Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rxn Rover: automation of chemical reactions with user-friendly, modular software†
Zachery
Crandall
 ab,
Kevin
Basemann
ab,
Kevin
Basemann
 ab,
Long
Qi
ab,
Long
Qi
 *a and
Theresa L.
Windus
*a and
Theresa L.
Windus
 *ab
*ab
aU.S. DOE Ames Laboratory, Iowa State University, Ames, Iowa 50011, USA. E-mail: lqi@iastate.edu; twindus@iastate.edu
bDepartment of Chemistry, Iowa State University, Ames, Iowa 50011, USA
First published on 29th November 2021
Abstract
The automation of chemical reactions in research and development can be an enabling technology to reduce cost and waste generation in light of technology transformation towards renewable feedstocks and energy in chemical industry. Automation of reaction optimization, in particular, would remove the need for expert input by designing algorithms to statistically analyze the reaction and automatically generate suggested results. In addition, automation can save time and resources, and reduce random human error. However, automation software is commonly coupled to a specific laboratory or device setup or not freely available for use. Rxn Rover is an open-source, modular automation platform for reaction discovery and optimization. Primarily targetting smaller research groups, it is designed using interchangeable plugins to be flexible and easy to integrate into a variety of laboratory environments. Using the Rxn Rover plugin architecture, novel optimization algorithms, analysis instrumentation, and reactor components can be used with minimal or no programming experience. The capability of Rxn Rover is demonstrated in the optimization of a reduction reaction of imine to amine, relevant to energy conversion and manufacturing of fine and commodity chemicals. The reaction was optimized separately using optimizer plugins for SQSnobFit, a Python implementation of the SNOBFIT global optimization algorithm, and Deep Reaction Optimizer (DRO), a deep reinforcement learning algorithm designed for reaction optimization. Using plugins designed for pumps, temperature controllers, and an online liquid chromatography system, the flow reaction was able to be controlled by each algorithm to automate reaction optimization for up to three days, at which point the results were gathered. A successful optimization was performed with SQSnobFit, achieving 70% yield and 95% selectivity, while no successful optimizations were achieved with DRO. Regardless of algorithm performance, Rxn Rover was able to successfully automate both multi-day optimization searches.
Introduction
With the advent of manufacturing transformation,1,2 many chemical processes require new designs with sustainable feedstocks3 and renewable energy.4,5 These new designs, especially those using heterogeneous catalytic processes, often require innovative reaction discovery and optimization techniques to ensure efficient conversion into desired products. However, newly discovered reactions and catalysts are rarely optimal immediately, so discovery is often followed by a long search for optimal reaction conditions to produce the highest desired product yield, optimal conditions for mechanistic studies, or chemo-, regio-, and/or stereo-selectivity.6 Multiple approaches have been taken to decrease this time-consuming task; and the use of efficient methods for reaction optimization can reduce the costs of research and development, minimize wastes, and increase the efficiency of manpower.These improvements are critical in low-budget research environments, where the resources may not be available to acquire and maintain, or rent, existing, high-cost robotics and automation equipment, such as the impressive equipment that is or is becoming standard to the pharmaceutical industry. Indeed, the pharmaceutical industry has taken the lead in developing and applying high-throughput, automated hardware and software to advance drug discovery and synthesis.7–15 While these advances have enabled significant improvements in time to solution, safety and reproducibility, the cost of this specialized instrumentation is well out of reach of most research groups. There is a need for delivering affordable and accessible automation solutions to these groups. Inspiration from the pharmaceutical efforts can be applied to many chemical research areas, including those related to energy and fine chemical discovery.
The traditional approach to explore and optimize a reaction space is one variable at a time (OVAT), where reaction conditions are individually and systematically varied, while all other parameters are held constant.16 OVAT can be particularly effective in mapping a reaction surface when there are minimal variables present. However, as the number of reaction parameters increases, a systematic search of the reaction space becomes impractical, and one must look to more advanced statistical analysis techniques to enable informed, efficient optimization. This is particularly challenging for sustainable applications with renewable feedstocks when feedstock composition varies in different batches.
One common statistical method applied to chemical reaction searches is design of experiments (DoE).17 DoE allows a user with expertise in the field to recognize high-impact factors of a chemical system through a set of controlled experiments. While DoE is widely used in industry and academia,18,19 it is still relatively uncommon for DoE to be incorporated into a chemical optimization algorithm. Such an algorithm may be capable of “mixed-variable” optimization, optimizing both discrete and continuous variables.20,21 For example, Jensen et al. used DoE to create one of the first chemical optimization algorithms capable of mixed-variable optimization.22–25 To further increase the productivity of research, it is advantageous to have search techniques that can effectively and automatically explore a large reaction space parameterized by many variables, which also requires little to no domain-specific knowledge to perform.
For a fully automated, generalized reaction optimization scheme, it is likely that no prior reaction or gradient information will be known. An effective optimization algorithm in this scenario will need to treat the objective function (production of preferred products) as a “black box” and be a direct search method.26 Various black box optimizers have been implemented in reaction optimization for single- and multi-objective optimization including SNOBFIT for global optimization, variations of the Nelder–Mead simplex method for both local and global optimization, and Bayesian approaches for multi-objective optimization.27 However, many of these algorithms fail to perform efficiently with a high number of variables as well.27
Machine learning (ML) is a promising alternative to the commonly used OVAT and DoE approaches of reaction optimization and generic, black box optimizers. However, some black box approaches, like Bayesian approaches, can be categorized as ML and can perform well on highly multi-variate problems, for example TS-EMO.28 ML has been shown to learn and efficiently optimize unknown, highly multi-variate, multi-objective functions.29–31 Once trained on a targeted data set, ML algorithms require little to no domain knowledge to operate. However, training can be difficult and data-intensive, and there is a risk of overfitting or underfitting. Additionally, the user usually needs knowledge of Python, MATLAB, or another programming language to integrate the ML algorithm into their laboratory situation.
The recent boom over the last decade using automatic reaction optimization, or self-optimization, in flow chemistry has seen an increased number of new optimization algorithms and reactor designs published.27,32 However, a method to incorporate, test, and design various optimization algorithms into arbitrary reactor designs freely and easily has not been realized. To accomplish this, sophisticated physical and software systems must be designed to incorporate automatic, algorithmic control of a reaction system. In many existing self-optimizing systems, automation software is designed for and tightly coupled to a specific physical hardware configuration. These hardware configurations may be designed for a specific reaction,33,34 to be general enough to be useful in many reactions,14,35 or to be reconfigurable.36 An optimization algorithm designed in this context takes considerable programming knowledge to apply to other reactors.
Ley et al. recently developed a generalized, modular software platform capable of integrating with any number of reactor setups and optimizers over the internet called LeyLab37 and demonstrated its generality by using LeyLab to control a custom reactor.38 LeyLab integrates a laboratory into the internet of things (IoT), a network of physical devices connected over the internet, in this case to coordinate reactions remotely. Like many self-optimization control codes before it, though, this internet-based, general control software is not available to freely download, install, and use. To provide a less complex, open-source alternative, Cherkasov et al. developed a generalized, modular system control platform using the LabVIEW programming language called OpenFlowChem.39 While some reactor hardware is supported, a considerable amount of work is required by the programmer to incorporate a new device into the system, and the tutorial process leaves much room for error and unexpected complexities to arise.40 OpenFlowChem code must also be modified to incorporate all reactor components used in an experiment. Also taking the IoT into consideration, OpenFlowChem can read files output from analysis instrumentation, which can be synchronized over the internet using cloud file sharing.
Rxn Rover (pronounced Reaction Rover) is introduced in this work as an open-source, general reaction automation software alternative to LeyLab and OpenFlowChem. Similarly, Rxn Rover connects an optimization algorithm – machine learning or non-machine learning – with a reactor and analysis instrumentation to create a self-optimizing flow system compatible with heterogeneous catalysts (Fig. 1). This system allows the optimizer to control and change the reaction parameters, perform the reaction, analyse the desired results, and choose the next search step with or without human intervention. Designed knowing that each research laboratory and experiment presents different challenges, Rxn Rover supports several optimization algorithms, a customizable, modular reactor setup to allow for arbitrary reactor designs, and has the potential to support an array of analysis tools using provided or user-created plugins. Rxn Rover and its plugins are designed to be easy to use, understand, and modify with little to no programming experience, but complex enough to support multi-variate reactions.
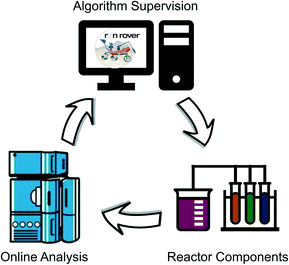 | ||
| Fig. 1 Rxn Rover creates a closed-loop optimization cycle, driving the reactions and exploring the reaction space. | ||
Architecture
Overview of Rxn Rover architecture
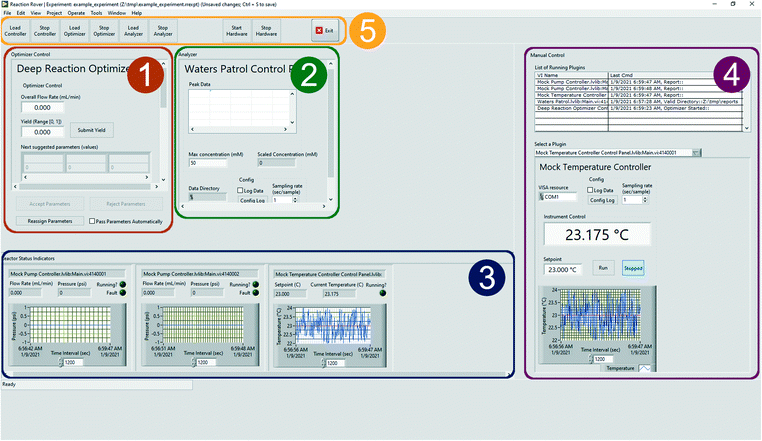
Rxn Rover is designed as the central control application to enable reaction self-optimization and provide a flexible platform to facilitate rapid testing, automation, and design of new reaction optimization algorithms in a variety of laboratory environments. Ease-of-use, modularity, and portability are at the core of the design. An overview of Rxn Rover will be provided from both a user and programming perspective in the following sections.The automated control section is used to configure an optimization algorithm for automated control over the reactor and is divided into three distinct subsections: optimizer control, analyzer, and reactor status indicators. The optimizer control section loads a control interface for the optimization plugin, lets the user decide when the next reaction parameters are sent to the reactor (either manually or automatically when new parameters are generated), and allows optimizer parameters to be associated with specific reactor components. The analyzer section loads the control interface for an analyzer plugin used to parse relevant reaction results for the optimizer, which may be from direct interaction with analysis instrumentation or from a text file. The reactor indicators section displays a small indicator panel for each reactor component, allowing the user to quickly monitor reactor component status, such as flow rates, temperatures, errors, and run state. The manual control section allows the user to take control of individual reactor components through the individual plugin user interfaces (discussed in more detail in the Plugins section below). Along the top of the window are the Rxn Rover control buttons to load/unload plugins at the start of an experiment, start/stop the entire reactor, and exit Rxn Rover.
While being flexible to different laboratory environments, some laboratories will not frequently change configuration. Rxn Rover uses an “experiment” file to save the current setup to be loaded later. An experiment file is created or loaded for each experiment using the Experiment Manager (Fig. 3), which is shown each time Rxn Rover is started. Using an experiment file mitigates the laborious and error-prone task of manually loading all plugins at the beginning of each experiment. Copies of an experiment file can also be used as a template to quickly accommodate new setups with only slight variations in the necessary plugins. By saving which plugins were loaded and the associations between the optimizer, analyzer, and reactor, experiment files can help to improve reproducibility of experimental results.
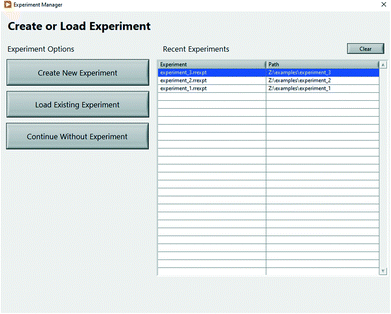 | ||
| Fig. 3 Experiment manager window of Rxn Rover. The experiment manager allows the user to create or load an experiment file when Rxn Rover is started. | ||
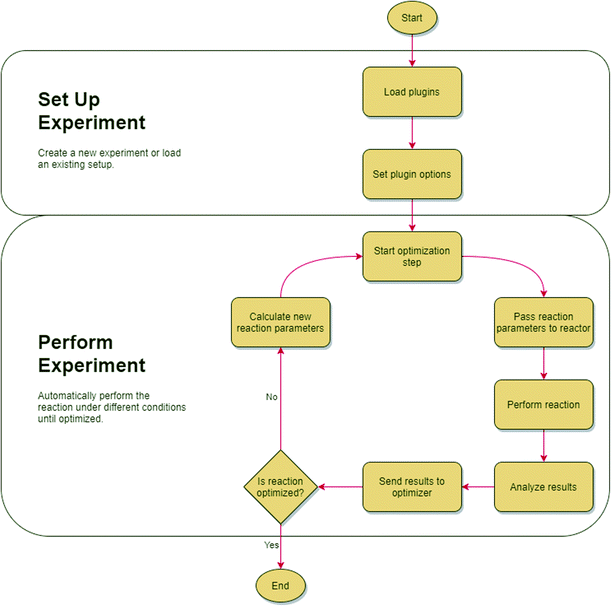
Once an experiment setup is configured in Rxn Rover, the self-optimization may begin. Optimization is started by passing initial conditions chosen by the optimization algorithm to the reactor. Parameter ranges are specified and enforced within the optimizer plugin or the optimization algorithm. The results of the reaction at the given conditions are analysed and parsed by an analyzer plugin that passes the processed results to the optimizer. The optimizer chooses new reaction conditions based on the results of the previous trial. This self-optimizing loop continues until optimal conditions are reached or a user manually stops the loop. The basic workflow of a Rxn Rover optimization is depicted in Fig. 4.
Toward the goals of modularity and portability, Rxn Rover is designed to use a plugin architecture, allowing an arbitrary number of optimizers, analyzers, and reactor components to be added or exchanged for any given process. The association between reaction parameters proposed by the optimizer and the reactor components controlling those reaction parameters can be customized. This modularity enables integration into a variety of laboratory environments with minimal adjustments.
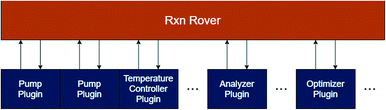
The Rxn Rover program is the overall system control program and acts as the all-in-one user interface. This user interface is used to load plugins for reactor hardware, analysis tools, and optimization algorithms, which are controlled through a communication layer between Rxn Rover and the plugins. The communication layer consists of servers in both the Rxn Rover and plugin which maintain the communication pathways of the system. The overall architecture and communication pathways of Rxn Rover and its plugins is illustrated in Fig. 5.
Plugins
A plugin design pattern was used to improve the modularity and flexibility of the Rxn Rover by allowing components of the system, i.e., reactor hardware, analyzers, and optimizers, to be represented by plugins which can be swapped at runtime. New plugins can also be designed by the user and readily used in Rxn Rover. A template plugin for hardware is provided to make the plugin design process more accessible.41 A plugin created with this template will also be able to run as a standalone control panel program for the given instrument.A plugin for Rxn Rover consists of a communication server object managed by Rxn Rover, a small indicator panel design, a plugin configuration file (essentially the identifier of the plugin), and the plugin source code. Altogether, the plugins serve as middleware, or software, between the primary Rxn Rover program and an “instrument” that monitors and reports state information. Any entity, software or hardware, that can send and/or receive commands regarding its state may be considered an instrument to Rxn Rover. For example, a new optimization algorithm can be considered an instrument, where the algorithm sends new reaction parameters and receives the relevant resulting value of the reaction. An instrument's specific commands are translated by the plugin to commands that Rxn Rover understands, and vice versa.
Rxn Rover will search for plugins in three places on a system. The first location is the “plugins” directory, located in the Rxn Rover installation directory. This location is reserved for pre-packaged plugins. The second and third locations are the Plugins and CustomPlugins directories, respectively. Both directories are found in the Rxn Rover subdirectory under the user's Documents directory. The “Plugins” directory is intended for curated, third-party plugins downloaded from the Rxn Rover plugin repository,41 while the “CustomPlugins” directory is a location to develop and test new plugins from the user.
The plugin configuration file (plugin.conf) is formatted as an INI file, which LabVIEW can read on all operating systems. An INI file is a text-based file comprised of key-value pairs separated into sections. Currently, this configuration file holds four plugin details: the plugin class, which accepts three distinct options: hardware, analyzer, and optimizer plugins; the name to be displayed for the plugin; the specific plugin type, such as a generic pump or temperature controller, company-specific hardware, a generic optimizer or specific algorithm, or a type of analyzer; and the relative path from the configuration file to the file that serves as the software entry point for the plugin, such as a Main.vi file used to launch the plugin. By convention, the plugin.conf file should exist at the root, or top, directory of the plugin, but by taking advantage of the relative path to the plugin entry point, it could be located elsewhere.
Fig. 6 shows the basic architecture for a plugin created using the provided plugin template. Each plugin created from this template consists of six control loops, the system control loop, remote connection manager (RCM), user interface manager, instrument manager loop (IML), acquisition loop, and data logging loop. The system control loop is the central communication hub inside the plugin and redirects messages received from one control loop to others. The RCM handles the connection to Rxn Rover, acting as an internal server for the plugin to send and receive commands to Rxn Rover, and will not be used if the plugin is used as a standalone control panel. The user interface manager handles user interactions with the individual plugin, as well as updating the data indicators of the plugin user interface as new data becomes available. The IML handles the connection to the instrument, sending and receiving commands about instrument state. The acquisition loop is a separate, more accurately timed loop which sends a series of data collection messages to the IML at the given sample rate. The data logging loop writes instrument data to a log file as it is acquired by the acquisition loop or if the instrument state changes.
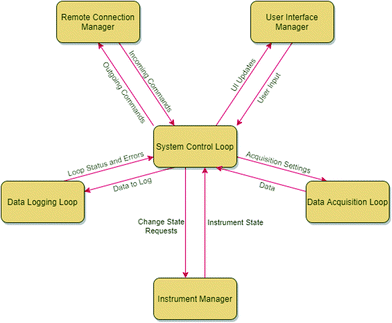 | ||
| Fig. 6 Main relationships between the various control loops of a Rxn Rover plugin. A description of the relationships between the different control loops is described in the text. | ||
It is important to note that for many simple plugins, only the instrument drivers in the IML, the specific acquisition messages of the acquisition loop, logging file headers in the data logging loop, the units and labels of the user interface, and the translation to more general, Rxn Rover-compliant data in the RCM need to be changed to create a new plugin. These modifications can take as little as five minutes if drivers are readily available, and the plugins are for the same type of instrument (creating a pump plugin by deriving it from another pump plugin). When creating a plugin from the generalized plugin template provided with Rxn Rover, it can take as little as 30 minutes to derive a new plugin for new types of instrumentation.
Communications
Rxn Rover is the central communication hub between the many complex components of a self-optimizing system. This communication must be fast, flexible, and reliable to support multi-day, fully autonomous optimizations. The three main communication methods in Rxn Rover are LabVIEW queues, ZeroMQ universal messaging library,50 and the implementation of the virtual instrument software architecture (VISA) standard by NI, NI-VISA.51LabVIEW provides a queue data structure as a useful data communication mechanism to pass data between parallel loops, like the loops used in the plugin architecture shown in Fig. 6. A queue is a container that maintains a first in/first out (FIFO) data sequence, where objects are added to one end of the sequence and removed from the other end. Using a queue allows messages to be processed in the order they are received and provides a buffer if message production exceeds consumption rates.52 Named LabVIEW queues are also used for communication between the Rxn Rover and plugins. A unique server object is maintained by Rxn Rover for each plugin, handling all communication to the plugin including parsing incoming messages, sending outgoing messages, and determining when the connection will be closed. At the heart of the Rxn Rover server object is the LabVIEW queue, named to be unique to the plugin. The RCM control loop is used to maintain the connection with Rxn Rover on the plugin side.
When inter-process and inter-language communication was required, ZeroMQ was used. ZeroMQ is a lightweight messaging library focused on removing complexity in message communication, fitting nicely with the ease-of-use goal in Rxn Rover. ZeroMQ also boasts implementations in 52 programming languages at the time of writing,53 ensuring flexibility of Rxn Rover. An example use case is the communication of an optimizer plugin, written in LabVIEW, with a machine learning algorithm, written in Python. ZeroMQ also supports TCP/IP messaging, enabling communication over the internet if an optimizer must be run on a remote computer cluster or other computer with the proper software installed.
Communication with hardware resources has been standardized in the test and measurement industry through the VISA standard.54 NI provides an implementation of this standard in the NI-VISA software, which provides an interface between LabVIEW and the hardware. NI-VISA is used across the NI product line. Taking advantage of NI-VISA, Rxn Rover plugins can be developed for many hardware resources using existing instrument drivers, such as the many NI and Eurotherm drivers available. New instrument drivers using NI-VISA communication can also be developed using NI's provided tools and resources for instrument driver development.55
Comparison with related software
It should be noted that Rxn Rover is similar to LeyLab and OpenFlowChem but improves upon and differentiates itself from the functionality of each software significantly due to differing design philosophies and approaches. Rxn Rover strives to strike a middle ground between the complex customization options of LeyLab and the restrictive simplicity of OpenFlowChem. Rxn Rover and OpenFlowChem are both open source, whereas LeyLab is not. While LeyLab is built to manage chemical reactions using devices anywhere in the world via the internet, Rxn Rover and OpenFlowChem may be installed on a computer in the laboratory and connect to devices via serial, USB, or Ethernet connections. When design of Rxn Rover began in 2018, the authors did not have knowledge of OpenFlowChem. Both programs are designed with a modular, plugin-like approach to device management, both are written in the LabVIEW programming language, and the device monitor plugins can run on their own.Besides specifics of how the user interacts with the end system and the types of devices/algorithms supported, there are significant design differences between Rxn Rover and OpenFlowChem. For example, OpenFlowChem has a default configuration that it starts with – 2 pumps and a heater. If the user's configuration is different from the default – say to accommodate a new reactor configuration or additional pumps – programming will be necessary. This requires the user to purchase a license to LabVIEW to change the code and to have programming experience. Rxn Rover, on the contrary, was designed with the philosophy that the user should not need to do any programming unless their device is not yet supported. Since Rxn Rover supports a customizable reactor setup by simply loading device plugins, only the free LabVIEW runtime environment is needed unless a component of the reactor is not supported. If a device is not supported, both OpenFlowChem and Rxn Rover provide templates to aid a user in creating proper supporting code. The OpenFlowChem template is deliberately simple in design to allow the code to be easily understood. This limits the extendibility of the device monitor template to simple device operation. Rxn Rover's plugin templates have been designed to accommodate many possible device operation patterns, and the same template has been used to design complex plugins for optimization algorithms, analysis tools, and reactor hardware (see Plugins subsection above). The necessary complexity of the template is hidden in back-end code of an easily understood architecture for beginners with ample documentation. It should also be noted that, while OpenFlowChem has been available since 2018 as one of the first of its kind, there has been little development activity on the project and no community adoption aside from continued work by Cherkasov56,57 to the author's knowledge at the time of writing. Rxn Rover is intended to be continuously developed in the future and collaborations will be actively pursued to foster a large community around the software.
Reaction optimization
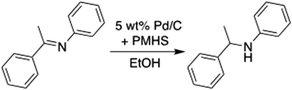
To test the automated optimization, Rxn Rover was used for the optimization of heterogeneously catalysed reduction of imine (Scheme 1) using the SQSnobFit58 and Deep Reaction Optimizer (DRO)30 algorithms. The reduction of imines is an important organic transformation for energy conversion and manufacturing of fine and commodity chemicals and is used as a relatively straightforward example to test the initial capabilities of Rxn Rover.59,60 For example, selective reduction of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N linkage in N-heterocycles is a critical transformation for hydrogen storage in liquid organic hydrogen storage molecules.61 The challenges for this reaction optimization lie in balancing the ratio of imine and reducing agent of choice and maintaining the proper reaction temperature to minimize the hydrolysis of imine, decomposition of amine, and over-reduction/hydrogenolysis of amine.62 Heterogeneous catalyst, in this case 5 wt% Pd/C, is desired over the homogeneous analogue in this work because solid catalysts are readily separated from the reaction streams and more robust towards a wide range of reaction conditions. Also, the use of robust, air-stable solid catalysts can eliminate the use of environmentally harmful reagents such as organo-tin or borohydride reagents,63,64 commonly used for the imine reduction. Poly(methylhydrosiloxane) (PMHS) is used as a mild reducing agent,65 in which the silane H can be readily activated by the Pd/C catalyst. Ethanol is chosen as the green solvent.
N linkage in N-heterocycles is a critical transformation for hydrogen storage in liquid organic hydrogen storage molecules.61 The challenges for this reaction optimization lie in balancing the ratio of imine and reducing agent of choice and maintaining the proper reaction temperature to minimize the hydrolysis of imine, decomposition of amine, and over-reduction/hydrogenolysis of amine.62 Heterogeneous catalyst, in this case 5 wt% Pd/C, is desired over the homogeneous analogue in this work because solid catalysts are readily separated from the reaction streams and more robust towards a wide range of reaction conditions. Also, the use of robust, air-stable solid catalysts can eliminate the use of environmentally harmful reagents such as organo-tin or borohydride reagents,63,64 commonly used for the imine reduction. Poly(methylhydrosiloxane) (PMHS) is used as a mild reducing agent,65 in which the silane H can be readily activated by the Pd/C catalyst. Ethanol is chosen as the green solvent.
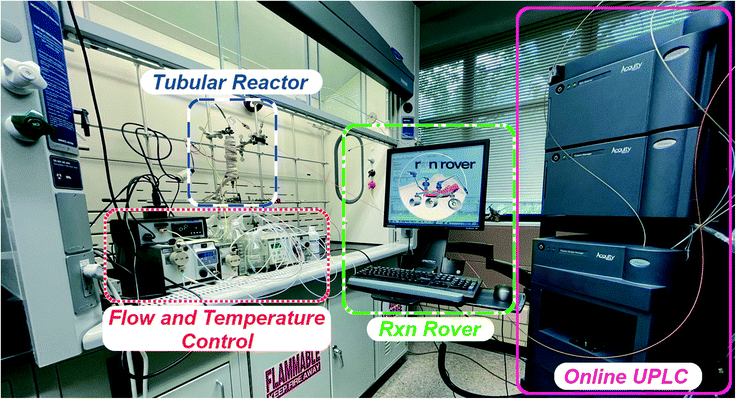
The reaction evaluation system includes three major components: flow reactor, controllers (temperature and flow), and online chromatographic analyzer (Fig. 7). To ensure generality of this work, Rxn Rover interfaces with and fully automates a standard plug-flow reactor; this type of reactor is most commonly used in R&D labs in both academia and industry. The plug-flow reactor is packed with 5 wt% Pd/C catalyst (100 mg). Three different liquid pump models (ChromTech P-MXT, Series III, and P-M110B) are used to deliver imine (5.0 mM in ethanol), PMHS (0.625 g L−1 in ethanol, containing 9.0 mM active silane H), and pure solvent (ethanol). Rxn Rover assigns the flow rates of all pumps to vary the overall concentration of imine or PMHS in the stream while maintaining a fixed total rate for the same residence time of the reaction mixtures. Three different models of liquid pumps are used, and Rxn Rover is fully compatible with all of the different pump drivers. This is an important feature for using Rxn Rover for different chemical applications and in different laboratory environments.
The reaction performance is directly quantified with the Waters™ PATROL™ system, an online ultrahigh pressure liquid chromatograph (UPLC™). Simultaneous detection and quantification can be carried out in one injection for starting imine, resulting amine, and side products (for example, acetophenone and aniline as hydrolysis products). A typical chromatogram is shown in Fig. S1.† The duration of each liquid chromatogram can be tuned between 1 and 5 minutes as necessitated by the setup of the reactor. While product quantification can also be achieved with other types of inline analyzers such as infrared spectroscopy or benchtop nuclear magnetic resonance spectroscopy,66 the use of UPLC™ allows the effective separation of different components and provides quantitative information for fairly complicated reactions in a timely manner. Rxn Rover automatically reads the reports generated by the Patrol system and delivers the analytical results to the optimization algorithm for decision making. For reliability of results and to better comply with current good manufacturing practice,67,68 Rxn Rover can either act on a single data report or on averaged results at steady state and the LC results are recorded and directly associated with the reactor operation conditions.
The maximal concentration of product amines in the stream is set as a more industrially relevant goal rather than highest conversion or yield. In the latter scenario, the optimization may lead to infinite low concentration of imine which is less meaningful. Starting conditions (0.6 mL min−1 total flow, comprised of 42% imine solution, 25% PMHS solution, and 33% ethanol; 30.0 °C) were chosen in a region guaranteed to yield product. The total flow rate was held constant over the course of each optimization, while the optimization algorithm could vary the percent composition of the flow rate and the temperature within a constrained region (0–50% imine solution, 0–50% PMHS solution, and 28–68 °C). The ethanol solvent pump was used as a compensator, ensuring the total flow rate always added to 100%. Although either reactant flow cannot exceed 50%, the higher concentration of PMHS (in respect to silane H, 9.0 mM) over imine (5.0 mM) allows the optimization to cover a broad reaction space. Default algorithm hyperparameter values were used since the goal of this study was to demonstrate the ability to automatically optimize a reaction for long periods of time with various optimization algorithms, not achieve the utmost optimal algorithm performance.
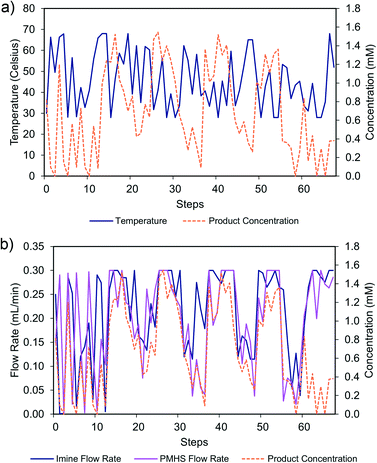
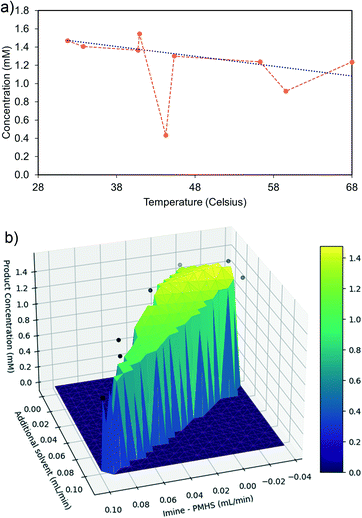
SQSnobFit was used as a state-of-the-art, black box, global optimizer. The reaction was optimized continuously using Rxn Rover and SQSnobFit for 68 reactions (204 UPLC injections; approx. 62 hours). A maximum concentration of 1.5 mM (50% imine, 50% PMHS, 40.9 °C) was found during the optimization (Fig. 8), which corresponds to 70% yield and 95% selectivity; the corresponding chromatograms are shown in Fig. S1.† This maximum concentration was found after approximately 20 hours (26 steps), although the optimization was run longer to explore other possible reaction conditions. The concentration of residual imine and quantifiable side products are also quantified and recorded, Fig. S2.†
The three independent reaction parameters (two independent flow rates and one temperature), together with product concentration, constitute a four-dimensional reaction space. To understand the effects of the different reaction parameters, cross-sections of the information-rich reaction space were constructed using the exploration data generated by SQSnobFit along reaction conditions that were frequently probed by SQSnobFit. The first cross-section was generated along 50% imine, 50% PMHS flow rates (Fig. 9a) and suggests a negative temperature effect on product concentration. Higher reaction temperature leads to the further hydrogenolysis62 of C–N bond in amine and thus causes decrease of imine yield. The hydrogenolysis products can be assigned in the LC chromatograms (Fig. S1†).
The second cross-section was generated along 28.0 °C (Fig. 9b), indicating a negative relationship between product concentration and the ethanol flow rate. This is obvious because higher ethanol percentage compromise the possible concentration of starting imine, which is the pre-requisite of maximal concentration of product amine. The reaction also benefits from a slightly higher PMHS flow relative to imine. This reflects that not all of the silane H in PMHS can be activated and thus a larger equivalence is often needed.62
A modified version of DRO was attempted which allows the number of parameters, parameter ranges, and parameter names to be modified through a configuration file. A modified objective function was injected to allow DRO to communicate and receive results remotely using ZeroMQ. These modifications occur at the interface of DRO with the user and no changes were made to affect the core algorithm driving DRO. The modified DRO was trained on nonconvex mixture Gaussian functions as described by Zhou et al.30 Unfortunately, DRO was unable to find the optimum reaction conditions in experiments performed for this work (Fig. S3†). This may be due to issues associated with the default parameters for the training.
Regardless of optimization algorithm performance, Rxn Rover was able to automate the optimization search without internal issues. The SQSnobFit optimization operated for 62 hours, while the DRO optimization operated for 22 hours, stopping early due to lack of progress toward an optimum.
Conclusion
Rxn Rover is presented as a flexible platform to enable automated reaction optimization in a modular, user-friendly, reproducible fashion for benchtop studies in independent research groups. Manual reactor control can also be accomplished by loading only the reactor components. Using plugins, Rxn Rover is highly customizable to meet the needs of a variety of laboratory configurations. If all hardware, analysis instrumentation, and optimization algorithm plugins exist, no programming knowledge is needed to configure and operate Rxn Rover. When plugins do not exist, a new plugin can be created quickly using a plugin template, programmed in LabVIEW. Rxn Rover, its plugins, and plugin templates are open source and freely available to download, modify, and use on GitHub.41,69 Using experiment files, Rxn Rover makes it easy to reproduce or modify existing experimental setups. Rxn Rover also boasts robust data logging for all plugins and will build a log of the attempted reaction conditions and results during an optimization.Reaction optimization of imine reduction was successfully carried out with Rxn Rover using a standard plug-flow reactor with a state-of-the-art online chromatography system. These optimizations were carried out by running the instrumentation continuously. Two optimization algorithms were applied to the same reaction. SNOBFIT was used to find optimal conditions for the reaction. Due to issues with training and usage, an optimum was not found for DRO. The coupling of Rxn Rover and a flow reactor with online analysis was able to save time routinely used for off-line data analysis, minimize the use of chemicals and catalyst in optimization, and improve safety by minimizing human and environmental exposure to chemicals.
Future development will focus on applying Rxn Rover to reaction discovery for more complex processes, involving multi-step synthesis and multi-objective problems, with additional optimizers and analyzers. With the emergence of new optimization algorithms, Rxn Rover also has potential as a platform for benchmarking these algorithms on different classes of reactions and reactor setups. Rxn Rover experiments can also be performed on simulated experiments using a virtual reactor and analysis instrumentation, which can reduce the cost of testing new reaction optimization algorithms by removing the need to perform potentially costly experiments in the laboratory.
Experimental
Chemicals and materials
Poly(methylhydrosiloxane) (PMHS, Mn = 1700–3200) was purchased from Sigma-Aldrich. PMHS contains 14.5 mol g−1 of active silane (quantified by solution NMR), which is very close to the calculated number (15.1 mol g−1) based on chemical formula, and therefore the PMHS solution (0.625 g L−1) contains 9.0 mmol L−1 transferable hydrogen. The 5 wt% Pd/C catalyst (27 micron) was purchased from Spectrum Chemical. Ethanol (200 proof) was purchased from Decon Labs. Acetonitrile (optima grade), water (optima grade), ammonium acetate (optima grade), toluene, methanol, acetophenone, aniline, and sodium borohydride were purchased from Fischer Scientific.(E)-N,1-Diphenylethan-1-imine, used as the model starting material for this study, was prepared in the typical manner through a condensation reaction between acetophenone and aniline.70 Reagents for this reaction were used as received from Fischer Scientific. The imine was purified via vacuum distillation giving a yellow oil that quickly solidified on cooling. N-(1-Phenylethyl)aniline was then synthesized via reduction of the imine with sodium borohydride in methanol.71
Reactor setup
A plug-flow controller is constructed using ¼′′ stainless steel tubing. Both glass wool and quartz beads are packed in the tubing to maintain the catalyst bed and minimize reactor dead volume. The pressure is controlled at 250 psi using a back pressure regulator (Equilibar). Liquid pumps (ChromTech P-MXT, Series III, and P-M110B) were used to flow liquid through the reactor with serial RJ12 6P6C RS232C (P-MXT and P-M110B) and USB-B (Series III) communications. The reaction temperature is managed using a temperature controller (OMEGA CN16DPt), with USB communications.Online stream analysis and quantification was achieved using a Waters™ PATROL™ system, an ultra-performance liquid chromatograph equipped with photo-diode array. Separation is achieved with a Waters™ ACQUITY™ phenyl column (1.7 μm, 2.1 mm × 150 mm). A gradient method (5 min per chromatogram) was used with solvent A (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 acetonitrile and water with 0.64 g L−1 ammonium acetate) and solvent B (10
10 acetonitrile and water with 0.64 g L−1 ammonium acetate) and solvent B (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 acetonitrile and water with 0.64 g L−1 ammonium acetate). The injection volume is fixed at 100 μL and an online dilution factor of 10 was accomplished with acetonitrile. Quantification of imine, amine, acetophenone, and aniline was achieved via external calibration, using the default online dilution function to minimize uncertainties (Fig. S4†). To allow sufficient time for the reactor to equilibrate to each new set of reaction parameters (flow rates and temperature), the instrument method injected three replicate samples after a pause of 30 min.
90 acetonitrile and water with 0.64 g L−1 ammonium acetate). The injection volume is fixed at 100 μL and an online dilution factor of 10 was accomplished with acetonitrile. Quantification of imine, amine, acetophenone, and aniline was achieved via external calibration, using the default online dilution function to minimize uncertainties (Fig. S4†). To allow sufficient time for the reactor to equilibrate to each new set of reaction parameters (flow rates and temperature), the instrument method injected three replicate samples after a pause of 30 min.
Automated reaction control
Rxn Rover was used to facilitate communication with the flow reactor components, parse analysis results, and allow automated control of the flow reactor by an optimization algorithm. Pump plugins (two instances of ChromTech MX-Class and one instance of ChromTech Series III) were loaded to establish communication with the corresponding pump models. A temperature controller plugin (OMEGA Platinum Series) was loaded for temperature controller communication. To parse report files generated by the UPLC, an analyzer plugin (Waters Patrol UPLC Average) was loaded. Optimizer plugins (SQSnobFit and Deep Reaction Optimizer for reactions) were loaded for the respective experiment that used each optimizer.SNOBFIT72 is a global optimization algorithm, originally written in MATLAB, which has found success in chemical optimization.34,35,39,73 A reimplementation of SNOBFIT in Python, called SQSnobFit,58 was used in lieu of the original SNOBFIT implementation to avoid the MATLAB language license pricing. Using the free, alternative implementation lowers the barrier of entry to replicate the reactions of this paper or expand upon this work in new chemical spaces with the same algorithms. To ensure SQSnobFit could perform for long periods of time, the maximum number of reactions allowed to find the optimal conditions (“budget”) was set to 1000.
Deep Reaction Optimizer (DRO) is a reinforcement learning algorithm for optimizing chemical reactions that was found to outperform state-of-the-art black box optimizers in mock reactions and real microdroplet reactions.30 A modified version of DRO was used which allows the number of parameters, parameter ranges, and parameter names to be modified through a configuration file. A modified objective function was injected to allow DRO to communicate and receive results remotely using ZeroMQ. These modifications occur at the interface of DRO with the user and no changes were made to affect the core algorithm driving DRO. The modified DRO was trained on a nonconvex mixture Gaussian functions as described by Zhou et al.30 Some issues were encountered while applying DRO to chemical reactions: some hyperparameters were not reported; default values in the code repository were different than those given in the publication, including a different default “mock reaction function” used to train the algorithm; and the code base did not contain sufficient documentation, making it difficult to understand and interpret the code.
Data statement
Rxn Rover software is available, and contributions are welcome, at: https://github.com/RxnRover/RxnRover. Documentation and tutorials are available at: https://rxnrover.github.io/RxnRover.Plugins and plugin templates for Rxn Rover are made available, and contributions are welcome, at: https://rxnrover.github.io/PluginCatalog.
Author contributions
Z. Crandall: conceptualization, data curation, formal analysis, investigation, methodology, software, validation, visualization, writing – original draft, writing – review & editing; K. Basemann: conceptualization, formal analysis, investigation, methodology, resources, validation, writing – original draft, writing – review & editing; L. Qi: conceptualization, formal analysis, funding acquisition, investigation, methodology, resources, validation, writing – original draft, writing – review & editing; T. L. Windus: conceptualization, funding acquisition, project administration, supervision, validation, writing – original draft, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was primarily supported by Ames Laboratory Directed Research & Development program. T. L. W., K. B. and Z. C. were also supported by the U.S. Department of Energy (U.S. DOE), Office of Science, Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences through the Ames Laboratory Chemical Physics program. L. Q. and the reactor system were in part supported by the U.S. DOE, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences through the Ames Laboratory Catalysis program. The Ames Laboratory is operated for the U.S. DOE by Iowa State University under Contract No. DE-AC02-07CH11358. We also acknowledge the useful conversion with Waters Corporation regarding the setup of the chromatography system.References
- J. B. Zimmerman, P. T. Anastas, H. C. Erythropel and W. Leitner, Science, 2020, 367, 397–400 CrossRef CAS PubMed.
- M. C. Bryan, P. J. Dunn, D. Entwistle, F. Gallou, S. G. Koenig, J. D. Hayler, M. R. Hickey, S. Hughes, M. E. Kopach, G. Moine, P. Richardson, F. Roschangar, A. Steven and F. J. Weiberth, Green Chem., 2018, 20, 5082–5103 RSC.
- R. Gérardy, D. P. Debecker, J. Estager, P. Luis and J.-C. M. Monbaliu, Chem. Rev., 2020, 120, 7219–7347 CrossRef.
- D. Cambié, C. Bottecchia, N. J. W. Straathof, V. Hessel and T. Noël, Chem. Rev., 2016, 116, 10276–10341 CrossRef.
- D. Pletcher, R. A. Green and R. C. D. Brown, Chem. Rev., 2018, 118, 4573–4591 CrossRef CAS.
- T. Yu, Z. Ding, W. Nie, J. Jiao, H. Zhang, Q. Zhang, C. Xue, X. Duan, Y. M. A. Yamada and P. Li, Chem. – Eur. J., 2020, 26, 5729–5747 CrossRef CAS.
- S. M. Mennen, C. Alhambra, C. L. Allen, M. Barberis, S. Berritt, T. A. Brandt, A. D. Campbell, J. Castañón, A. H. Cherney, M. Christensen, D. B. Damon, J. Eugenio De Diego, S. García-Cerrada, P. García-Losada, R. Haro, J. Janey, D. C. Leitch, L. Li, F. Liu, P. C. Lobben, D. W. C. Macmillan, J. Magano, E. Mcinturff, S. Monfette, R. J. Post, D. Schultz, B. J. Sitter, J. M. Stevens, I. I. Strambeanu, J. Twilton, K. Wang and M. A. Zajac, Org. Process Res. Dev., 2019, 23, 1213–1242 CrossRef CAS.
- E. Garcia-Egido, V. Spikmans, S. Y. F. Wong and B. H. Warrington, Lab Chip, 2003, 3, 73–76 RSC.
- A. Vasudevan, A. R. Bogdan, H. F. Koolman, Y. Wang and S. W. Djuric, in Progress in Medicinal Chemistry, ed. D. R. Witty and B. Cox, Elsevier, 2017, vol. 56, pp. 1–35 Search PubMed.
- R. V. Jones, L. Godorhazy, N. Varga, D. Szalay, L. Urge and F. Darvas, J. Comb. Chem., 2006, 8, 110–116 CrossRef CAS PubMed.
- R. Grainger and S. Whibley, Org. Process Res. Dev., 2021, 25, 354–364 CrossRef CAS.
- A. Buitrago Santanilla, E. L. Regalado, T. Pereira, M. Shevlin, K. Bateman, L.-C. Campeau, J. Schneeweis, S. Berritt, Z.-C. Shi, P. Nantermet, Y. Liu, R. Helmy, C. J. Welch, P. Vachal, I. W. Davies, T. Cernak and S. D. Dreher, Science, 2015, 347, 49–53 CrossRef CAS.
- T. Ahneman Derek, G. Estrada Jesús, S. Lin, D. Dreher Spencer and G. Doyle Abigail, Science, 2018, 360, 186–190 CrossRef CAS.
- D. Perera, J. W. Tucker, S. Brahmbhatt, C. J. Helal, A. Chong, W. Farrell, P. Richardson and N. W. Sach, Science, 2018, 359, 429–434 CrossRef CAS.
- J. W. Sawicki, A. R. Bogdan, P. A. Searle, N. Talaty and S. W. Djuric, React. Chem. Eng., 2019, 4, 1589–1594 RSC.
- O. W. Gooding, Curr. Opin. Chem. Biol., 2004, 8, 297–304 CrossRef CAS.
- S. A. Weissman and N. G. Anderson, Org. Process Res. Dev., 2015, 19, 1605–1633 CrossRef CAS.
- R. J. Del Vecchio, Understanding Design of Experiments: A Primer for Technologists, Hanser/Gardner Publications, New York, 1997 Search PubMed.
- R. L. Mason, R. F. Gunst and J. L. Hess, Statistical Design and Analysis of Experiments, J. Wiley & Sons, Hoboken, N.J., 2nd edn, 2003 Search PubMed.
- C. Zhang, Y. Amar, L. Cao and A. A. Lapkin, Org. Process Res. Dev., 2020, 24, 2864–2873 CrossRef CAS.
- B. J. Shields, J. Stevens, J. Li, M. Parasram, F. Damani, J. I. M. Alvarado, J. M. Janey, R. P. Adams and A. G. Doyle, Nature, 2021, 590, 89–96 CrossRef CAS.
- B. J. Reizman and K. F. Jensen, Chem. Commun., 2015, 51, 13290–13293 RSC.
- B. J. Reizman and K. F. Jensen, Acc. Chem. Res., 2016, 49, 1786–1796 CrossRef CAS.
- B. J. Reizman, Y.-M. Wang, S. L. Buchwald and K. F. Jensen, React. Chem. Eng., 2016, 1, 658–666 RSC.
- H.-W. Hsieh, C. W. Coley, L. M. Baumgartner, K. F. Jensen and R. I. Robinson, Org. Process Res. Dev., 2018, 22, 542–550 CrossRef CAS.
- D. Cortés-Borda, K. V. Kutonova, C. Jamet, M. E. Trusova, F. Zammattio, C. Truchet, M. Rodriguez-Zubiri and F.-X. Felpin, Org. Process Res. Dev., 2016, 20, 1979–1987 CrossRef.
- A. D. Clayton, J. A. Manson, C. J. Taylor, T. W. Chamberlain, B. A. Taylor, G. Clemens and R. A. Bourne, React. Chem. Eng., 2019, 4, 1545–1554 RSC.
- A. M. Schweidtmann, A. D. Clayton, N. Holmes, E. Bradford, R. A. Bourne and A. A. Lapkin, Chem. Eng. J., 2018, 352, 277–282 CrossRef CAS.
- N. Peremezhney, E. Hines, A. Lapkin and C. Connaughton, Eng. Optim., 2014, 46, 1593–1607 CrossRef.
- Z. Zhou, X. Li and R. N. Zare, ACS Cent. Sci., 2017, 3, 1337–1344 CrossRef CAS.
- Z. Fu, X. Li, Z. Wang, Z. Li, X. Liu, X. Wu, J. Zhao, X. Ding, X. Wan, F. Zhong, D. Wang, X. Luo, K. Chen, H. Liu, J. Wang, H. Jiang and M. Zheng, Org. Chem. Front., 2020, 7, 2269–2277 RSC.
- A. Sivo, R. S. Galaverna, G. R. Gomes, J. C. Pastre and G. Vilé, React. Chem. Eng., 2021, 6, 756–786 RSC.
- S. Krishnadasan, R. J. C. Brown, A. J. deMello and J. C. deMello, Lab Chip, 2007, 7, 1434–1441 RSC.
- N. Holmes, G. R. Akien, R. J. D. Savage, C. Stanetty, I. R. Baxendale, A. J. Blacker, B. A. Taylor, R. L. Woodward, R. E. Meadows and R. A. Bourne, React. Chem. Eng., 2016, 1, 96–100 RSC.
- J. P. Mcmullen and K. F. Jensen, Org. Process Res. Dev., 2010, 14, 1169–1176 CrossRef CAS.
- A.-C. Bédard, A. Adamo, K. C. Aroh, M. G. Russell, A. A. Bedermann, J. Torosian, B. Yue, K. F. Jensen and T. F. Jamison, Science, 2018, 361, 1220–1225 CrossRef.
- D. E. Fitzpatrick, C. Battilocchio and S. V. Ley, Org. Process Res. Dev., 2016, 20, 386–394 CrossRef CAS.
- D. E. Fitzpatrick and S. V. Ley, React. Chem. Eng., 2016, 1, 629–635 RSC.
- N. Cherkasov, Y. Bai, A. J. Expósito and E. V. Rebrov, React. Chem. Eng., 2018, 3, 769–780 RSC.
- N. Cherkasov, OpenFlowChem Wiki: Create Device Monitor, https://sourceforge.net/p/openflowchem/wiki/Create%20Device%20Monitor/, (accessed May 23, 2021) Search PubMed.
- ReactionRover Organization, ReactionRover/plugins, https://github.com/ReactionRover/plugins, (accessed May 23, 2021) Search PubMed.
- National Instruments Corporation, NI Driver Downloads, https://www.ni.com/en-us/support/downloads/drivers.html, (accessed May 23, 2021) Search PubMed.
- National Instruments Corporation, Instrument Driver Network (IDNet), https://www.ni.com/en-us/support/downloads/instrument-drivers.html, (accessed May 23, 2021) Search PubMed.
- R. A. Bourne, R. A. Skilton, A. J. Parrott, D. J. Irvine and M. Poliakoff, Org. Process Res. Dev., 2011, 15, 932–938 CrossRef CAS.
- A. J. Parrott, R. A. Bourne, G. R. Akien, D. J. Irvine and M. Poliakoff, Angew. Chem., Int. Ed., 2011, 50, 3788–3792 CrossRef CAS.
- D. J. Kim and Z. Fisk, Rev. Sci. Instrum., 2012, 83, 123705 CrossRef CAS PubMed.
- V. Sans, L. Porwol, V. Dragone and L. Cronin, Chem. Sci., 2015, 6, 1258–1264 RSC.
- A. Adamo, R. L. Beingessner, M. Behnam, J. Chen, T. F. Jamison, K. F. Jensen, J.-C. M. Monbaliu, A. S. Myerson, E. M. Revalor, D. R. Snead, T. Stelzer, N. Weeranoppanant, S. Y. Wong and P. Zhang, Science, 2016, 352, 61–67 CrossRef CAS PubMed.
- M. Rubens, J. H. Vrijsen, J. Laun and T. Junkers, Angew. Chem., Int. Ed., 2019, 58, 3183–3187 CrossRef CAS PubMed.
- The ZeroMQ authors, ZeroMQ, 2021.
- National Instruments Corporation, NI-VISA, 2019.
- National Instruments Corporation, What Is a Queue in LabVIEW?, https://knowledge.ni.com/KnowledgeArticleDetails?id=kA00Z000000P7OfSAK&l=en-US, (accessed May 19, 2021) Search PubMed.
- ØMQ Language Bindings, http://wiki.zeromq.org/bindings:_start, (accessed May 23, 2021).
- IVI Foundation, IVI Specifications, https://www.ivifoundation.org/specifications/default.aspx, (accessed May 19, 2021).
- National Instruments Corporation, Instrument Driver Development Tools and Resources, https://www.ni.com/devzone/idnet/development.htm, (accessed May 23, 2021) Search PubMed.
- N. Cherkasov, A. J. Expósito, Y. Bai and E. V. Rebrov, React. Chem. Eng., 2019, 4, 112–121 RSC.
- N. Cherkasov, P. Denissenko, S. Deshmukh and E. V. Rebrov, Chem. Eng. J., 2020, 379, 122292 CrossRef CAS.
- W. Lavrijsen and Scikit-quant, SQSnobFit, https://github.com/scikit-quant/scikit-quant/tree/master/opt/snobfit, (accessed April 30, 2021) Search PubMed.
- B. Chen, U. Dingerdissen, J. G. E. Krauter, H. G. J. Lansink Rotgerink, K. Möbus, D. J. Ostgard, P. Panster, T. H. Riermeier, S. Seebald, T. Tacke and H. Trauthwein, Appl. Catal., A, 2005, 280, 17–46 CrossRef CAS.
- L. Qi, J. Chen, B. Zhang, R. Nie, Z. Qi, T. Kobayashi, Z. Bao, Q. Yang, Q. Ren, Q. Sun, Z. Zhang and W. Huang, ACS Catal., 2020, 10, 5707–5714 CrossRef CAS.
- R. H. Crabtree, Energy Environ. Sci., 2008, 1, 134–138 RSC.
- A. Volkov, K. P. J. Gustafson, C.-W. Tai, O. Verho, J.-E. Bäckvall and H. Adolfsson, Angew. Chem., Int. Ed., 2015, 54, 5122–5126 CrossRef CAS PubMed.
- J. M. Blackwell, E. R. Sonmor, T. Scoccitti and W. E. Piers, Org. Lett., 2000, 2, 3921–3923 CrossRef CAS PubMed.
- R. M. Lopez and G. C. Fu, Tetrahedron, 1997, 53, 16349–16354 CrossRef CAS.
- J. M. Lavis and R. E. Maleczka Jr, Encyclopedia of Reagents for Organic Synthesis, 2001 Search PubMed.
- S. Chatterjee, M. Guidi, P. H. Seeberger and K. Gilmore, Nature, 2020, 579, 379–384 CrossRef CAS.
- U.S. Food & Drug Administration, Current Good Manufacturing Practice (CGMP) Regulations, https://www.fda.gov/drugs/pharmaceutical-quality-resources/current-good-manufacturing-practice-cgmp-regulations, (accessed May 23, 2021) Search PubMed.
- I. E. W. Group, ICH harmonised tripartite guideline, 2005, vol. 25, p. 781 Search PubMed.
- RxnRover Organization, RxnRover, https://github.com/RxnRover/RxnRover, (accessed May 23, 2021) Search PubMed.
- P.-S. Lai, J. A. Dubland, M. G. Sarwar, M. G. Chudzinski and M. S. Taylor, Tetrahedron, 2011, 67, 7586–7592 CrossRef CAS.
- Y. Xing, S. Wu, M. Dong, J. Wang, L. Liu and H. Zhu, Tetrahedron, 2019, 75, 130495 CrossRef.
- W. Huyer and A. Neumaier, ACM Trans. Math. Softw., 2008, 35, 1–25 CrossRef.
- M. I. Jeraal, N. Holmes, G. R. Akien and R. A. Bourne, Tetrahedron, 2018, 74, 3158–3164 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1re00265a |
| This journal is © The Royal Society of Chemistry 2022 |