Open Access Article
Open Access ArticleRetaining the structural integrity of disulfide bonds in diphtheria toxoid carrier protein is crucial for the effectiveness of glycoconjugate vaccine candidates†
Filippo
Carboni‡
a,
Annabel
Kitowski‡
 b,
Charlotte
Sorieul
b,
Charlotte
Sorieul
 a,
Daniele
Veggi
a,
Marta C.
Marques
b,
Davide
Oldrini
a,
Evita
Balducci
a,
Barbara
Brogioni
a,
Linda
Del Bino
a,
Alessio
Corrado
a,
Francesca
Angiolini
a,
Lucia
Dello Iacono
a,
Immaculada
Margarit
a,
Maria Rosaria
Romano
a,
Gonçalo J. L.
Bernardes
a,
Daniele
Veggi
a,
Marta C.
Marques
b,
Davide
Oldrini
a,
Evita
Balducci
a,
Barbara
Brogioni
a,
Linda
Del Bino
a,
Alessio
Corrado
a,
Francesca
Angiolini
a,
Lucia
Dello Iacono
a,
Immaculada
Margarit
a,
Maria Rosaria
Romano
a,
Gonçalo J. L.
Bernardes
 *bc and
Roberto
Adamo
*bc and
Roberto
Adamo
 *a
*a
aGSK, Via Fiorentina 10, 53100 Siena, Italy. E-mail: roberto.x.adamo@gsk.com
bInstituto de Medicina Molecular, Faculdade de Medicina da Universidade de Lisboa, Lisboa, Portugal. E-mail: gbernardes@medicina.ulisboa.pt
cYusuf Hamied Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK. E-mail: gb453@cam.ac.uk
First published on 10th February 2022
Abstract
The introduction of glycoconjugate vaccines marks an important point in the fight against various infectious diseases. The covalent conjugation of relevant polysaccharide antigens to immunogenic carrier proteins enables the induction of a long-lasting and robust IgG antibody response, which is not observed for pure polysaccharide vaccines. Although there has been remarkable progress in the development of glycoconjugate vaccines, many crucial parameters remain poorly understood. In particular, the influence of the conjugation site and strategy on the immunogenic properties of the final glycoconjugate vaccine is the focus of intense research. Here, we present a comparison of two cysteine selective conjugation strategies, elucidating the impact of both modifications on the structural integrity of the carrier protein, as well as on the immunogenic properties of the resulting glycoconjugate vaccine candidates. Our work suggests that conjugation chemistries impairing structurally relevant elements of the protein carrier, such as disulfide bonds, can have a dramatic effect on protein immunogenicity.
Introduction
Vaccines are considered as one of the most cost-effective interventions to prevent morbidity and mortality from infectious diseases.1 Among the different approaches to vaccine design, glycoconjugate vaccines have been proven efficacious and cost-effective in the prevention of Haemophilus influenzae type b (Hib), Streptococcus pneumoniae (23 serotypes), Neisseria meningitidis (A, C, W135 and Y) and Salmonella typhi.2 Conjugate vaccines are obtained by the covalent linkage of bacterial polysaccharides to immunogenic carrier proteins, and have been demonstrated to overcome the limitations frequently exhibited by unconjugated polysaccharide vaccines.3 The T-cell help provided by the protein epitopes of glycoconjugates imparts to the carbohydrates – which are per se T-cell independent antigens – the capacity to induce long-lasting and boostable IgG antibody production. Six proteins are currently used as carriers in licensed vaccines, including tetanus toxoid (TT), diphtheria toxoid (DT), Cross-Reactive Material 197 (CRM197), the outer membrane protein complex of Meningococcus B (OMPC), protein D from H. influenzae, and the recombinant exotoxin A of Pseudomonas aeruginosa.Carbohydrates can be linked to proteins by using a variety of approaches. Amino acid residues that are most suitable for chemical linkage to sugars are those well-exposed onto the protein surface and whose side chains have reactive functional groups, such as primary amino groups of lysines and carboxylic groups of glutamic, or aspartic acid residues.4–6 Generally, linking of a polysaccharide (PS) to a carrier protein results invariably in a random display of the carbohydrate on the protein surface, although some selectivity homogeneity can be obtained by modulating the carbohydrate to protein stoichiometry. A variety of factors that are associated with the conjugation methodology (e.g. conjugation chemistry, multiple attachment versus single-point attachment of carbohydrates, presence/absence of linkers) have an impact on the chemical and biological properties of different glycoconjugate constructs.6 This poses a serious limitation in the comparison of different glycoconjugates and in understanding their mechanism of action. Therefore, there is a need for site-selective glycoconjugation methods and effort has been recently devoted to the preparation of glycoconjugates with defined attachment.7 High regioselectivity has been achieved by targeting amino acids naturally present in the protein carrier, such as cysteine,8,9 cysteine disulfide bridges10,11 or tyrosines.12–18 Alternatively, amino acid tags for enzyme mediated conjugation19,20 or incorporation of unnatural amino acids21 have been exploited. The development of site-selective conjugation strategies on naturally occurring amino acids, such as cysteine, is appealing for the simplicity of these approaches, as more complicated and time-consuming steps like sequence engineering become unnecessary.22 Selectivity of glycoconjugation is achieved by targeting either amino acids with sufficiently high reactivity during chemical reactions, or amino acid patterns within the protein structure that allow selective reactivities. In this context, disulfide bridges are an optimal target for site-selective reactions, as they are usually present in limited numbers and, upon reduction, show nucleophilic properties that can be used for chemical reactions.23,24 Disulfide modifications have been successfully used for incorporation of small molecule payloads into antibodies and have attracted specific attention for the development of glycoconjugate vaccines, because of the selectivity that can be achieved on these groups on the carrier protein CRM197.5,25,26
CRM197 is an enzymatically inactive and nontoxic form of diphtheria toxin, which has been detoxified through a single G52E mutation. CRM197 is a well-characterized protein, whose X-ray structure has been elucidated.27 It is synthesized as a single-chain holoprotein, which comprises two domains, fragment A (catalytic domain) and fragment B (transmembrane domain). Two disulfide bridges are present in the intact holoprotein: one bridge joins C186 to C201, linking fragment A to fragment B, while a second bridge joins C461 to C471 within fragment B.28 Polysaccharide conjugates of CRM197 are components of vaccine formulations protecting against important bacterial pathogens including Streptococcus pneumoniae (Prevnar), Haemophilus influenzae type b (HibTITER, Vaxem-Hib) and Neisseria meningitidis serogroup A, C, Y and W-135 (e.g. Menveo and Menjugate).29 Recently, disulfide re-bridging with acetone was developed for site-selective conjugation of Salmonella O-antigen to CRM197 and improved immunogenicity of the protein as an antigen was observed after grafting one of the two disulfide bonds with oxetane.9,30
In this study, we investigated the scope and limitations of two different site-selective modification methods on the disulfide functionalities of the carrier protein CRM197. By choosing two opposing strategies, we sought to elaborate the importance of retaining the structural integrity of a disulfide functionality through acetone re-bridging or, in contrast, the effect of opening and converting a disulfide bridge into two dehydroalanine residues, on the properties of a resulting glycoconjugate vaccine candidate. In depth structural characterization of the modified CRM197 was carried out to unravel differences between the two methods, and the X-ray crystallographic structure of acetone modified CRM197 was successfully resolved. Capsular polysaccharides from group B Streptococcus (GBS) serotypes Ia and III, as well as capsular polysaccharide from S. pneumoniae (Sp) serotype 14, were chosen as model antigens and the generation of immune responses after administration of these vaccine candidates was evaluated.
Results
Selective glycoconjugation at C186-201 of CRM197
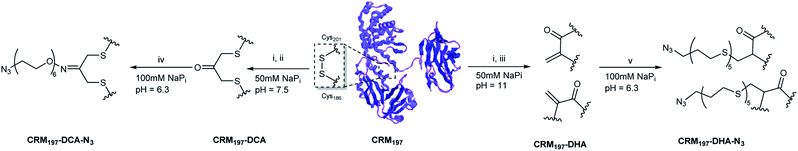
From the two disulfide bridges present in the carrier protein CMR197, the C461–C471 bond appeared to be buried inside the protein, while the C186–201 is surface-exposed. We have already reported that selective modification of the latter disulfide bridge can be achieved by partial reduction of CRM197 to release C186 and C201 in the presence of tris(2-carboxyethyl) phosphine (TCEP).9 We envisaged to modify the C186–C201 bond using two different strategies for the subsequent two-step conjugation of large polysaccharides (Scheme 1). First, re-bridging of C186–C201 with 1,3-dichloroacetone (DCA) would allow insertion of a ketone handle, which can be further modified with a bifunctional linker bearing an aminoxy to form an aminooxy derivative, and an azide group useful for the conjugation of polysaccharides derivatized with alkyne functionalities via azido-alkyne Huisgen (3 + 2) cycloaddition (click chemistry). This method enables incorporation of a single carbohydrate moiety per protein. Also, preliminary experiments with sugar modified with aminoxy linkers proved the direct conjugation challenging, due to the size of GBS polysaccharides (MW ∼150–200 kDa).The multivalent presentation of sugar antigens on a protein carrier is known to be an important factor for the efficient generation of an immunogenic response. To enable selective conjugation of a higher number of sugar molecules on the protein, a second strategy was developed, relying on the opening of C186–C201 and subsequent conversion of the free cysteine residue into dehydroalanine (Dha). In this approach, further conjugation can be achieved through thiol Michael addition on the introduced Dha groups of a bifunctional thiol linker bearing an azido moiety for the subsequent conjugation reaction to alkyne derivatized polysaccharides by click chemistry (Scheme 1). These two methods appeared very attractive to be compared in terms of the construction of site-selective glycoconjugates and immunogenicity of the resulting biomolecules, since the first strategy retains the covalent connection between cysteine residues C186 and C201, whereas the second strategy results in the opening of this disulfide bridge.
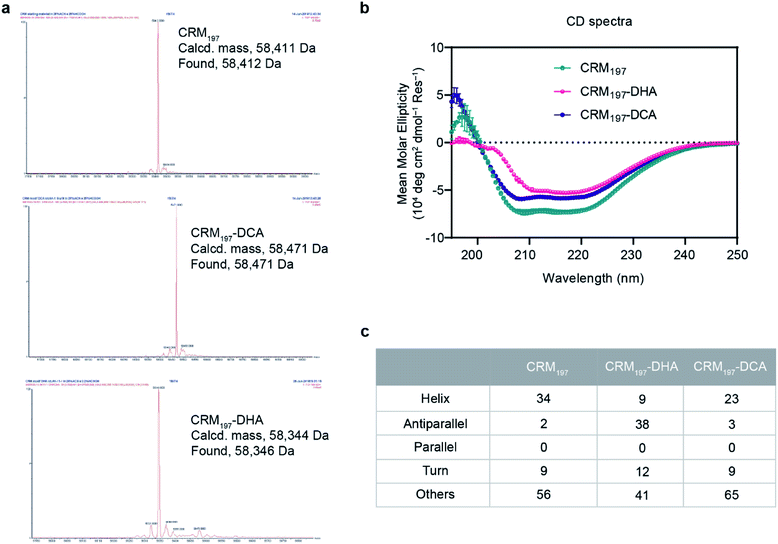
For the generation of CRM197-DCA, after selective reduction of C186–C201 in the presence of tris(2-carboxyethyl) phosphine (TCEP, 12 equiv.) at pH 7.5 for three hours, the protein was incubated with 1,3-dichloroacetone (10 equiv.) for 3.5 hours yielding the modified CRM197. Size exclusion chromatography allowed the removal of small molecules and exchange of buffer to 100 mM sodium phosphate at pH 6.3 for acid catalyzed reaction with the aminoxy linker (Scheme 1). Mass spectrometry revealed virtually complete derivatization of the starting material (Fig. 1). The impact of the installation of the DCA moiety into CRM197 on its structure was also studied by CD (Fig. 1B) and dynamic light scattering (DLS) analysis (Fig. S4, ESI†) and compared with the native protein. We found that CD and DLS spectra of CRM197-DCA were nearly identical to those of CRM197, which indicates that the secondary structure was preserved upon the chemical stapling. The CRM197-DCA was used for condensation with an excess of aminoxy-PEG5-azido linker (600 equiv.) for quantitative insertion of an azido moiety for further polysaccharide conjugation (Scheme 1), as confirmed by LC-MS analysis (Fig. S1, ESI†).
The second strategy was intended to open the disulfide bond C186–C201 in a selective manner and convert each of the two cysteine residues into the amino acid Dha. Similar to the previous approach, CRM197 was first selectively reduced at C186–C201,9 and then introduction of Dha was achieved by treatment with 500 equiv. of methyl 2,5-dibromopentanoate over a period of 5 hours at pH 11. Residual small molecules were removed by size exclusion chromatography with 100 mM sodium phosphate at pH 6.3 for elution. The reaction with methyl 2,5-dibromopentanoate works through a bisalkylation mechanism, followed by an elimination step.21 This second part was the critical step, as reactions at lower pH values were found to result in incomplete elimination products with the pentanoate molecule attached to the protein. A concentration of 5 mg mL−1 protein was key to achieve a high level of modification. Higher concentrations resulted in protein precipitation during the reaction. The introduction of two Dha residues (Scheme 1) and complete conversion of the starting material was confirmed by LC-MS analysis (Fig. 1A and S2, ESI†). Despite the ring opening, a minimal level of protein aggregation was observed when freshly prepared samples were used for the subsequent modifications (Fig. S3, ESI†). The impact of the opening of the disulfide bridge and subsequent conversion of cysteine in Dha residues on the CRM197 structure was evaluated by CD (Fig. 1B) and DLS (Fig. S4, ESI†) experiments using the native protein as the control. We found a few differences both in CD and DLS spectra corresponding probably to a less stable structure for CRM197-DHA related to the loss of the disulfide bond that seems important to maintain the 3D structure of the protein. The selective modification of only C186 and C201 was further confirmed by using peptide mapping and LC-MS analysis (Fig. S5, ESI†). Importantly, the CD spectrum, which is indicative of helical conformation, showed that CRM197-DHA displays a more pronounced minimum at 218 nm compared to CRM197-DCA, typical of the β-sheet conformation. We estimated quantitatively the helix content and twist angle distribution for the antiparallel and parallel β-sheets using the BeStSel server31,32 for both the modified forms (Fig. 1C). The data indicated that the CRM197 variants have lower helical content than the wild type, with the DHA variant showing a higher degree of antiparallel β-sheets that could be related to some level of aggregation. Next, the two dehydroalanine residues of CRM197-DHA were used for thiol Michael addition of the bis(11-azidoundecyl) disulfide linker (Scheme 1). The successful insertion of two linker moieties was confirmed by MS experiments (ESI, Fig. 2†). This reaction provided, as for CRM197-DCA, azido functionalities ready for click chemistry with alkyne derivatized glycan antigens.
X-ray crystallography of CRM197-DCA
Considering the higher similarity observed between CRM197-DCA and the native form, further structural insights were obtained by X-ray crystallography. To this end, we solved the structure of the apo form (NAD-free) of CRM197-DCA at 2 Å resolution (data collection and refinement statistics are reported in Table S1, ESI†). The asymmetric unit (ASU) contains one molecule of CRM197-DCA, which interacts with a second (symmetry-related) molecule by “domain swapping” (Fig. S6, ESI†), as observed in noncovalently associated dimeric native forms previously solved.25 The structure assumes the canonical Y shape of both wild type and mutant forms of diphtheriae toxin, whose arms are composed of C (Catalytic domain) and R (Receptor domain) domains while the T domain (Transmembrane domain) is located at the base30 (Fig. S6, ESI†). The overall fold of CRM197-DCA is superimposable to the NAD-free CRM197 (PDB ID 4AE0) with an overall root mean square deviation (rmsd) of 1.1 Å for 454 equiv. Cα atoms (Fig. 2A). All the regions appear clearly overlapping except for the loops 37–50 in the C domain, 348–353 in the T domain and 516–520 in the R domain, which are in general better resolved in this CRM197-DCA structure compared to apo CRM197 (Fig. 2A). Indeed, the electron density maps of the CRM197-DCA are overall of high quality, allowing the modelling of the entire molecule apart from the region 187–198, connecting the T and C domains and quite close in space to the S–S bond C186–C201, and the residues 42–44 of the catalytic loop 37–50 which appeared completely disordered in the apo CRM197 structure.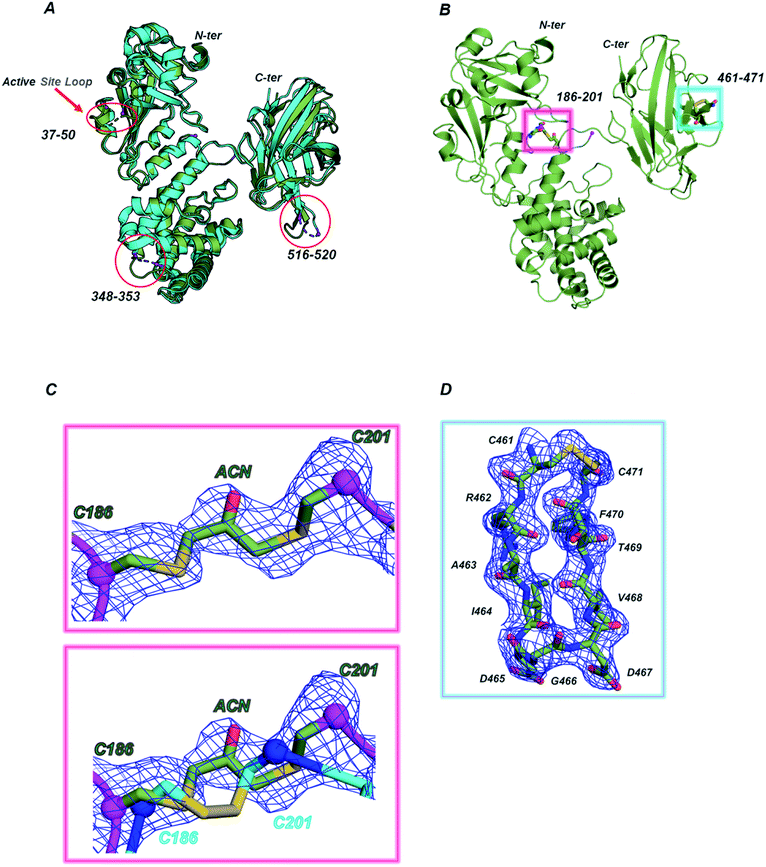 | ||
| Fig. 2 (A) Superimposition between CRM197-DCA (PDB 7O4W, green smudge) and CRM197 (PDB 4AE0, cyan). The loop connecting the C and T domains (187–198) is absent in both structures, the loops 516–520 and 348–353, lacking in the CRM197, are present in the CRM197-DCA while the active site loop 37–50 is still partially incomplete in the CRM197-DCA structure (42–44 residues are missing). (B) Magenta and cyan boxes show the position of the two S–S bonds (C186–C201 and C461–471) in the structure of CRM197-DCA. The C186–C201 is the modified disulfide bond, carrying an acetone moiety, whereas the C461–C471 is still an intact disulfide bridge. (C) Top, 1σ 2Fo–Fc (blue mesh) electron density map of the modified S–S bridge. Bottom, superimposition of the derivatized S–S bridge (green C sticks) with the unmodified one (PDB 4AE0, cyan C sticks). The Cα atoms of C186 and C201 are highlighted as spheres. (D) 1σ 2Fo–Fc (blue mesh) electron density map of the second and still intact S–S bridge (C461–C471), located at the tip of a loop in the R domain. | ||
After the first cycles of refinement, clear extra electron density nearby the S–S bond C186–C201 appeared, confirming the successful and selective insertion of a single acetone molecule (Fig. 2B and C). The distances within Cα atoms of C186 and C201, angles and length bonds values are in agreement with the geometry of Cys–acetone–Cys bridges. Moreover, the flexibility of the loop accommodating the modified S–S bridge allows this acetone moiety to be solvent-exposed and potentially accessible to the conjugation of glycan antigens. In contrast, no modification of the C461–C471 disulfide bond was found (Fig. 2C). Remarkably, our structural data expand what previously shown by entire mass analysis,9,28 indicating the unambiguous presence of selective acetone insertion in the S–S bond C186–C201 of the CRM197-DCA and showing no major impact of this modification on the tertiary structure of the protein.
Glycoconjugation with CRM197-DCA and CRM197-DHA
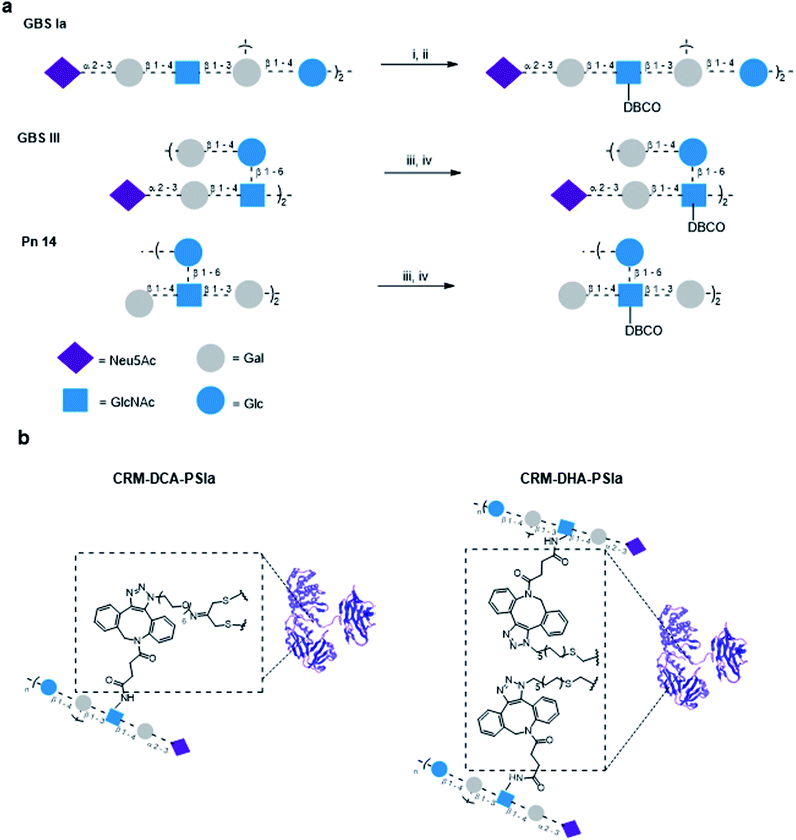
To compare the influence of the introduced protein modifications on the immunogenicity of resulting glycoconjugate constructs, GBS and Sp polysaccharide antigens were chosen for conjugation.Structurally similar polysaccharides from two different bacteria (GBS serotypes types III and Ia and Sp serotype 14) were partially de-N-acetylated and reacted by reaction with dibenzocyclooctyne-N-hydroxysuccinimidyl (DBCO) ester to insert a handle suited for azide–alkyne cycloaddition, which we have shown to be particularly efficient with large polysaccharides.11 To minimize the impact on the polysaccharide structure and preserve sugar epitopes, a limited number of repeating units were modified. The relative ratio of DBCO units per polysaccharide molecule was determined by NMR spectrometry considering the ratio of the peak intensity of the aromatic protons of the linker and the H-3equatorial or H-3axial of the sialic acid for GBS PS or the H-2 of the Glc residue Sp14 PS, respectively (Fig. S7, ESI†). Typically one out of 20 and 40 repeating units was modified, for GBS PS and Sp14, respectively. Azide moieties were incorporated into the protein using either a PEG or an alkyl linker (Fig. 3). Based on our previous experience with strain promoted click chemistry,15–17 it was found that while the alkyl or PEG chain is immunosilent, rigid aromatic systems like DBCO are not; however this type of linker does not shift the immune response away from the carbohydrate. Therefore, although not identical, we considered them not to impact the immunogenicity outcome.
Through the inserted DBCO moiety GBS types Ia and III PS and Sp type 14 were conjugated to CRM197-DCA and CRM197-DHA (Fig. 3) at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polysaccharide/protein w/w ratio and a protein concentration of 2 mg mL−1. Larger amounts of sugar did not further improve the course of conjugation. After removal of the unconjugated polysaccharide, the level of sugar incorporation was estimated by HPAEC-PAD and the protein content was assessed by colorimetric assay (bicinchoninic acid assay). The characteristics of the synthesized site-selective conjugates are summarized in Table 1. The conversion of cysteine residue from C186–C201 into Dha was already shown to be more impactful on the protein 3D structure as compared to the disulfide rebridging. To further assess that the structural integrity of CRM197-DCA was preserved upon glycoconjugation, CD spectra were recorded for CRM197-PSIa and CRM197-DCA-PSIa, using unmodified CRM197 and CRM197-DCA as controls. The CD spectra of the two conjugates (which exhibited a similar size in the DLS analysis) showed that both random and selective conjugation caused a slight shift of the minimum at 218 nm to higher molar ellipticity values, with respect to CRM197 and CRM197-DCA, which appeared almost overlapping (Fig. S8, ESI†).
1 polysaccharide/protein w/w ratio and a protein concentration of 2 mg mL−1. Larger amounts of sugar did not further improve the course of conjugation. After removal of the unconjugated polysaccharide, the level of sugar incorporation was estimated by HPAEC-PAD and the protein content was assessed by colorimetric assay (bicinchoninic acid assay). The characteristics of the synthesized site-selective conjugates are summarized in Table 1. The conversion of cysteine residue from C186–C201 into Dha was already shown to be more impactful on the protein 3D structure as compared to the disulfide rebridging. To further assess that the structural integrity of CRM197-DCA was preserved upon glycoconjugation, CD spectra were recorded for CRM197-PSIa and CRM197-DCA-PSIa, using unmodified CRM197 and CRM197-DCA as controls. The CD spectra of the two conjugates (which exhibited a similar size in the DLS analysis) showed that both random and selective conjugation caused a slight shift of the minimum at 218 nm to higher molar ellipticity values, with respect to CRM197 and CRM197-DCA, which appeared almost overlapping (Fig. S8, ESI†).
| Glycoconjugate | Glycosylation ratio (w/w) | Free saccharide (%) |
|---|---|---|
| CRM197-PSIa | 2.5 | 16 |
| CRM197-DHA-PSIa | 1.1 | <6 |
| CRM197-DCA-PSIa | 4.8 | 11.9 |
| CRM197-PSIIII | 1.2 | <2 |
| CRM197-DCA-PSIII | 1.4 | Nd |
| CRM197-PN14PS | 1.0 | Nd |
| CRM197-DCA-PN14PS | 1.9 | <5 |
In vivo results
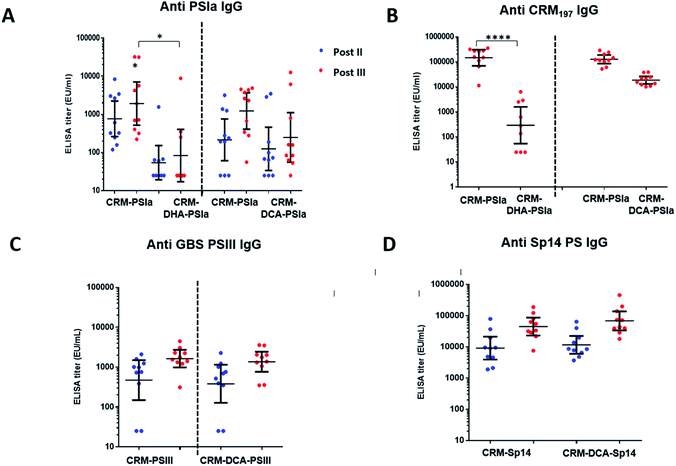
Immune responses induced by site-selective CRM197-DCA-PSIII and CRM197-DHA-PSIa conjugates were compared to the ones obtained with random conjugates where polysaccharide is attached randomly to lysines on the surface of CRM197. These conjugates were prepared as previously reported33 and are considered viable candidates for clinical studies.34,35 In the immunization experiments, groups of ten mice received three doses of the prepared conjugates adjuvanted with alum hydroxide. Two weeks after the second and third vaccine doses, individual IgG titers were measured by ELISA using full-length PSIII or PSIa conjugated to Human Serum Albumin as the coating agent.The immunogenicity of the CRM197-DCA-PSIa conjugate was 3 fold higher than that of the CRM197-DHA counterpart and statistically comparable to that of the random CRM197-PSIa conjugate, although a trend to be higher for the latter was observed. The functional activity of the elicited antibodies was estimated on pooled sera by OPKA, an assay that mimics in vivo GBS killing by effector cells in the presence of complement and specific antibodies, and correlates with mouse protection.36 In agreement with the ELISA outcome, OPKA titers of the pooled sera after the second and third doses were comparable for CRM197-DCA-PSIa and CRM197PSIa conjugates (Table 2), while they were lower for the CRM197-DHA-PSIa conjugate.
In addition, the anti-protein antibodies for CRM197-DCA were comparable to those for CRM197 and more than 10-fold higher as compared to those for CRM197-DHA (Fig. 4B and ESI, Fig. S9†), possibly as a result of protein stabilization.30 It is noteworthy that the protein dose for CRM197-DCA was 2 fold lower than that for the random conjugate.
Given the positive results obtained with the CRM197-DCA conjugate, we assessed whether strong immunogenicity could be elicited through this conjugation approach despite the type of polysaccharide used. As shown in Fig. 4C, the immune response after two and three injections was comparable for CRM197-PSIII and the site selective CRM197-DCA-PSIII conjugate. The functional activity elicited by site selective CRM197-DCA-PSIII (67 and 283 are average OPK titer of three different experiments from the pool of mouse sera obtained after two and three doses, respectively) was also comparable with the one obtained by immunizing mice with the reference random conjugate (143 and 154, respectively). Finally, a site-selective CRM197-DCA conjugate of Sp14 PS, which is part of commercial pneumococcal vaccines,37 was prepared and tested in mice. Again, in vivo immunogenicity on 10 mice showed after two and three vaccine doses a level of elicited specific anti-Sp14 PS antibodies comparable to those for the respective random conjugate (Fig. 4D).
Discussion
The rational modification of the structure of peptides and proteins offers a wide range of opportunities for the modulation of their biological activity.Glycoconjugates present in licensed vaccines are generally generated by classic lysine random conjugation of carbohydrates on the surface of carrier protein. These conjugates have often heterogeneous compositions, which cause batch-to-batch variability of structure and activity, often leading to an incomplete understanding of their mechanism of action. Site-selective conjugation is a powerful method to direct polysaccharide conjugation at predetermined sites of the protein and ensure higher batch-to-batch consistency in comparison to classic nonspecific conjugation procedures, particularly when the protein is used with the dual role of antigen and carrier.
Stefanetti et al.9 demonstrated that specific site-selective single or double attachment of glycan antigens to carrier protein CRM197 is sufficient to induce high levels of anti-Salmonella typhimurium O-antigen IgG specific antibodies with serum bactericidal activity. Conjugation at the C186–C201 bond resulted in high anti O-antigen bactericidal antibody titers.
Starting from this discovery, we generated two different vaccines with a defined conjugation point, by modifying the same C186–C201 disulfide bridge in CRM197. In the first approach, a DCA graft was installed in CRM197. The second strategy involved the conversion of cysteine residues in dehydroalanine. Both methods offer the possibility to attach through condensation reaction or thiol Michael addition bifunctional linkers for click reactions with carbohydrates. Particularly, here we incorporated azido moieties, ready for strain promoted click chemistry glycoconjugation with DBCO-derivatized polysaccharides from group B Streptococcus and S. pneumoniae.
In vivo data highlighted that CRM197-DCA conjugates elicited an immune response comparable to the reference random CRM197-GBS PSIa and PSIII conjugates, which are vaccine candidates under clinical development, while CRM197-DHA resulted in poor immunogenicity. Moreover, antibodies elicited by CRM197-DCA-PSIa and PSIII are functional with an OPK titer comparable to the reference vaccines. Also, CRM197-DCA conjugation of S. pneumoniae equally provided a strongly immunogenic conjugate. This observation converges with the work done by Martínez-Sáez et al.30 according to which oxetane graft installation on protein through the regioselective disulfide stapling of the protein carrier CRM197 enables stabilization of folded structures and results in an enhanced bioactivity, e.g. a significant increase in its immunogenicity in vivo.
It has been reported that the 3D structure of CRM197 is more altered by random conjugation of glycans in comparison to the one of formaldehyde treated proteins, including DT or CRM197 itself.38 The loss of the CRM197 tertiary structure with potential detrimental impact on certain conformational epitopes could partially explain the lower propensity of this protein to be subjected to immune interference in the presence of pre-existing anti-protein antibodies.38,39 Of note, the modification induced by glycoconjugation might appear more evident in CRM197 with respect to formaldehyde treated proteins because the marked unfolding caused by chemical detoxication might result in negligible further structural impact caused by glycan coupling.
Our data complement this information and suggest that slight modifications of the protein tertiary structure, such as the ones induced by a random reaction of surface exposed lysine residues, are compatible with a strong anti-carbohydrate immune response.
Among the two cysteine directed chemistries herein tested, disulfide re-bridging of CRM197 with DCA is shown to aid preservation of the 3D structure of the protein, as demonstrated by combined CD and DLS and X-ray experiments.
Importantly, the crystal structure of CRM197-DCA presented herein clearly shows the selective presence of one disulfide bridge at C185–C201, while the second disulfide bridge (C461–C471) remains untouched, and preservation of the 3D protein structure compared to the native form. Minimal alterations of the 3D structure caused by glycoconjugation and detectable by CD did not impair its carrier properties, resulting in a robust anti-carbohydrate response.
Conversely, a modification based on ring opening (CRM197-DHA) resulted in structural changes that strongly impacted the immunogenicity of the conjugated glycan.
Overall this study underpins that selective protein modifications based on bridging of disulfide bonds are optimal to preserve the structural integrity and stability, and consequently the immunogenicity of the glycoconjugates.
Conclusions
This work underscores the impact of protein modification on the stability and immunogenicity of glycoconjugates and highlights disulfide stapling as an effective strategy for selective protein conjugation, widely applicable to different polysaccharides. In addition, it opens the path for the use of highly selective chemical methods for the preparation of glycoconjugate vaccines with advantages in terms of consistency of production and characterization, and a better understanding of their immunological mechanism of action.Ethical statement
Animal experiments were performed in accordance with the regulations of the Directive 2010/63/EU and GSK ethical guidelines, under the approval of the Italian Ministry of Health (Italian Legislative Decree no. 26/2014). All mice were housed under specific pathogen-free conditions at the GSK Vaccines Animal Resource Centre in compliance with the relevant guidelines.Data availability
All relevant data are reported as ESI.†Author contributions
FC, AK, GJLB and RA conceived the study; FC, AK, MCM, DV, LDI, EB, BB, FA, and LDB performed experimental work; FC, AK, MCM, IM, MRR, GJLB and RA analyzed results; FC, AK, GJLB and RA wrote the manuscript; all revised the manuscript.Conflicts of interest
FC, DV, LDI, DO, EB, BB, FA, LDB, IM, MRR, and RA are employees of the GSK group of companies. AK was hosted in a secondment in GSK during her PhD programme. Prevnar and HibTITER are trademarks from Pfizer; Vaxem-Hib, Menveo and Menjugate are trademarks from GSK.Acknowledgements
This work was sponsored by GlaxoSmithKline Biologicals SA and has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 675671. CS is recipient of the grant no. 861194 (PAVax) under the EU Horizon 2020 program. Dr Werner Pansegrau is acknowledged for support in CD experiments. We are grateful to the staff at beamlines ID23-1 of the European Synchrotron Radiation Facility (ESRF, France) for their assistance in collecting X-ray diffraction data.Notes and references
- R. Rosini, S. Nicchi, M. Pizza and R. Rappuoli, Front. Immunol., 2020, 11, 1048 CrossRef CAS PubMed.
- R. Rappuoli, Sci. Transl. Med., 2018, 9, eaat4615 CrossRef PubMed.
- R. Rappuoli, E. De Gregorio and P. Costantino, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 14–16 CrossRef CAS PubMed.
- F. Berti and R. Adamo, Chem. Soc. Rev., 2018, 47, 9015–9025 RSC.
- V. Chudasama, A. Maruani and S. Caddick, Nat. Chem., 2016, 8, 114–119 CrossRef CAS PubMed.
- R. Adamo, Acc. Chem. Res., 2017, 50, 1270–1279 CrossRef CAS PubMed.
- O. Boutureira and G. J. L. Bernardes, Chem. Rev., 2015, 115, 2174–2195 CrossRef CAS PubMed.
- G. J. Bernardes, D. P. Gamblin and B. G. Davis, Angew. Chem., Int. Ed. Engl., 2006, 54, 13198–13203 Search PubMed.
- A. Pillot, A. Defontaine, A. Fateh, A. Lambert, M. Prasanna, M. Fanuel, M. Pipelier, N. Csaba, T. Violo, E. Camberlein and C. Grandjean, Front. Chem., 2019, 7(726) DOI:10.3389/fchem.2019.00726.
- G. Stefanetti, Q. Y. Hu, A. Usera, Z. Robinson, M. Allan, A. Singh, H. Imase, J. Cobb, H. Zhai, D. Quinn, M. Lei, A. Saul, R. Adamo, C. A. MacLennan and F. Micoli, Angew. Chem., Int. Ed. Engl., 2015, 54, 13198–13203 CrossRef CAS PubMed.
- O. Boutureira, N. Martinez-Saez, K. M. Brindle, A. A. Neves, F. Corzana and G. J. L. Bernardes, Chem. Eur. J., 2017, 23, 6483–6489 CrossRef CAS PubMed.
- A. Nilo, M. Allan, B. Brogioni, D. Proietti, V. Cattaneo, S. Crotti, S. Sokup, H. Zhai, I. Margarit, F. Berti, Q. Y. Hu and R. Adamo, Bioconjugate Chem., 2014, 25, 2105–2111 CrossRef CAS PubMed.
- A. Nilo, I. Passalacqua, M. Fabbrini, M. Allan, A. Usera, F. Carboni, B. Brogioni, A. Pezzicoli, J. Cobb, M. R. Romano, I. Margarit, Q.-Y. Hu, F. Berti and R. Adamo, Bioconjugate Chem., 2015, 26, 1839–1849 CrossRef CAS PubMed.
- Q.-Y. Hu, M. Allan, R. Adamo, D. Quinn, H. Zhai, G. Wu, K. Clark, J. Zhou, S. Ortiz, B. Wang, E. Danieli, S. Crotti, M. Tontini, G. Brogioni and F. Berti, Chem. Sci., 2013, 4, 3827–3832 RSC.
- R. Adamo, A. Nilo, B. Castagner, O. Boutureira, F. Berti and G. J. L. Bernardes, Chem. Sci., 2013, 4, 2995–3008 RSC.
- R. Adamo, Q.-Y. Hu, A. Torosantucci, S. Crotti, G. Brogioni, M. Allan, P. Chiani, C. Bromuro, D. Quinn, M. Tontini and F. Berti, Chem. Sci., 2014, 5, 4302–4311 RSC.
- A. Nilo, L. Morelli, I. Passalacqua, B. Brogioni, M. Allan, F. Carboni, A. Pezzicoli, F. Zerbini, D. Maione, M. Fabbrini, M. R. Romano, Q. Y. Hu, I. Margarit, F. Berti and R. Adamo, ACS Chem. Biol., 2015, 10, 1737–1746 CrossRef CAS PubMed.
- P. A. Szijj, K. A. Kostadinova, R. J. Spears and V. Chudasama, Org. Biomol. Chem., 2020, 18(44), 9018–9028 RSC.
- G. Garufi, Y. T. Wang, S. Y. Oh, H. Maier, D. M. Missiakas and O. Schneewind, Vaccine, 2012, 30, 3435–3444 CrossRef CAS PubMed.
- Z. Wu, X. Guo, Q. Wang, B. M. Swarts and Z. Guo, J. Am. Chem. Soc., 2010, 132, 1567–1571 CrossRef CAS PubMed.
- N. Kapoor, I. Vanjak, J. Rozzelle, A. Berges, W. Chan, G. Yin, C. Tran, A. K. Sato, A. R. Steiner, T. P. Pham, A. J. Birkett, C. A. Long, J. Fairman and K. Miura, Biochemistry, 2018, 57, 516–519 CrossRef CAS PubMed.
- E. A. Hoyt, P. M. S. D. Cal, B. L. Oliveira and G. J. L. Bernardes, Nat. Rev. Chem., 2019, 3, 147–171 CrossRef CAS.
- J. M. Chalker, S. B. Gunnoo, O. Boutureira, S. C. Gerstberger, M. Fernández-González, G. J. L. Bernardes, L. Griffin, H. Hailu, C. J. Schofield and B. G. Davis, Chem. Sci., 2011, 2, 1666–1676 RSC.
- J. M. Chalker, G. J. L. Bernardes, Y. A. Lin and B. G. Davis, Chem.–Asian J., 2009, 4, 630–640 CrossRef CAS PubMed.
- G. J. L. Bernardes, G. Casi, S. Trüssel, I. Hartmann, K. Schwager, J. Scheuermann and D. Neri, Chem. Sci., 2012, 51, 941–944 CAS.
- R. P. Lyon, D. L. Meyer, J. R. Setter and P. D. Senter, in Methods in Enzymology, ed. K. D. Wittrup and G. L. Verdine, Academic Press, 2012, vol. 502, pp. 123–138 Search PubMed.
- E. Malito, B. Bursulaya, C. Chen, P. Lo Surdo, M. Picchianti, E. Balducci, M. Biancucci, A. Brock, F. Berti, M. J. Bottomley, M. Nissum, P. Costantino, R. Rappuoli and G. Spraggon, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 5229–5234 CrossRef CAS PubMed.
- M. Bröker, P. Costantino, L. DeTora, E. D. McIntosh and R. Rappuoli, Biologicals, 2011, 39, 195–204 CrossRef PubMed.
- M. E. Pichichero, Hum. Vaccines Immunother., 2013, 9, 2505–2523 CrossRef CAS PubMed.
- N. Martínez-Sáez, S. Sun, D. Oldrini, P. Sormanni, O. Boutureira, F. Carboni, I. Compañón, M. Deery, M. Vendruscolo, F. Corzana, R. Adamo and G. J. L. Bernardes, Angew. Chem., Int. Ed., 2017, 56(47), 14963–14967 CrossRef PubMed.
- A. Micsonai, F. Wien, E. Bulyaki, J. Kun, E. Moussong, Y. H. Lee, Y. Goto, M. Refregiers and J. Kardos, Nucleic Acids Res., 2018, 46, W315–W322 CrossRef CAS PubMed.
- S. Choe, M. Bennett, G. Fujii, P. Curmi, K. Kantardjieff, R. Collier and D. Eisenberg, Nature, 1992, 357, 216–222 CrossRef CAS PubMed.
- F. Carboni, R. Adamo, M. Fabbrini, R. De Ricco, V. Cattaneo, B. Brogioni, D. Veggi, V. Pinto, I. Passalacqua, D. Oldrini, R. Rappuoli, E. Malito, I. Y. R. Margarit and F. Berti, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5017–5022 CrossRef CAS PubMed.
- J. Absalon, N. Segall, S. L. Block, K. J. Center, I. L. Scully, P. C. Giardina, J. Peterson, W. J. Watson, W. C. Gruber, K. U. Jansen, Y. Peng, S. Munson, D. Pavliakova, D. A. Scott and A. S. Anderson, Lancet Infect. Dis., 2020, 21, 263–274 CrossRef PubMed.
- S. A. Madhi, C. L. Cutland, L. Jose, A. Koen, N. Govender, F. Wittke, M. Olugbosi, A. S. Meulen, S. Baker, P. M. Dull, V. Narasimhan and K. Slobod, Lancet Infect. Dis., 2016, 16, 923–934 CrossRef CAS PubMed.
- S. Crotti, H. Zhai, J. Zhou, M. Allan, D. Proietti, W. Pansegrau, Q.-Y. Hu, F. Berti and R. Adamo, ChemBioChem, 2014, 15, 836–843 CrossRef CAS PubMed.
- E. Delgleize, O. Leeuwenkamp, E. Theodorou and N. Van de Velde, BMJ Open, 2016, 6, e010776 CrossRef PubMed.
- S. Pecetta, P. Lo Surdo, M. Tontini, D. Proietti, C. Zambonelli, M. J. Bottomley, M. Biagini, F. Berti, P. Costantino and M. R. Romano, Study Group, Vaccine, 2015, 33(2), 314–320 CrossRef CAS PubMed.
- S. Pecetta, M. Tontini, E. Faenzi, R. Cioncada, D. Proietti, A. Seubert, S. Nuti, F. Berti and M. R. Romano, Vaccine, 2016, 34(20), 2334–2341 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Procedures for linker incorporation and glycoconjugation, mass spectrometry, X-ray crystallography and in vivo studies are reported as ESI. CCDC PBD ID 7O4W. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1sc01928g |
| ‡ F. C. and A. K. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |