Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Design of millidevices to expedite apparent solubility measurements
Maria del Carme
Pons Royo
 abc,
Jean-Luc
Beulay
a,
Eric
Valery
a,
Alois
Jungbauer
abc,
Jean-Luc
Beulay
a,
Eric
Valery
a,
Alois
Jungbauer
 *bc and
Peter
Satzer
b
*bc and
Peter
Satzer
b
aDepartment of Innovation, Novasep, 81 Boulevard de la Moselle, 54340 Pompey, France
bDepartment of Biotechnology, University of Natural Resources and Life Sciences Vienna, 1190 Vienna, Austria. E-mail: alois.jungbauer@boku.ac.at
cAustrian Centre of Industrial Biotechnology (ACIB), Muthgasse 18, 1190 Vienna, Austria
First published on 17th June 2022
Abstract
Protein solubility is a critical attribute in the development and production of monoclonal antibodies. Available solubility data refer to pure solutes which do not consider solvents and impurities which have a significant effect on solubility. Thus, solubility curves need to be determined experimentally. Previously established methods to determine the apparent solubility of proteins are based on manual assays which are time-consuming and labor-intensive. We present the design of simple and adaptable millidevices for fast solubility curve determination. Such a device in the form of a tubular reactor was manufactured from polymethylmethacrylate by laser cutting. The reactors had multiple injection points for the precipitant that allowed a controlled and precise addition of the precipitating agent at different concentrations. Hence, antibodies could be directly harvested at different concentrations of precipitating agents. The simple and flexible design allowed the number of pumps required to be reduced to only one for each solution and the distribution of the precipitating agent at different concentrations without valves. To demonstrate the wide applicability of the prototype in determining solubility curves, we used 2 industrially relevant precipitating agents, PEG6000 and ZnCl2, to measure the apparent solubilities of 4 antibodies and CaCl2 to measure the apparent solubility curves for dsDNA. In all cases, the data obtained were consistent between the device and manual assays with good reproducibility. This millidevice can be used for fast characterization of protein solutions such as solubility, degradation or stability of the antibodies under different conditions.
Introduction
Protein solubility is a key parameter during development and production of therapeutic proteins. It is especially important during identification of drug candidates or formulation at high protein concentrations.1–3 Detailed data on solubility are necessary to avoid issues with solubility such as aggregation, which impact negatively on protein stability, immunogenicity and efficacy. Furthermore, protein solubility plays an important role in the development of precipitation processes, which have received renewed interest as an alternative to protein A chromatography for the purification of antibodies.4–13 Solubility curves are defined as the thermodynamic equilibrium between the solid and the liquid phase. This equilibrium can be affected by several parameters, including external factors like solution composition, pH and ionic strength1,14 as well as intrinsic factors and properties of the protein itself, such as size, charge distribution, overall charge, etc.15 For the process design of protein precipitation, it is essential to determine the solubility curve of each component in the harvest, including impurities. To predict the process performance and to evaluate the separation behavior, it would be necessary to collect the solubility data on each component from the cell culture supernatant under certain buffer conditions and concentrations.14 However, published solubility data refer to pure solutes and solvents while impurities can have a significant effect on solution properties. Previous studies16,17 determined that curves obtained from purified antibodies and total protein impurities present different features and can result in different apparent solubilities. Therefore, the solubility in solution is typically referred to as “apparent solubility”.4 Typically, antibodies present higher solubility and steeper curves than impurities, with lower solubility and flatter curves. These differences in solubility can be exploited to precipitate mAbs avoiding coprecipitation of impurities. For that reason, apparent solubility curves need to be determined experimentally.18 As described by Juckes,19 the apparent solubility of purified antibodies and impurities can be derived from a linear regression following a semi-logarithmic behavior, where the apparent maximum solubility will be extrapolated from the intercept in the absence of a precipitating agent such as PEG.20 From these studies, a model for non-ionic precipitation was established. Hence, a common method to determine the relative apparent solubility of each component is using non-ionic polymers as precipitating agents like PEG (polyethylene glycol) to decrease protein solubility.1,21 This methodology has been extensively used to determine the apparent solubility curves of various proteins.22–31Nevertheless, all these methodologies for determining the relative apparent solubility require a vast stock of reagents at different pH values, ionic strength, viscosity, etc. as well as several manipulation steps such as filtration, centrifugation or additional mixing systems, which complicate the utilization of handling systems.22,29,32–34 The emergence of microfluidic technology provides a new method to investigate protein solubilization under several solution conditions. Analysis with microdevices can be performed with high precision, high speed, and high throughput, while reducing the required experimental time and the amount of protein required.35 Furthermore, it is important to highlight that microdevices can lead to more automated labs and miniaturized analyses.36 Characterization of protein solutions using microdevices has been already described,37,38 to select flocculating agents39 or investigate protein phase behavior.40 Unfortunately, these devices are not widely used in laboratories.36 Most of these microdevices require a clean-room environment for their fabrication,41 which is not available in most laboratories. Besides the lack of appropriate facilities, the cost and expertise to develop and fabricate such microdevices are another drawback. For example, the integration of microvalves or micropumps to achieve better flow control will increase the complexity and the cost of the device.42,43 Additionally, valves or pumps cannot be easily integrated in several materials such as glass or silicon, because of their rigidity. Thus, these materials must be combined with more flexible materials such as PDMS, which will allow the integration of valves or connection to pumps.41,44 Another difficulty is the implementation of appropriate kinetics into microfluidic devices. As all flows are laminar in such devices, achieving proper mixing between highly viscous solutions like a PEG solution and low viscous solutions like the harvest from a monoclonal fermentation is not always easy.45 All in all, going to such small scales allows for a reduction in protein solution consumption, but comes with a large cost in terms of necessary equipment and expertise, not present in many labs.
In this work, we have developed a millifluidic device to determine apparent solubility profiles to circumvent the issues typical for already presented microfluidic devices. We demonstrated the feasibility of apparent solubility curve determination using these new millidevices in various industrially relevant buffers for protein and DNA purification. We compared our methodology with a typical manual assay to evaluate the accuracy and precision of our millidevices. In addition, we demonstrated a proof of concept to develop more automated devices for accelerating downstream processing.
Materials and methods
CHO cell culture supernatants
Experiments were performed with 4 different clarified Chinese hamster ovary (CHO) cell culture supernatants of antibodies labeled mAbA, mAbB, mAbC and mAbD. The concentration of the antibodies was, respectively, 1.4 g L−1, 1.2 g L−1, 1.15 g L−1 and 1.3 g L−1. Respectively, the HCP concentration was 3.5 g L−1, 8.3 g L−1, 8.6 g L−1 and 10.25 g L−1, and for dsDNA, 17.74 ng mL−1, 10.37 ng mL−1, 5.21 ng mL−1, and 4.56 ng mL−1. The supernatants were filtered with a 0.22 μm membrane (Merck KGaA, Darmstadt, Germany) before use.Protein A chromatography
Protein A chromatography for protein concentration was performed using an affinity column POROS A 20 μm Column (2.1 × 30 mm, 0.1 mL; Thermo Scientific, MA, USA), as previously described.46 The column was equilibrated with 50 mM phosphate buffer with 150 mM NaCl, pH 7.0, and eluted with 100 mM glycine buffer, pH 2.4.dsDNA assay
Quantification of double-stranded DNA was performed using a Quant-iT PicoGreen dsDNA reagent (Invitrogen, Waltham, MA, United States). Samples were diluted in serial 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 dilutions in a 96-well plate using 1× TE buffer (10 mM Tris-HCl, 1 mM ethylenediaminetetraacetic acid, pH 7.5). 100 μL of sample dilution was transferred to a black microtiter plate and mixed with 100 μL of diluted PicoGreen reagent. The intensity was measured at 480 nm excitation and with an emission filter of 520 nm. The concentration was calculated using a standard lambda DNA solution (Invitrogen, Waltham, MA, United States).
2 dilutions in a 96-well plate using 1× TE buffer (10 mM Tris-HCl, 1 mM ethylenediaminetetraacetic acid, pH 7.5). 100 μL of sample dilution was transferred to a black microtiter plate and mixed with 100 μL of diluted PicoGreen reagent. The intensity was measured at 480 nm excitation and with an emission filter of 520 nm. The concentration was calculated using a standard lambda DNA solution (Invitrogen, Waltham, MA, United States).
Prototype fabrication
Millidevices were designed using CorelDRAW 2020 (Corel Co., Canada) and a laser cutter (Laser Trotec Speedy 100, Trotec GmbH, Austria). The microchannels were 1 mm wide and 3 mm deep. The main mixing channels and injection microchannels were cut in 3 mm, and the inlets and outlets in 4 mm poly(methyl methacrylate) (PMMA) sheets (Acrylstudio GmbHW, Austria). The PMMA sheets were heated to 165 °C for at least 45 min and cooled down at room temperature. Prior to the bonding process, PMMA sheets were cleaned by rinsing with 70% ethanol.Apparent solubility curve determination – manually
Manual apparent solubility curve determination was performed in 1.5 mL vials (Eppendorf, Germany), by adding each stock solution, 30 mM ZnCl2, 40% PEG6000 and 300 mM CaCl2, in a determined ratio into 1 mL cell culture supernatant to obtain the desired final concentrations, 2.7 mM to 8.6 mM ZnCl2, 3.6% to 11.4% PEG600 and 27.3 mM to 85.7 mM CaCl2. After 15 min of incubation at room temperature on an end-over-end shaker, the samples were centrifuged at 4.000g for 10 min. The supernatant was withdrawn and analysed with protein A affinity chromatography for mAb concentration or with the PicoGreen assay for dsDNA concentration.Apparent solubility curve determination – devices
Continuous apparent solubility curve determination was performed with the millidevice directly connected to an ÄKTA pure 25 system (Cytiva, Uppsala, Sweden). The flow rate was set at 4 mL min−1 for the mAb supernatant and 1 mL min−1 for the precipitating solution to obtain the desired final concentrations, 2.7 mM to 8.6 mM ZnCl2, 3.6% to 11.4% PEG600 and 27.3 mM to 85.7 mM CaCl2. The collected precipitated solution was continuously mixed for 15 minutes to ensure complete precipitation. After that, the samples were centrifuged at 4.000g for 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) min. The supernatant was withdrawn and analysed with protein A affinity chromatography for mAb concentration or with the PicoGreen assay for dsDNA concentration.
min. The supernatant was withdrawn and analysed with protein A affinity chromatography for mAb concentration or with the PicoGreen assay for dsDNA concentration.
Results and discussion
Design of precipitation reactors
Although several methodologies have been developed to determine apparent solubility, manual methods are time-consuming and labor-intensive due to the large amounts of buffer solutions and concentrations that need to be managed: sample, precipitant, buffer, etc. To facilitate this procedure, we designed millifluidic devices that can be used in an automated system such as an Äkta system to determine the protein solubility. The millidevices were designed using CorelDRAW and manufactured from polymethylmethacrylate (PMMA) by laser cutting. PMMA sheets were stacked and adhered to each other by heat binding at 165 °C to ensure that they are fully sealed and there is no leakage. The millidevice was composed of a main inlet path for the protein solution (1) and a second path for the selected precipitating agent (2), both connected through a valve-free dispenser with different injection points (3) depending on the required concentration of the precipitating agent to be added (Fig. 1). To ensure proper mixing even for viscous liquids, the millidevice was designed with a complex serpentine pattern as a passive mixer, 1 mm wide and 3 mm deep (4) (Fig. 1), and with several outlets for each tested condition (5). Our millidevices were designed to be microvalve-free to reduce the manufacturing costs and the complexity of the design and to simplify the experimental setup by reducing the number of pumps required to only 2, one for the supernatant and one for the precipitating agent (Fig. 1). The dispenser was composed of several outlets grouped in different injection zones, which regulate the addition of precipitating agents. In this case, the device was composed of 4 zones with 1, 2, 3 and 4 holes of 0.1 mm in diameter. The total precipitant flow running in the main channel was divided in different fractions, which will result in different concentrations of the precipitating agent. For example, at 1 ml min−1 flow of 40% PEG6000 stock solution, 1 hole corresponds to 3.6 ± 0.4% PEG6000 in 1 mL antibody solution, 2 holes to 6.6 ± 0.5%, 3 holes to 9.2 ± 0.5% and 4 holes to 11.4% ± 0.5%. The same mechanism was used to distribute equally the main antibody solution in 4 different mixing channels, but in that case, the number of holes was kept constant (Fig. 1c) to obtain the same flow in the mixing channels, 1 mL min−1. The mixing channels were also coupled to the dispenser for precipitating agents, in which a determined amount of precipitating agent was added onto the number of holes connected to the selected mixing channel with the antibody solution. Therefore, antibodies can be harvested at different precipitating agent concentrations in the outlet of each mixing channel. Furthermore, the device integrated a standardized female luer lock system (Fig. 1) in each inlet and outlet, which allows the device to be directly connected to commonly used lab equipment such as peristaltic pumps or more complex equipment such as an Äkta system, without the need for flexible materials such as polydimethylsiloxane (PDMS) (Fig. 2).The female luer lock fitting was achieved by drilling with an M6 broach the PMMA to obtain the same mold as a female luer lock. Therefore, the male luer lock from the laboratory equipment can be simply connected by rotating the tube connector. In addition, as PMMA is barely permeable to gasses, sample evaporation can be neglected.41 The automation of the precipitation methodology allowed the screening of a larger number of conditions and solutions, identifying rapidly and accurately the necessary solubility information for molecule selection or the design of a precipitation process at an early stage. Additionally, the flexibility of the design and the mixing system allows the developed prototypes to be customized and modified for any precipitation protocol or precipitation strategy.
Apparent solubility curves
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8,55 or CA.9,56 To demonstrate the application of the millidevice to measure the apparent solubility of different dsDNA concentrations, the experiment was performed with CaCl2 with the 4 different mAb supernatants. The apparent solubility curves were determined over a range of 27.3 mM to 85.7 mM CaCl2 with mixing a flow rate of 4 mL min−1 for the supernatant and 1 mL min−1 for 300 mM CaCl2 solution. The samples were taken from the devices and filtered, prior to dsDNA quantification. Afterwards, the dsDNA concentration in the liquid phase was plotted against the precipitating agent concentration (Fig. 5). The addition of CaCl2 resulted in the precipitation of dsDNA (Fig. 5). Due to the differences in the initial concentration of dsDNA of each supernatant, each apparent solubility curve showed a particular shape, different also from the required CaCl2 concentration to obtain acceptable dsDNA concentrations, comparable to currently used purification methods. Impurities such as dsDNA in solution decreased with the increase of CaCl2 concentration. For mAbA, mAbB and mAbD, lower CaCl2 concentration was necessary to achieve almost complete dsDNA precipitation, almost no dsDNA was found at 85.7 mM CaCl2 (>0.3 ng mL−1), while for mAbC higher concentration, 0.5 ng mL−1, was found.
8,55 or CA.9,56 To demonstrate the application of the millidevice to measure the apparent solubility of different dsDNA concentrations, the experiment was performed with CaCl2 with the 4 different mAb supernatants. The apparent solubility curves were determined over a range of 27.3 mM to 85.7 mM CaCl2 with mixing a flow rate of 4 mL min−1 for the supernatant and 1 mL min−1 for 300 mM CaCl2 solution. The samples were taken from the devices and filtered, prior to dsDNA quantification. Afterwards, the dsDNA concentration in the liquid phase was plotted against the precipitating agent concentration (Fig. 5). The addition of CaCl2 resulted in the precipitation of dsDNA (Fig. 5). Due to the differences in the initial concentration of dsDNA of each supernatant, each apparent solubility curve showed a particular shape, different also from the required CaCl2 concentration to obtain acceptable dsDNA concentrations, comparable to currently used purification methods. Impurities such as dsDNA in solution decreased with the increase of CaCl2 concentration. For mAbA, mAbB and mAbD, lower CaCl2 concentration was necessary to achieve almost complete dsDNA precipitation, almost no dsDNA was found at 85.7 mM CaCl2 (>0.3 ng mL−1), while for mAbC higher concentration, 0.5 ng mL−1, was found.
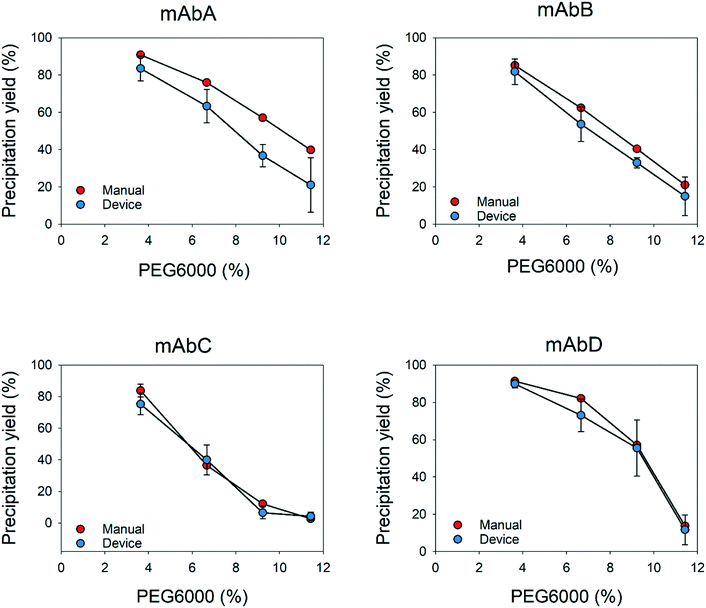
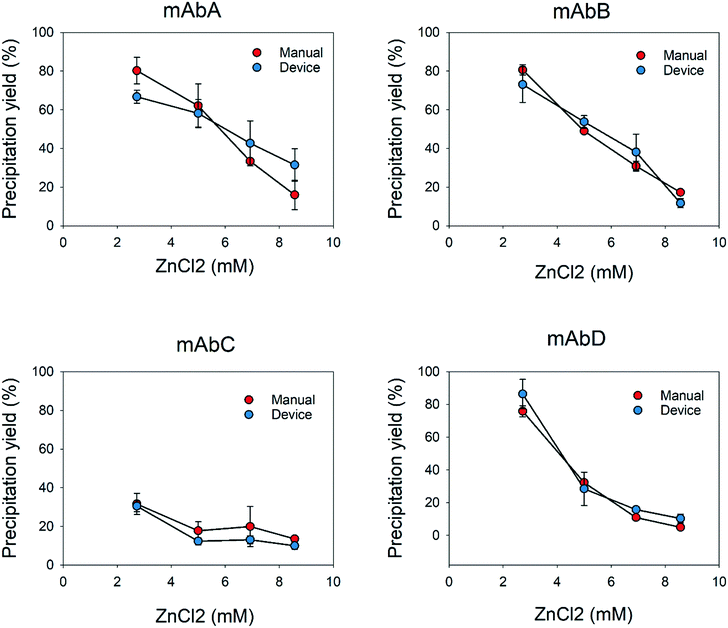
The different behaviors and concentrations of the impurities used in this study show that the device is capable of accurately measure the apparent solubility for impurities such as dsDNA and using another precipitating agent. Afterwards, apparent solubility measurements were repeated manually. We compared both methods to determine the effectiveness and accuracy of the prototypes. The use of millidevices resulted in comparable shapes of the precipitation curves to those from experiments performed manually. Since results of dsDNA of mAbA did not show similar deviations as with PEG6000 and ZnCl2, that may confirm that previous variation on the solubility curves were caused by shear stress.
In addition, the results showed that the flow distribution was very accurate and precise, similar to the results obtained manually for the different supernatants and precipitating agents. The use of millidevices resulted in comparable shapes of the precipitation curves to those from experiments performed manually. There was a clear reduction in the experimental time and sample manipulation handling by a factor of 5 (Table 1), since intermediate steps, such as centrifugation to remove precipitates or transferring the supernatants to 96-well plates for the analysis, can be skipped. The above-mentioned issues can be overcome by using a more sophisticated prototyping tool such as a 3D printer, where more soft and complex geometries can be obtained. Hence, it will provide proper mixing with lower shear stress and volume reduction, which will lead to a higher reduction of material consumption compared to the manual method.57 In comparison with similar devices previously reported, several precipitant concentrations can be tested simultaneously with a single solution in a couple of minutes, without extra sample handling steps, as observed in previous devices, where samples need to be manipulated and transferred to other devices,22,57–59 or methods to determine the apparent solubility of proteins.14,17,60
| Millidevices | Manual | |
|---|---|---|
| Antibody solution | 2.5 mL | 12 mL |
| 40% PEG6000 | 0.2 mL | 1 mL |
| 30 mM ZnCl2 | 0.2 mL | 1 mL |
Additionally, the flexibility of the design and the mixing system allows the developed prototypes to be customized and modified for any precipitation protocol or precipitation strategy. An example of an alternative process could be protein resolubilization, which is often neglected during precipitation studies. Commonly, samples are diluted in different buffers and concentrations to determine the optimal dilution ratio. Millidevices can be used to screen resolubilization buffers at different concentrations and dilutions. Other interesting uses could be for formulation development for protein stability and to assess protein aggregation under certain conditions. Furthermore, our millidevices provide enhanced mixing which accelerates mAb resolubilization and reduces experimental times, as compared to previous resolubilization studies.17 In addition, the automation of the experiments will allow the screening of larger experimental conditions in shorter time and will reduce operator effects.61
Conclusions
In this study, we showed a proof of concept of millidevices for determining apparent solubility curves for antibodies and impurities. The developed prototype showed consistent performance and very good reproducibility compared to conventional techniques such as manual apparent solubility determination. Further advantages of using these millidevices are the reduction of reagents, sample consumption and experimental time by reducing the experimental steps, such as centrifugation. A much higher number of experimental conditions can be screened with the automation of the PEG precipitation methodology while removing further human-prone errors such as operator effects. In addition, the easy, flexible and manufacturable in-house screening tools can be modified depending on the experimental needs, for example, determination of degradation or stability of antibodies at different concentrations during formulation. Millidevices are an interesting tool to develop and optimize processes, which should start being implemented for screening and down-scaling processes.Author contributions
Carme Pons Royo: designed the devices, conducted the experiments and wrote the manuscript. Jean Luc Beulay: managed and coordinated the research plan. Eric Valery: designed of the mixing device. Alois Jungbauer: drafted the research project and reviewed and edited the manuscript. Peter Satzer: supervised the project and reviewed and edited the manuscript.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
This work has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 812909 CODOBIO, within the Marie Skłodowska-Curie International Training Networks framework.References
- S. R. Trevino, J. M. Scholtz and C. N. Pace, J. Pharm. Sci., 2008, 97, 4155–4166 CrossRef CAS PubMed.
- V. M. Toprani, S. B. Joshi, L. A. Kueltzo, R. M. Schwartz, C. R. Middaugh and D. B. Volkin, J. Pharm. Sci., 2016, 105, 2319–2327 CrossRef CAS PubMed.
- D. G. Lim, J. C. Lee, D. J. Kim, S. J. Kim, H. W. Yu and S. H. Jeong, J. Pharm. Invest., 2020, 50, 493–503 CrossRef CAS.
- D. Burgstaller, A. Jungbauer and P. Satzer, Biotechnol. Bioeng., 2019, 116, 1053–1065 CrossRef CAS PubMed.
- G. Dutra, D. Komuczki, A. Jungbauer and P. Satzer, Eng. Life Sci., 2020, 20, 265–274 CrossRef CAS PubMed.
- N. Hammerschmidt, B. Hintersteiner, N. Lingg and A. Jungbauer, Biotechnol. J., 2015, 10, 1196–1205 CrossRef CAS PubMed.
- N. Hammerschmidt, S. Hobiger and A. Jungbauer, Process Biochem., 2016, 51, 325–332 CrossRef CAS.
- R. Sommer, P. Satzer, A. Tscheliessnig, H. Schulz, B. Helk and A. Jungbauer, Process Biochem., 2014, 49, 2001–2009 CrossRef CAS.
- R. Sommer, A. Tscheliessnig, P. Satzer, H. Schulz, B. Helk and A. Jungbauer, Biochem. Eng. J., 2015, 93, 200–211 CrossRef CAS.
- M. Bhat, A. Mullerpatan, J. Chen, M. Holstein, S. Ghose, Z. J. Li and S. Cramer, J. Biotechnol., 2021, 338, 1–4 CrossRef CAS PubMed.
- V. Warikoo and R. Godawat, Biotechnol. J., 2015, 10, 1101–1102 CrossRef CAS PubMed.
- M. Zelger, S. Pan, A. Jungbauer and R. Hahn, Process Biochem., 2016, 51, 1610–1621 CrossRef CAS.
- N. Kateja, D. Kumar, S. Sethi and A. S. Rathore, J. Chromatogr. A, 2018, 1579, 60–72 CrossRef CAS PubMed.
- R. M. Kramer, V. R. Shende, N. Motl, C. N. Pace and J. M. Scholtz, Biophys. J., 2012, 102, 1907–1915 CrossRef CAS PubMed.
- S.-J. Wu, G. Gilliland and Y. Feng, Solubility and Early Assessment of Stability for Protein Therapeutics, 2014, pp. 65–91, DOI:10.1002/9781118354698.ch3.
- R. Sommer, Continuous Precipitation of Therapeutic Proteins, with an Emphasis on Monoclonal Antibodies, 2013 Search PubMed.
- S. Großhans, S. Suhm and J. Hubbuch, Bioprocess Biosyst. Eng., 2019, 42, 1039–1051 CrossRef PubMed.
- P. M. Doran, Bioprocess engineering principles, 2nd edn, 2012 Search PubMed.
- I. R. Juckes, Biochim. Biophys. Acta, 1971, 229, 535–546 CrossRef CAS.
- L. Li, A. Kantor and N. Warne, Protein Sci., 2013, 22, 1118–1123 CrossRef CAS PubMed.
- S. L. Sim, T. He, A. Tscheliessnig, M. Mueller, R. B. Tan and A. Jungbauer, J. Biotechnol., 2012, 157, 315–319 CrossRef CAS PubMed.
- M. Oeller, P. Sormanni and M. Vendruscolo, Sci. Rep., 2021, 11, 21932 CrossRef CAS PubMed.
- D. H. Atha and K. C. Ingham, J. Biol. Chem., 1981, 256, 12108–12117 CrossRef CAS PubMed.
- R. N. Haire, W. A. Tisel, J. G. White and A. Rosenberg, Biopolymers, 1984, 23, 2761–2779 CrossRef CAS PubMed.
- F. Haskó, R. Vaszileva and L. Halász, Biotechnol. Bioeng., 1982, 24, 1931–1939 CrossRef PubMed.
- H. Mahadevan and C. K. Hall, Fluid Phase Equilib., 1992, 78, 297–321 CrossRef CAS.
- C. Middaugh, W. Tisel, R. Haire and A. Rosenberg, J. Biol. Chem., 1979, 254, 367–370 CrossRef CAS PubMed.
- T. J. Gibson, K. McCarty, I. J. McFadyen, E. Cash, P. Dalmonte, K. D. Hinds, A. A. Dinerman, J. C. Alvarez and D. B. Volkin, J. Pharm. Sci., 2011, 100, 1009–1021 CrossRef CAS PubMed.
- Q. Chai, J. Shih, C. Weldon, S. Phan and B. E. Jones, mAbs, 2019, 11, 747–756 CrossRef CAS PubMed.
- V. M. Toprani, S. B. Joshi, L. A. Kueltzo, R. M. Schwartz, C. R. Middaugh and D. B. Volkin, J. Pharm. Sci., 2016, 105, 2319–2327 CrossRef CAS PubMed.
- L. Li, A. Kantor and N. Warne, Protein Sci., 2013, 22, 1118–1123 CrossRef CAS PubMed.
- H. Zhao, O. Graf, N. Milovic, X. Luan, M. Bluemel, M. Smolny and K. Forrer, J. Pharm. Sci., 2010, 99, 2279–2294 CrossRef CAS PubMed.
- T. J. Gibson, K. McCarty, I. J. McFadyen, E. Cash, P. Dalmonte, K. D. Hinds, A. A. Dinerman, J. C. Alvarez and D. B. Volkin, J. Pharm. Sci., 2011, 100, 1009–1021 CrossRef CAS PubMed.
- L. J. Lohmann and J. Strube, Processes, 2021, 9, 488 CrossRef CAS.
- R. Bhambure, K. Kumar and A. S. Rathore, Trends Biotechnol., 2011, 29, 127–135 CrossRef CAS PubMed.
- M. N. Säo Pedro, T. C. Silva, R. Patil and M. Ottens, Biotechnol. Bioeng., 2021, 118, 3275–3286 CrossRef PubMed.
- M. R. G. Kopp and P. Arosio, J. Pharm. Sci., 2018, 107, 1228–1236 CrossRef CAS PubMed.
- M. Martinez, G. Mannall, M. Spitali, E. L. Norrant and D. G. Bracewell, J. Chem. Technol. Biotechnol., 2020 DOI:10.1002/jctb.6652.
- A. N. P. Radhakrishnan, M. P. C. Marques, M. J. Davies, B. O'Sullivan, D. G. Bracewell and N. Szita, Lab Chip, 2018, 18, 585–594 RSC.
- C. L. Hansen, M. O. A. Sommer and S. R. Quake, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 14431 CrossRef CAS PubMed.
- K. Ren, J. Zhou and H. Wu, Acc. Chem. Res., 2013, 46, 2396–2406 CrossRef CAS PubMed.
- J. Melin and S. R. Quake, Annu. Rev. Biophys. Biomol. Struct., 2007, 36, 213–231 CrossRef CAS PubMed.
- G. T. Roman and R. T. Kennedy, J. Chromatogr. A, 2007, 1168, 170–188 CrossRef CAS PubMed , discussion 169.
- X. Hou, Y. S. Zhang, G. T.-d. Santiago, M. M. Alvarez, J. Ribas, S. J. Jonas, P. S. Weiss, A. M. Andrews, J. Aizenberg and A. Khademhosseini, Nat. Rev. Mater., 2017, 2, 17016 CrossRef CAS.
- A. Afzal and K.-Y. Kim, in Analysis and Design Optimization of Micromixers, Springer Singapore, Singapore, 2021, pp. 1–10, DOI:10.1007/978-981-33-4291-0_1.
- G. Dutra, D. Komuczki, A. Jungbauer and P. Satzer, Eng. Life Sci., 2020, 20, 265–274 CrossRef CAS PubMed.
- L. Lohmann and J. Strube, Processes, 2020, 8, 58 CrossRef CAS.
- P. Sormanni, L. Amery, S. Ekizoglou, M. Vendruscolo and B. Popovic, Sci. Rep., 2017, 7, 8200 CrossRef PubMed.
- Q. Gu, Z. Li, J. L. Coffman, T. M. Przybycien and A. L. Zydney, Biotechnol. Prog., 2020, 36, e3041 CrossRef CAS PubMed.
- E. Byrne, J. J. Fitzpatrick, L. W. Pampel and N. J. Titchener-Hooker, Chem. Eng. Sci., 2002, 57, 3767–3779 CrossRef CAS.
- Q. Gu, Z. Li, J. Coffman, T. Przybycien and A. Zydney, Biotechnol. Prog., 2020, 36(6), e3041 CrossRef CAS PubMed.
- Z. Li, Q. Gu, J. Coffman, T. Przybycien and A. Zydney, Biotechnol. Prog., 2019, 35(6), e2886 CrossRef CAS PubMed.
- P. Satzer, D. Burgstaller, W. Krepper and A. Jungbauer, Eng. Life Sci., 2020, 20, 67–78 CrossRef CAS PubMed.
- C. J. England, T. C. Gray, S. R. L. Malla, S. A. Oliveira, B. R. Martin, G. W. Beall and L. K. Lewis, Anal. Biochem., 2021, 616, 114099 CrossRef CAS PubMed.
- P. Satzer, A. Tscheließnigg, R. Sommer and A. Jungbauer, Eng. Life Sci., 2014, 14, 477–484 CrossRef CAS.
- Y. Wan, T. Zhang, T. Chen, Y. Wang and Y. Li, Protein Expression Purif., 2019, 164, 105460 CrossRef CAS PubMed.
- G. Peybernès, R. Grossier, F. Villard, P. Letellier, M. Lagaize, N. Candoni and S. Veesler, Org. Process Res. Dev., 2018, 22, 1856–1860 CrossRef.
- V. J. Sieben, A. K. Tharanivasan, S. I. Andersen and F. Mostowfi, Energy Fuels, 2016, 30, 1933–1946 CrossRef CAS.
- M. Ildefonso, N. Candoni and S. Veesler, Org. Process Res. Dev., 2012, 16, 556–560 CrossRef CAS.
- S. R. Trevino, J. M. Scholtz and C. N. Pace, J. Pharm. Sci., 2008, 97, 4155–4166 CrossRef CAS PubMed.
- A. Christler, E. Felföldi, M. Mosor, D. Sauer, N. Walch, A. Dürauer and A. Jungbauer, Bioprocess Biosyst. Eng., 2020, 43, 753–764 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |