Fig. 4d, blue). The most extreme deviations concerned ER probes with maximal Δτ1 responsiveness for tracker 4 and minimal Δτ1 responsiveness for Halo flipper 5E. For dehydration Δdh, both probes showed high responsiveness. These extremes could reflect the diverse membrane properties of the ER, with τ = 4.1, 3.5 and 3.4 ns reported previously for different flipper mechanophores in tubular, sheet, and nuclear membranes of COS7 cells, respectively.15,77 Although less resolvable in HK cells, this heterogeneity of ER membranes is also visible in the FLIM images with 4 (Fig. 3). Tracker 4 and Halo flipper 5E both react covalently with membrane proteins and report on the respective surrounding ER membrane, which differs significantly according to the two HydroFlipper probes. The extreme values for Halo flipper 5E suggested that other factors like fractions of mispositioned flipper in more hydrophilic environment could also contribute to the global outcome (Fig. 4b, Table 1, entry 8).
The result of the mostly MOI-independent Δτ1 and the more MOI-dependent Δdh naturally afforded inconsistent trends. For Δτ1, the response to tension decreased with 4 > 2, 5G, 3, 1 > 5E, while Δdh followed the order 5E > 3, 4 > 5G > 2 > 1. In general, Δdh values (Fig. 4d, blue) increased with membranes disorder characterized by shorter τ1i and low dhi (Fig. 4b), while Δτ1 remained more constant until the possible onset of decreases at very high hydration (5E, Fig. 4d, red). Both observations - independence of mechanical flipper planarization and dependence of dynamic covalent hydrate formation on the water concentration in the surrounding membrane - were chemically meaningful.
The validity of these conclusions was tested by removing cholesterol with methyl-β-cyclodextrin (MβCD). As expected for the increased hydration level and decreased order of cholesterol depleted membranes, Δdh and Δτ1 of 1 and 4 increased by MβCD treatment compared to those obtained on the same day without the treatment (Fig. 4e, C). Stronger response of ER HydroFlipper 4 to the cholesterol removal can be attributed to the poorer cholesterol content in ER membranes than in PM.82 Consistent with the overall trend, Δdh was more significantly affected by changes of the MOI by MβCD treatment than by tension change (Fig. 4e, blue, C vs. σ), while Δτ1 responded better to membrane tension than MOI change (Fig. 4e, red, C vs. σ).
Taken together, these results reveal HydroFlippers as first dual mode fluorescent membrane tension probe, reporting on membrane hydration and membrane compression at the same time. Mechanical compression is reported as shift in τ, while tension-induced hydration is reported as change in relative photon counts for hydrated and dehydrated probes in the reconvoluted FLIM histograms. The response of flipper deplanarization to membrane tension is robust and less dependent on the nature of the MOI, including plasma membrane, ER, mitochondria, lysosomes and Golgi. In contrast, the responsiveness of flipper hydration to membrane tension depends strongly on the nature of the MOI, generally increasing with increasing intrinsic disorder, that is hydration, already under iso-osmotic conditions. These results validate the flipper probes as most reliable to routinely image membrane tension in cells, while the simultaneous information provided on membrane dehydration provides attractive possibilities for biological applications.
Experimental section
See ESI.†Data availability
Data for this paper are available at Zenodo at https://doi.org/10.5281/zenodo.5801916.Author contributions
J. G.-C. synthesized and characterized all probes, J. L.-A. developed the new method, V. M., C. R., A. R., J. M. and A. F. contributed to probe characterization, N. S. and S. M. directed the study, and all co-authors contributed to experiment design, data analysis and interpretation, and manuscript writing.Conflicts of interest
The authors declare the following competing financial interest: The University of Geneva has licensed four Flipper-TR® probes to Spirochrome for commercialization.Acknowledgements
We thank K. Strakova for imaging, the NMR, MS, Bioimaging and ACCESS platforms for services, J. A. Kritzer (Tufts University), S. J. Marciniak (University of Cambridge), D. Toomre (Yale University), M. Hensel (Osnabrück University), and M. A. Lampson (University of Pennsylvania) for providing materials, and the University of Geneva, the National Centre for Competence in Research (NCCR) Chemical Biology, the NCCR Molecular Systems Engineering and the Swiss NSF for financial support. AR acknowledges funding from the Swiss National Fund for Research Grants N°31003A_149975 and N°31003A_173087, and Synergia Grant N° CRSII5_189996, and the European Research Council Synergy Grant N°951324-R2- TENSION. AF acknowledges financial support by the Société Académique de Genève.Notes and references
- R. J. Leiphart, D. Chen, A. P. Peredo, A. E. Loneker and P. A. Janmey, Langmuir, 2019, 35, 7509–7519 CrossRef CAS PubMed.
- B. Pontes, P. Monzo and N. C. Gauthier, Semin. Cell Dev. Biol., 2017, 71, 30–41 CrossRef CAS PubMed.
- P. Roca-Cusachs, V. Conte and X. Trepat, Nat. Cell Biol., 2017, 19, 742–751 CrossRef CAS PubMed.
- M. Krieg, G. Fläschner, D. Alsteens, B. M. Gaub, W. H. Roos, G. J. L. Wuite, H. E. Gaub, C. Gerber, Y. F. Dufrêne and D. J. Müller, Nat. Rev. Phys., 2019, 1, 41–57 CrossRef.
- A. Diz-Muñoz, O. D. Weiner and D. A. Fletcher, Nat. Phys., 2018, 14, 648–652 Search PubMed.
- C. Grashoff, B. D. Hoffman, M. D. Brenner, R. Zhou, M. Parsons, M. T. Yang, M. A. McLean, S. G. Sligar, C. S. Chen, T. Ha and M. A. Schwartz, Nature, 2010, 466, 263–266 CrossRef CAS PubMed.
- J. H. R. Hetmanski, H. de Belly, I. Busnelli, T. Waring, R. V. Nair, V. Sokleva, O. Dobre, A. Cameron, N. Gauthier, C. Lamaze, J. Swift, A. del Campo, T. Starborg, T. Zech, J. G. Goetz, E. K. Paluch, J.-M. Schwartz and P. T. Caswell, Dev. Cell, 2019, 51, 460–475 CrossRef CAS PubMed.
- G. A. Shamsan and D. J. Odde, Curr. Opin. Chem. Biol., 2019, 53, 125–130 CrossRef CAS PubMed.
- E. J. Aird, K. J. Tompkins, M. P. Ramirez and W. R. Gordon, ACS Sens., 2020, 5, 34–39 CrossRef CAS PubMed.
- R. Jöhr, M. S. Bauer, L. C. Schendel, C. Kluger and H. E. Gaub, Nano Lett., 2019, 19, 3176–3181 CrossRef PubMed.
- A. Saric and S. A. Freeman, Front. Cell Dev. Biol., 2021, 8, 611326 CrossRef PubMed.
- M. C. Piontek, R. B. Lira and W. H. Roos, Biochim. Biophys. Acta, Gen. Subj., 2021, 1865, 129486 CrossRef CAS PubMed.
- E. Sitarska and A. Diz-Muñoz, Curr. Opin. Cell Biol., 2020, 66, 11–18 CrossRef CAS PubMed.
- Y. Liu, K. Galior, V. P.-Y. Ma and K. Salaita, Acc. Chem. Res., 2017, 50, 2915–2924 CrossRef CAS PubMed.
- L. Assies, J. García-Calvo, F. Piazzolla, S. Sanchez, T. Kato, L. Reymond, A. Goujon, A. Colom, J. López-Andarias, K. Straková, D. Mahecic, V. Mercier, M. Riggi, N. Jiménez-Rojo, C. Roffay, G. Licari, M. Tsemperouli, F. Neuhaus, A. Fürstenberg, E. Vauthey, S. Hoogendoorn, M. Gonzalez-Gaitan, A. Zumbuehl, K. Sugihara, J. Gruenberg, H. Riezman, R. Loewith, S. Manley, A. Roux, N. Winssinger, N. Sakai, S. Pitsch and S. Matile, Chimia, 2021, 75, 1004–1011 CrossRef CAS PubMed.
- K. Oglęcka, P. Rangamani, B. Liedberg, R. S. Kraut and A. N. Parikh, eLife, 2014, 3, e03695 CrossRef PubMed.
- M. Páez-Pérez, I. López-Duarte, A. Vyšniauskas, N. J. Brooks and M. K. Kuimova, Chem. Sci., 2020, 12, 2604–2613 RSC.
- A. Colom, E. Derivery, S. Soleimanpour, C. Tomba, M. D. Molin, N. Sakai, M. González-Gaitán, S. Matile and A. Roux, Nat. Chem., 2018, 10, 1118–1125 CrossRef CAS PubMed.
- J. C. S. Ho, P. Rangamani, B. Liedberg and A. N. Parikh, Langmuir, 2016, 32, 2151–2163 CrossRef CAS PubMed.
- R. S. Petruzielo, F. A. Heberle, P. Drazba, J. Katsaras and G. W. Feigenson, Biochim. Biophys. Acta, 2013, 1828, 1302–1313 CrossRef CAS PubMed.
- T. Hamada, Y. Kishimoto, T. Nagasaki and M. Takagi, Soft Matter, 2011, 7, 9061 RSC.
- D. Chen and M. M. Santore, Biochim. Biophys. Acta, 2014, 1838, 2788–2797 CrossRef CAS PubMed.
- K. Oglecka, J. Sanborn, A. N. Parikh and R. S. Kraut, Front. Physiol., 2012, 3, 120 CAS.
- M. Yanagisawa, M. Imai and T. Taniguchi, Phys. Rev. Lett., 2008, 100, 148102 CrossRef PubMed.
- R. Koynova and M. Caffrey, Biochim. Biophys. Acta, 1998, 1376, 91–145 CrossRef CAS.
- A. Fin, A. Vargas Jentzsch, N. Sakai and S. Matile, Angew. Chem., Int. Ed., 2012, 51, 12736–12739 CrossRef CAS PubMed.
- M. Dal Molin, Q. Verolet, A. Colom, R. Letrun, E. Derivery, M. Gonzalez-Gaitan, E. Vauthey, A. Roux, N. Sakai and S. Matile, J. Am. Chem. Soc., 2015, 137, 568–571 CrossRef CAS PubMed.
- G. Licari, K. Strakova, S. Matile and E. Tajkhorshid, Chem. Sci., 2020, 11, 5637–5649 RSC.
- D. I. Danylchuk, P.-H. Jouard and A. S. Klymchenko, J. Am. Chem. Soc., 2021, 143, 912–924 CrossRef CAS PubMed.
- N. Li, W. Zhang, H. Lin and J.-M. Lin, Chin. Chem. Lett., 2021 DOI:10.1016/j.cclet.2021.08.060.
- L. Jin, A. C. Millard, J. P. Wuskell, X. Dong, D. Wu, H. A. Clark and L. M. Loew, Biophys. J., 2006, 90, 2563–2575 CrossRef CAS PubMed.
- D. M. Owen, P. M. P. Lanigan, C. Dunsby, I. Munro, D. Grant, M. A. A. Neil, P. M. W. French and A. I. Magee, Biophys. J., 2006, 90, L80–L82 CrossRef CAS PubMed.
- J. H. Lorent, K. R. Levental, L. Ganesan, G. Rivera-Longsworth, E. Sezgin, M. Doktorova, E. Lyman and I. Levental, Nat. Chem. Biol., 2020, 16, 644–652 CrossRef CAS PubMed.
- E. Sezgin, T. Sadowski and K. Simons, Langmuir, 2014, 30, 8160–8166 CrossRef CAS PubMed.
- T. Baumgart, G. Hunt, E. R. Farkas, W. W. Webb and G. W. Feigenson, Biochim. Biophys. Acta, 2007, 1768, 2182–2194 CrossRef CAS PubMed.
- A. S. Klymchenko, Acc. Chem. Res., 2017, 50, 366–375 CrossRef CAS PubMed.
- A. S. Klymchenko and A. P. Demchenko, J. Am. Chem. Soc., 2002, 124, 12372–12379 CrossRef CAS PubMed.
- A. S. Klymchenko, S. Oncul, P. Didier, E. Schaub, L. Bagatolli, G. Duportail and Y. Mély, Biochim. Biophys. Acta, 2009, 1788, 495–499 CrossRef CAS PubMed.
- J. A. Robson, M. Kubánková, T. Bond, R. A. Hendley, A. J. P. White, M. K. Kuimova and J. D. E. T. Wilton-Ely, Angew. Chem., Int. Ed., 2020, 59, 21431–21435 CrossRef CAS PubMed.
- A. Vyšniauskas, M. Balaz, H. L. Anderson and M. K. Kuimova, Phys. Chem. Chem. Phys., 2015, 17, 7548–7554 RSC.
- A. S. Kashirina, I. López-Duarte, M. Kubánková, A. A. Gulin, V. V. Dudenkova, S. A. Rodimova, H. G. Torgomyan, E. V. Zagaynova, A. V. Meleshina and M. K. Kuimova, Sci. Rep., 2020, 10, 14063 CrossRef CAS PubMed.
- J. E. Chambers, M. Kubánková, R. G. Huber, I. López-Duarte, E. Avezov, P. J. Bond, S. J. Marciniak and M. K. Kuimova, ACS Nano, 2018, 12, 4398–4407 CrossRef CAS PubMed.
- Y. Zheng, Y. Ding, J. Ren, Y. Xiang, Z. Shuai and A. Tong, Anal. Chem., 2020, 92, 14494–14500 CrossRef CAS PubMed.
- L. Yu, J. F. Zhang, M. Li, D. Jiang, Y. Zhou, P. Verwilst and J. S. Kim, Chem. Commun., 2020, 56, 6684–6687 RSC.
- R. Guo, J. Yin, Y. Ma, Q. Wang and W. Lin, J. Mater. Chem. B, 2018, 6, 2894–2900 RSC.
- Y. Zheng, Y. Ding, X. Zheng, C. Zhang, Y. Zhang, Y. Xiang and A. Tong, Anal. Chem., 2021, 93, 10272–10281 CrossRef CAS PubMed.
- I. E. Steinmark, A. L. James, P.-H. Chung, P. E. Morton, M. Parsons, C. A. Dreiss, C. D. Lorenz, G. Yahioglu and K. Suhling, PLoS One, 2019, 14, e0211165 CrossRef CAS PubMed.
- T. Liu, X. Liu, D. R. Spring, X. Qian, J. Cui and Z. Xu, Sci. Rep., 2014, 4, 5418 CrossRef CAS PubMed.
- L. Wang, Y. Xiao, W. Tian and L. Deng, J. Am. Chem. Soc., 2013, 135, 2903–2906 CrossRef CAS PubMed.
- Z. Yang, Y. He, J.-H. Lee, N. Park, M. Suh, W.-S. Chae, J. Cao, X. Peng, H. Jung, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2013, 135, 9181–9185 CrossRef CAS PubMed.
- K. T. Fam, L. Saladin, A. S. Klymchenko and M. Collot, Chem. Commun., 2021, 57, 4807–4810 RSC.
- X. Peng, Z. Yang, J. Wang, J. Fan, Y. He, F. Song, B. Wang, S. Sun, J. Qu, J. Qi and M. Yan, J. Am. Chem. Soc., 2011, 133, 6626–6635 CrossRef CAS PubMed.
- D. I. Danylchuk, E. Sezgin, P. Chabert and A. S. Klymchenko, Anal. Chem., 2020, 92, 14798–14805 CrossRef CAS PubMed.
- C.-H. Wu, Y. Chen, K. A. Pyrshev, Y.-T. Chen, Z. Zhang, K.-H. Chang, S. O. Yesylevskyy, A. P. Demchenko and P.-T. Chou, ACS Chem. Biol., 2020, 15, 1862–1873 CrossRef CAS PubMed.
- Z. Zhang, G. Sun, W. Chen, J. Su and H. Tian, Chem. Sci., 2020, 11, 7525–7537 RSC.
- H. V. Humeniuk, A. Rosspeintner, G. Licari, V. Kilin, L. Bonacina, E. Vauthey, N. Sakai and S. Matile, Angew. Chem., Int. Ed., 2018, 57, 10559–10563 CrossRef CAS PubMed.
- W. Nakanishi, S. Saito, N. Sakamoto, A. Kashiwagi, S. Yamaguchi, H. Sakai and K. Ariga, Chem.–Asian J., 2019, 14, 2869–2876 CrossRef CAS PubMed.
- O. A. Kucherak, S. Oncul, Z. Darwich, D. A. Yushchenko, Y. Arntz, P. Didier, Y. Mély and A. S. Klymchenko, J. Am. Chem. Soc., 2010, 132, 4907–4916 CrossRef CAS PubMed.
- G. M'Baye, Y. Mély, G. Duportail and A. S. Klymchenko, Biophys. J., 2008, 95, 1217–1225 CrossRef PubMed.
- B. Baumeister and S. Matile, Chem.–Eur. J., 2000, 6, 1739–1749 CrossRef CAS.
- A. Bauzá, T. J. Mooibroek and A. Frontera, ChemPhysChem, 2015, 16, 2496–2517 CrossRef PubMed.
- M. Macchione, A. Goujon, K. Strakova, H. V. Humeniuk, G. Licari, E. Tajkhorshid, N. Sakai and S. Matile, Angew. Chem., Int. Ed., 2019, 58, 15752–15756 CrossRef CAS PubMed.
- J. García-Calvo, J. Maillard, I. Fureraj, K. Strakova, A. Colom, V. Mercier, A. Roux, E. Vauthey, N. Sakai, A. Fürstenberg and S. Matile, J. Am. Chem. Soc., 2020, 142, 12034–12038 CrossRef PubMed.
- D. Drahoňovský and J.-M. Lehn, J. Org. Chem., 2009, 74, 8428–8432 CrossRef PubMed.
- R.-C. Brachvogel and M. von Delius, Eur. J. Org. Chem., 2016, 2016, 3662–3670 CrossRef CAS.
- I. V. Kolesnichenko and E. V. Anslyn, Chem. Soc. Rev., 2017, 46, 2385–2390 RSC.
- L. You and E. V. Anslyn, Org. Lett., 2009, 11, 5126–5129 CrossRef CAS PubMed.
- R. Caraballo, H. Dong, J. P. Ribeiro, J. Jiménez-Barbero and O. Ramström, Angew. Chem., Int. Ed., 2010, 49, 589–593 CrossRef CAS PubMed.
- M. Sakulsombat, Y. Zhang and O. Ramström, Chem.–Eur. J., 2012, 18, 6129–6132 CrossRef CAS PubMed.
- Y. Zhang, L. Hu and O. Ramström, Chem. Commun., 2013, 49, 1805–1807 RSC.
- E. Mertz and S. C. Zimmerman, J. Am. Chem. Soc., 2003, 125, 3424–3425 CrossRef CAS PubMed.
- Y. Qian, J. Karpus, O. Kabil, S.-Y. Zhang, H.-L. Zhu, R. Banerjee, J. Zhao and C. He, Nat. Commun., 2011, 2, 1–7 Search PubMed.
- Y. Xu, S. Yu, Y. Wang, L. Hu, F. Zhao, X. Chen, Y. Li, X. Yu and L. Pu, Eur. J. Org. Chem., 2016, 2016, 5868–5875 CrossRef CAS.
- G. J. Mohr, F. Lehmann, U.-W. Grummt and U. E. Spichiger-Keller, Anal. Chim. Acta, 1997, 344, 215–225 CrossRef CAS.
- G. J. Mohr, Sens. Actuators, B, 2003, 90, 31–36 CrossRef CAS.
- S. Sasaki, Y. Kotegawa and H. Tamiaki, Tetrahedron Lett., 2006, 47, 4849–4852 CrossRef CAS.
- A. Goujon, A. Colom, K. Straková, V. Mercier, D. Mahecic, S. Manley, N. Sakai, A. Roux and S. Matile, J. Am. Chem. Soc., 2019, 141, 3380–3384 CrossRef CAS PubMed.
- K. Straková, J. López-Andarias, N. Jiménez-Rojo, J. E. Chambers, S. J. Marciniak, H. Riezman, N. Sakai and S. Matile, ACS Cent. Sci., 2020, 6, 1376–1385 CrossRef PubMed.
- L. Peraro, K. L. Deprey, M. K. Moser, Z. Zou, H. L. Ball, B. Levine and J. A. Kritzer, J. Am. Chem. Soc., 2018, 140, 11360–11369 CrossRef CAS PubMed.
- R. S. Erdmann, S. W. Baguley, J. H. Richens, R. F. Wissner, Z. Xi, E. S. Allgeyer, S. Zhong, A. D. Thompson, N. Lowe, R. Butler, J. Bewersdorf, J. E. Rothman, D. St Johnston, A. Schepartz and D. Toomre, Cell Chem. Biol., 2019, 26, 584–592 CrossRef CAS PubMed.
- J. García-Calvo, J. López-Andarias, N. Sakai and S. Matile, Chem. Commun., 2021, 57, 3913–3916 RSC.
- G. van Meer, D. R. Voelker and G. W. Feigenson, Nat. Rev. Mol. Cell Biol., 2008, 9, 112–124 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed procedures and results for all reported experiments. See DOI: 10.1039/d1sc05208j |
| ‡ These two authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2022 |