Open Access Article
Open Access ArticleFormulation of water-dispersible hydrophobic compound nanocomplexes with polypeptides via a supramolecular approach using a high-speed vibration milling technique†
Shogo Kawamura‡
a,
Riku Kawasaki‡ *a,
Shodai Hino‡
*a,
Shodai Hino‡ ab,
Keita Yamanaa,
Masafumi Okunoa,
Takuro Etoa and
Atsushi Ikeda
ab,
Keita Yamanaa,
Masafumi Okunoa,
Takuro Etoa and
Atsushi Ikeda *a
*a
aProgram of Applied Chemistry, Graduate School of Advanced Science and Engineering, Hiroshima University, 1-4-1 Kagamiyama, Higashi Hiroshima, 739-8527, Japan. E-mail: riku0528@hiroshima-u.ac.jp; aikeda@hiroshima-u.ac.jp
bBiomedical Research Institute, National Institute of Advanced Industrial Science and Technology, AIST, 1-8-31 Midorigaoka, Ikeda, 563-8577, Japan
First published on 8th November 2022
Abstract
Polypeptides were used to solubilize functional hydrophobic molecules via a high-speed vibrational milling method. Poly-L-lysine and poly-γ-glutamic acid, which are polypeptides, were able to prepare more highly concentrated water-dispersible complexes of hydrophobic compounds, including fullerenes, organic dyes, and porphyrin derivatives, than conventional water solubilizers, such as cyclodextrins and pullulan. In addition, the polypeptide systems endowed the complexes with long-term stability and resistance against thermal stress, which is advantageous for industrial applications. Furthermore, complexes of polypeptides and porphyrin derivatives showed a photodynamic activity against cancer cells, and the current system improved the dispersibility and storability of guest molecules without compromising their functionality.
Introduction
The development of water solubilization platforms opens the applicability of hydrophobic compounds as pharmaceutical drugs1–5 and provides eco-friendly methodologies to handle organic dyes without utilizing organic solvents to process organic electrolytes6,7 and resins8 in casts. Although surfactants are used as water solubilizers,9,10 these can contribute to water pollution11,12 and do harmful damage to ecosystems.11,12 Therefore, water solubilization techniques have been developed using amphiphilic calixarenes13 and cyclodextrins (CDxs).14–18Recently, we also addressed the water solubilization of hydrophobic compounds, such as carbon nanotubes,18 porphyrin derivatives,15,16,19,20 fullerene derivatives,21 and phthalocyanine derivatives,17 by formulating supramolecular complexes with biocompatible water solubilizers, including CDxs and polysaccharides, via mechanochemical ball milling technique called high-speed vibratory milling (HSVM). With easiness in handling, scalability, and unique preservation properties of crystal information, the HSVM technique is expected to be applicable for various fields, such as pharmaceuticals,15–17,19–21 organic electrolytes,18 and organic crystals.22 Although the technique using CDxs and polysaccharides is a powerful means to fabricate monodisperses in water and water-dispersible nanocomplexes, there are still several limitations in the solubilization capacity, long-term stability, and thermal stability of complexes, depending on guest molecules. As new platforms for water solubilizers, we focused on polypeptides due to their excellent biocompatibility, biodegradability, thermal stability, and functionality.23–34 More recently, using polypeptides as solubilizers, we succeeded in solubilizing paclitaxel, a hydrophobic anticancer drug, in larger quantities compared to that using polysaccharides, which are conventional solubilizers.35 The results encouraged us to investigate the applicability of polypeptides as water solubilizers.
With an excellent biocompatibility23–35 and responsiveness to external stimuli, including enzymes,23,24 pH,25,26 and reactive oxygen species,27 polypeptides have been used in various fields, including the biomedical field, as drug delivery systems23,27,29 and hydrogels.24,27–29
For example, poly-L-lysine (PLL), which is traditionally used as a coating agent for culture media to enhance the attachment of cells30 and manipulate neuronal cellular fate,31 has a primary amino group in its side chain, which contributes to structural changes associated with pH changes. These features enable the establishment of pH-responsive materials.25 Furthermore, PLL has an excellent surface attachment and surface adhesion properties, making it a promising anti-reflection coating for photovoltaic applications.32 On the other hand, poly-γ-glutamic acid (PGA), the main component of natto strings,33 induces structural changes with pH changes due to the carboxyl group in the side chain, and pH-responsive nanomaterials have been designed using this property.26 Moreover, the carboxyl and amide groups of PGA can be used for characteristic coordination bonds with metal ions, making it a promising and advanced adsorbent material for the removal of heavy metal ions.34 In addition, collagen (Col), the major component of structural proteins that are abundantly distributed in the human dermis and tendons, is degraded by an enzyme called matrix metalloproteinase23,24 and is expected to be applied as a biomaterial for cancer metastasis.23
In this study, we demonstrated the applicability of polypeptides as water solubilizers for the inclusion of hydrophobic functional guest molecules by HSVM (Fig. 1a). Polypeptides, including PLL, PGA, and Col, were used as solubilizers. γ-Cyclodextrin (γ-CDx) or 2,3,6-tri-O-methyl-β-cyclodextrin (TMe-β-CDx) and pullulan (Pul), were also employed for comparisons. In addition, guest molecules and their physicochemical properties are shown in Fig. 1b. Moreover, we compared the long-term stability of the dispersion prepared with polypeptides in aqueous media against thermal stimuli with those prepared with CDxs and Pul.
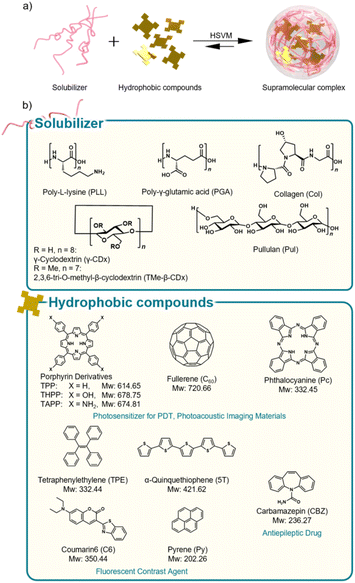 | ||
| Fig. 1 (a) Schematic illustration of supramolecular nanostructure formation by HSVM. (b) Solubilizers and hydrophobic compounds used in this study. | ||
Finally, we demonstrated the applicability of polypeptide complexes with porphyrin derivative, 5,10,15,20-tetrakis(4-hydroxyphenyl)porphyrin (THPP), as drugs for photo-cancer therapy, including photodynamic therapy (PDT).
Results and discussion
Preparation and characterization of supramolecular complexes
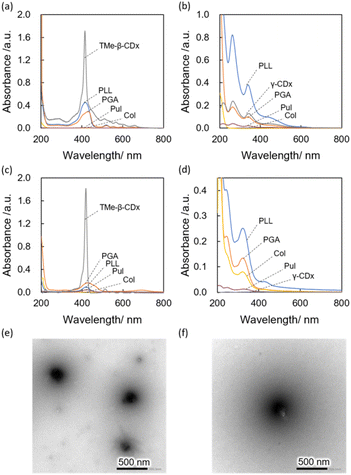
Complexes of hydrophobic compounds with solubilizers were prepared via HSVM, as previously reported.15–22,35 Briefly describing the procedure, solid mixtures of solubilizers and hydrophobic compounds were shaken with two agate mixing balls using a high-speed vibrating mill (25 Hz, 30 min), suspended in 2.0 mL of aqueous solution, and then centrifuged to collect the supernatant. Polypeptides, including PLL, PGA, and Col, were used as solubilizers. γ-CDx, TMe-β-CDx, and Pul were also used as conventional solubilizers.15–17,19,22 Porphyrin derivatives tetraphenylporphyrin (TPP), THPP, 5,10,15,20-tetrakis(4-aminophenyl)porphyrin (TAPP), [C60]fullerene (C60), tetraphenylethylene (TPE), α-quinquethiophene (5T), carbamazepin (CBZ), coumarin6 (C6), pyrene (Py), and phthalocyanine (Pc) were used as functional hydrophobic guest molecules. After centrifugation to remove the precipitates, the UV-Vis absorption spectra of each supernatant aqueous solution were measured to confirm the solubilization of guest molecules (Fig. 2a–d and S1†). Col could not dissolve TPP and C60 in water (Fig. 2a and b, yellow line). The UV-Vis absorption spectra of porphyrin derivatives complexed with the polypeptides showed broad absorption bands assignable to Soret and Q bands compared with those with porphyrin complexed with TMe-β-CDx (Fig. 2a and c: blue, PLL; orange, PGA; yellow, Col; gray, TMe-β-CDs), indicating that the porphyrin derivatives self-aggregated in the polypeptides. Similar results were obtained when the porphyrins were complexed with Pul (Fig. 2a and c, purple).The concentrations of hydrophobic compounds that resulted in dispersion were quantified by measuring the absorbance of solution redissolving the guest molecules in an organic solvent, and the concentration of each compound was summarized in Table1 and S1.† The concentrations of TPP dissolved with PLL, PGA, Col, Pul, and TMe-β-CDx were 1020, 650, 0, 40, and 2900 μM, respectively. Among these biopolymers, PLL showed the most excellent capability to dissolve TPP. PLL and PGA were able to disperse TPP at high concentrations than Pul, which is used as a conventional water solubilizer. These differences may be caused by the flexibility of polymer backbones.35–37 Similar trends were found in other functional hydrophobic molecules.
| Conc. (μM) | Dhy (nm) | PDI | ζ-Potential (mV) | |
|---|---|---|---|---|
| a All the samples were measured in water (pH, 7.4; 25 °C). PDI value was calculated by cumulant method. ζ-Potential was measured with capillary cells. | ||||
| PLL/TPP | 1000 | 650 ± 20 | 0.40 | +74 ± 1 |
| PGA/TPP | 650 | 430 ± 6 | 0.21 | −72 ± 1 |
| Col/TPP | 0 | — | — | — |
| Pul/TPP | 40 | 290 ± 10 | 0.28 | −13 ± 1 |
| TMe-β-CDx/TPP | 2900 | — | — | — |
| PLL/C60 | 1900 | 270 ± 4 | 0.21 | +65 ± 1 |
| PGA/C60 | 440 | 530 ± 20 | 0.18 | −68 ± 2 |
| Col/C60 | 0 | — | — | — |
| Pul/C60 | 95 | 120 ± 10 | 0.23 | −21 ± 1 |
| γ-CDx/C60 | 560 | — | — | — |
| PLL/THPP | 200 | 250 ± 4 | 0.26 | +70 ± 2 |
| PGA/THPP | 580 | 280 ± 2 | 0.19 | −43 ± 1 |
| Col/THPP | 6 | 200 ± 10 | 0.31 | +2 ± 1 |
| Pul/THPP | 130 | 230 ± 10 | 0.19 | +15 ± 1 |
| TMe-β-CDx/THPP | 1300 | — | — | — |
| PLL/TPE | 1300 | 580 ± 20 | 0.19 | +60 ± 1 |
| PGA/TPE | 640 | 690 ± 8 | 0.26 | −61 ± 1 |
| Col/TPE | 450 | 70 ± 8 | 0.58 | +0.7 ± 0.1 |
| Pul/TPE | 150 | 380 ± 10 | 0.30 | −11 ± 1 |
| TMe-β-CDx/TPE | 0 | — | — | — |
The hydrodynamic diameter (Dhy) of each complex was examined by dynamic light scattering (DLS) measurements (Table1 and S1†), and all complexes using biopolymers were prepared with a diameter of 100–700 nm, with a relatively narrow dispersity. Furthermore, the zeta potential of major complexes was measured, and the electrical characteristics of the complexes were highly dependent on the electrical properties of the polymer backbone, which is consistent with previous reports19,35 (Table 1). We also conducted a morphological observation of the C60 complexed with PLL and PGA by transmission electron microscopy with staining (Fig. 2e and f). Spherical morphologies were found in each sample, and their size distribution corresponded to the results obtained from DLS.
Stability of water-dispersible supramolecular complexes
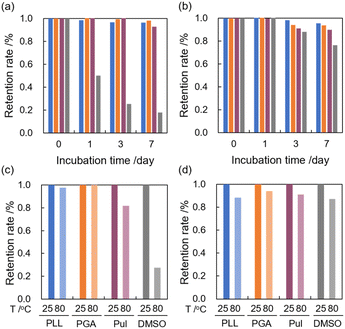
The stability of the complexes after preparation is significant for the practical use of functional compounds, such as pharmaceuticals,38 dietary supplements,39,40 and dye coating.41 The long-term dispersibility of guest molecules, TPP and C60, was examined by examining the UV-Vis absorption spectral changes. When the complexes in aqueous media were incubated at room temperature, the absorbance at the maximum absorption wavelength of polypeptide/TPP complexes and Pul/TPP complexes did not change for seven days without forming undesirable precipitates (Fig. 3a). The retention ratio of TPP dissolved by PLL and PGA in water was estimated to be 96% and 98%, respectively. In contrast, over 80% of TPP/TMe-β-CDx complexes precipitated out by forming aggregations. Similar trends were found in C60 complexes with a water solubilizer (Fig. 3b). The retention ratios of PLL/C60 and PGA/C60 complexes after seven days were 95% and 94%, respectively, which were more stable than those of Pul/C60 and γ-CDx/C60 complexes (90% and 77%, respectively). These results suggest that biopolymer-based water solubilization is advantageous to prepare a highly concentrated dispersion of guest molecules with long-term stability. In addition, we examined the thermal stability of the complexes based on spectral changes. The absorption spectra of PLL/TPP, PGA/TPP, Pul/TPP, and TMe-β-CDx/TPP complexes were measured after heating the dispersion at 80 °C for 2 h. In case of PLL and PGA, almost 100% of TPP was still dispersed in water (Fig. 3c, blue and orange). On the other hand, heating decreased the concentration of TPP complexed with Pul and TMe-β-CDx to 80% and 30%, respectively. In contrast, C60 complexes with these water solubilizers were relatively stable against thermal stimuli (Fig. 3d). These results suggest that current systems using polypeptides improved their thermal stability, and the feature is attractive for fabrication in industry.Applications with polypeptide/THPP complexes for photodynamic therapy
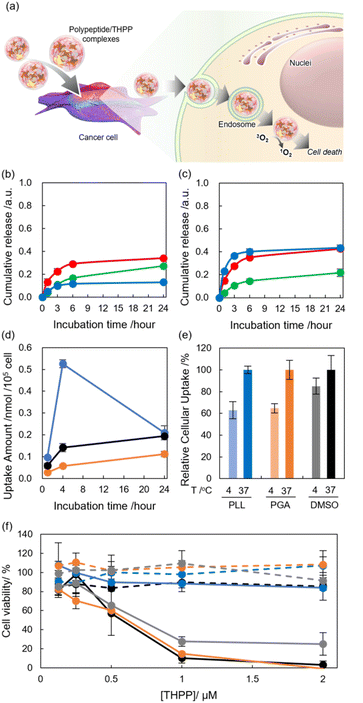
Finally, we demonstrated the applicability of the current peptide-based system for pharmaceutics, and we focused on photo-triggered cancer therapy, which is photodynamic PDT.15–17,19–21 PDT is achieved by cytotoxic reactive oxygen species generated by an excited photosensitizer42 via irradiation of light (Fig. 4a). Here we employed THPP as a photosensitizer.We initially tried to address the loading capacity of THPP using polypeptides and by manipulating the size of the complex, resulting in the formulation of highly concentrated dispersion using PLL and PGA with a diameter of 200–300 nm (Table S2†). TEM revealed the spherical morphology of THPP/polypeptide complexes, which corresponded to the results from DLS (Fig. S2†). In addition, the diameter of the complexes was reasonable for meeting tumor passive targeting, which has an enhanced permeation and retention effect.43,44
As PLL and PGA were used as pH-responsive materials,25,26 a pH-triggered cargo release was initially examined using PLL/THPP and PGA/THPP complexes. After incubation at pH 5, 7, and 9, released THPP complexes were isolated by centrifugation, and the amount of THPP was quantified by measuring the UV-Vis absorption spectra of solutions redissolved in DMSO. In the case of PLL/THPP complexes, incubation with an acidic pH enhanced the release, while incubation with a basic pH inhibited the release (Fig. 4b). To address the differences in release profile, the Dhy of the complexes was examined. In the case of PLL complexes, secondary structural changes25 triggered by surrounding pH did not significantly induce Dhy changes (Fig. S3a†), which might not contribute to the release. As the nitrogen in pyrrole ring got protonated,45 the acceleration of THPP release at pH 5 should be caused by electro repulsion effects. In contrast, the Dhy of THPP complexes with PGA was drastically changed by pH. The complexes shrunk at both acidic and basic pH. At an acidic pH, the release was suggested to be due to a rapid decrease in Dhy associated with conformational changes in the peptide.26 At a basic pH, the release was suggested to be due to a rapid decrease in Dhy associated with electrostatic repulsion (Fig. S3b†). After the shrink of the complexes, the loading capacity should decrease, suggesting that pH stimuli manipulated the release profile of cargo molecules (Fig. 4c). These results suggest that current systems can work as pH-responsive payload release systems.
We investigated the capability of free THPP, PLL/THPP, and PGA/THPP to convert dissolved oxygen into singlet oxygen molecules (1O2) using 9,10-anthracenediyl-bis(methylene)dimalonic acid (ABDA) (Fig. S4a†).46 The consumption of ABDA was monitored based on absorption changes at 380 nm (i.e., maximum absorption for ABDA) following the irradiation of 15 μM samples of PLL/THPP complexes, PGA/THPP complexes, and DMSO/THPP solution (Fig. S4b–d†). Among these three systems, free THPP exhibited the highest photodynamic activity, which was almost the same as that in PGA and PLL systems. This should be caused by the formulation of self-aggregation of THPP within polymer matrix. In addition, PLL (23 μg mL−1) was added to the aqueous solution of THPP prepared by DMSO injection method, resulting in a significantly lessened conversion. This suggests that PLL can work as an 1O2 scavenger (Fig. S4e and f†).47
To confirm the deliverability of the polypeptide systems, we quantified the cellular uptake of THPP toward murine colon carcinoma cells (Colon26 cells) by measuring the fluorescence (Fig. 4d).19 The cells were co-incubated with DMSO/THPP, PLL/THPP, and PGA/THPP (THPP, 4.7 μM), and the cells were collected at each time point (1, 4, and 24 h). Results showed that the PLL system exhibited the highest efficiency in cellular uptake in the initial 4 h among the three systems, suggesting that cationic PLL systems interacted with negatively charged cancer cell membrane. Fluorescence signals from delivered THPP were also detected within cells incubated with all systems by confocal laser scanning microscope (CLSM) (Fig. S5a–c†). In addition, the cellular uptake of THPP was significantly lessened by maintaining the cells at 4 °C, which can inhibit energy-dependent cellular events, including endocytosis. This result indicates that the polypeptide systems were dominantly accumulated in the cells via endocytosis (Fig. 4e). In contrast, cellular uptake changes were faint in free THPP prepared in DMSO, indicating that THPP is internalized by cells mainly via diffusion.
Next, the photodynamic activity of polypeptide/THPP complexes toward Colon26 cells was investigated. After incubation with 0.125–2.0 μM of porphyrin derivatives of these complexes for 4 h, the cells were exposed to light (610–740 nm), which can travel into the deeper sites of our body (Fig. 4f).
In the absence of light, no cytotoxicity was found even at the highest concentrations used (i.e., [THPP] = 2.0 μM), indicating that our systems are non-toxic photosensitizers in a dark condition (Fig. 4f blue lines), and the PLL system could not induce photo-triggered cytotoxicity even in the presence of light. This might be caused by the 1O2 scavenging effects of PLL. Conversely, the photodynamic activity was dose-dependently induced to Colon26 cells in all other systems. We also compared the photodynamic activity using THPP by half-maximal inhibitory concentration (IC50) values. The IC50 values of PGA/THPP, TMe-β-CDx, and DMSO/THPP under photoirradiation were estimated to be 0.61, 0.71, and 0.58 μM, respectively (Fig. 4f orange, gray, and black lines). These results suggest that our systems using PGA improved the dispersibility and storability of model pharmaceuticals without impairing the functionality of drugs.
Conclusion
In conclusion, we demonstrated the use of HSVM for the preparation of water-dispersible nanocomplexes of hydrophobic compounds using polypeptides, PLL, PGA, and Col. The systems dissolved functional molecules, including C60, TPE, porphyrin derivatives, Pc, and CBZ, in aqueous media, and the concentration of guest molecules was higher than that prepared using conventional water solubilizers like Pul and CDxs. Moreover, the complexes of guest molecules with polypeptides exhibited a better stability for long-term storage and thermal stress than those with Pul and CDxs, indicating that the systems are advantageous in manipulating functional molecules for medical, dietary supplemental, and industrial application. Finally, we demonstrated the applicability of water-dispersible, peptide-based nanosystems comprising porphyrin derivative (THPP), which has been employed as a pharmaceutical drug for photo-triggered cancer therapy, which is PDT. The systems worked as pH-triggered payload release systems due to the nature of polypeptides. Moreover, the PGA system exhibited a photodynamic activity against cancer cells at the same level as free THPP, which was prepared with DMSO injection method. These results indicate that the current system improved the dispersibility and storability of the guest molecules without compromising their functionality. We believe that our water solubilization techniques provide solutions for pharmaceutical, dietary, and semiconductor industry by manipulating functional hydrophobic molecules as sustainable approach.Experimental section
Materials
Poly-L-lysine (PLL was purchased from Peptide Institute, Inc. (Osaka, Japan). PGA, γ-Cyclodextrin (γ-CDx), 2,3,6-tri-O-methyl-β-cyclodextrin (TMe-β-CDx), and pyrene (Py) were purchased from FUJIFILM Wako Pure Chemical Industries Ltd (Tokyo, Japan). Col was purchased from Nacalai Tesque (Kyoto, Japan). Pullulan (Pul), tetraphenylporphyrin (TPP), 5,10,15,20-tetrakis(4-hydroxyphenyl)porphyrin (THPP), 5,10,15,20-tetrakis(4-aminophenyl)porphyrin (TAPP), tetraphenylethylene (TPE), α-quinquethiophene (5T), carbamazepin (CBZ), and phthalocyanine (Pc) were purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan). [C60]fullerene (C60) was purchased from Frontier Carbon (Tokyo, Japan). Coumarin 6 (C6) and 9,10-anthracenediyl-bis(methylene)dimalonic acid (ABDA) were purchased from Sigma-Aldrich (St Louis, MO, USA). Colon26 cells were maintained in Dulbecco's Modified Eagle Medium containing 10% fetal bovine serum and 1% penicillin–streptomycin.Apparatus
UV-Vis absorption was determined using a Shimadzu 3600 UV-Vis-NIR spectrometer (Shimadzu Corporation, Tokyo, Japan). Fluorescence spectra were recorded on a Hitachi F-2700 fluorescence spectrometer (Hitachi Ltd, Tokyo, Japan).Preparation of the complex via HSVM
A solubilizer (PLL, PGA, Col, Pul, γ-CDx, or TMe-β-CDx; 10 mg) and hydrophobic compound (TPP, THPP, TAPP, C60, or Pc; 2.0 μmol or TPE, 5T, CBZ, C6, or Py; 5.0 μmol) were placed in a vial for processing via HSVM (25 Hz, 30 min). For PLL/THPP complexes used in the cell experiments, PLL (10 mg) and THPP (10 μmol) were treated via HSVM (25 Hz, 60 min). The mixtures were suspended in 2.0 mL of Milli-Q and were centrifuged (4500 rpm, 20 min) to collect the supernatant. For PLL/THPP and PGA/THPP complexes in the cell experiments, the sizes of the complexes were controlled by a probe-type sonicator. The solubility of hydrophobic compounds with solubilizers was confirmed by measuring the UV-Vis absorption spectrum of supernatant solutions. The concentrations of hydrophobic compounds in each resulting dispersion were evaluated by redissolving these compounds in an organic solvent and measuring the absorbance of the solutions.Characterization of the complexes
The hydrodynamic diameter (Dhy) of the complexes was measured using a DLS instrument (Zeta-sizer Nano ZS; Malvern, Malvern, UK). The PDI value was calculated by cumulant fitting. The zeta potential of the complexes was measured by Zeta-sizer Nano ZS using capillary cells. Morphological observations were carried out using a transmission electron microscope (JEM-1400, JEOL Ltd Co., Tokyo, Japan). The samples were cast on a hydrophilized, ultrathin, carbon-deposited Cu grid and incubated for 10 s. Afterward, the samples were stained with ammoniummolybdate (3 wt%). The stained samples were observed using a transmission electron microscope (acceleration voltage, 100 keV).Evaluation of the long-term and thermal stability of the complexes
The complex dispersions obtained by HSVM (PLL/TPP, PGA/TPP, Pul/TPP, TMe-β-CDx/TPP, PLL/C60, PGA/C60, Pul/C60, and γ-CDx/C60) were allowed to stand, and UV-Vis absorption spectra were measured after 0, 1, 3, and 7 days. UV-Vis absorption spectra were also measured before and after incubation at 80 °C for 2 h. The long-term stability and thermal stability were evaluated based on λmax absorption changes.Evaluation of pH-triggered cargo release
PLL/THPP and PGA/THPP complex dispersions were incubated in water at pH 5, 7, and 9 prepared with hydrochloric acid or sodium hydroxide solutions ([THPP] = 15 μM). After incubation for 1, 3, 6, and 24 h, the precipitates were centrifuged (3500 rpm, 15 min) and collected. The precipitates were redissolved in DMSO, and the ratio of THPP release was quantified by measuring the UV-Vis absorption spectrum of the solution.Generation of a singlet oxygen
TheA singlet oxygen (1O2) generated by ABDA bleaching (Sigma-Aldrich Corp.) was detected using a previously reported method.15–17 Here, ABDA was used as a DMSO solution ([ABDA] = 2.50 mM). PLL/THPP, PGA/THPP, DMSO/THPP, and mixtures of DMSO/THPP and PLL were used in the experiments, and the final concentrations of THPP and ABDA were 15 μM and 25 μM, respectively. Here, oxygen was bubbled through all sample solutions for 30 min before photoirradiation to generate aerobic conditions. Photoirradiation was achieved using a 500 W SX-UID500X xenon lamp (Ushio Inc., Tokyo, Japan) equipped with a long-pass filter with a cut-off at 620 nm. The light was cooled by passing it through a water filter. The power of the light was 16 mW cm−2 (over 620 nm) at the sample level.Cellular uptake of THPP complexes with polypeptides
Colon26 cells (1.0 × 105 cells) were seeded in 12-well culture plates (1000 μL media per well) and cultured for 24 h at 37 °C and 5% CO2. Then, they were incubated (at 37 °C for 1 h, 4 h, or 24 h and at 4 °C for 4 h) in a medium with PLL/THPP, PGA/THPP, or DMSO/THPP complexes ([THPP] = 4.7 μM). The cells were washed twice with PBS and lysed with 500 μL of RIPA lysis buffer. The precipitates were removed by centrifugation (3500 rpm, 5 min, 25 °C). The THPP in the supernatant solution was extracted using ethyl acetate (800 μL), and the fluorescence spectrum (excitation wavelength, 540 nm) of the extract was measured.Deliverability of the complexes to Colon26 cells
Colon26 cells (1.0 × 105 cells) were seeded in glass-bottom dishes (Iwaki, Tokyo, Japan) (1000 μL of medium) and cultured for 24 h at 37 °C and 5% CO2. Then, they were incubated in a medium with PLL/THPP, PGA/THPP, or DMSO/THPP complexes for 4 h in the dark ([THPP] = 1.0 μM). The cells were washed twice with PBS, and lysosomes were stained using LysoTracker Green DND-26 (Life Technologies, USA). The samples were observed by CLSM (LSM700, Carl Zeiss, Germany).Photodynamic activity experiments
Colon26 cells (8.55 × 104 cells) were seeded in 48-well culture plates (200 μL media per well) and cultured for 24 h at 37 °C and 5% CO2. Then, they were incubated in a medium with PLL/THPP, PGA/THPP, TMe-β-CDx/THPP, or DMSO/THPP complexes for 4 h in the dark. The cells were washed twice with PBS and exposed to light for 30 min at room temperature. Irradiation was achieved using a 300 W MAX-301 xenon lamp (Asahi Spectra Co., Ltd, Tokyo, Japan) equipped with a Vis mirror module (385–740 nm) and a long-pass filter with a cut-off at 610 nm. The power of the light was 9 mW cm−2 (610–740 nm) at the cell level. The viability of the cells was measured as a ratio (%) of the number of viable cells in the treated and untreated groups. WST-8 assay was conducted 24 h after photoirradiation using Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan), following the manufacturer's instructions. Data are expressed as the mean percentage of absorbance of treated cells relative to that of untreated control cells ± SD.Author contributions
S. H., R. K. and A. I. designed this work. S. K., R. K., and S. H. conducted all the experiments and analyzed the data. K. Y. and M. O., and T. E. conducted experiments using cells. S. K., R. K., and A. I. wrote this manuscript. All authors agreed to submit the manuscript in current form.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Japan Society for the Promotion of Science, KAKENHI (R. K., JP19K15401 and JP22K18196) and Research Fellowship for Young Scientist (K. Y., 22J20442). Experiments related to CLSM and TEM were carried out in N-BARD Hiroshima University.Notes and references
- P. Nkansah, A. Antipas, Y. Lu, M. Varma, C. Rotter, B. Rago, A. El-Kattan, G. Taylor, M. Rubio and J. Litchfield, J. Controlled Release, 2013, 169, 150–161 CrossRef CAS PubMed.
- X. Zhang, R. Zhang, J. Huang, M. Luo, X. Chen, Y. Kang and J. Wu, J. Mater. Chem. B, 2019, 7, 3537–3545 RSC.
- X. Shi, S. Bai, C. Yang, X. Ma, M. Hou, J. Chen, P. Xue, C. M. Li, Y. Kang and Z. Xu, J. Mater. Chem. B, 2018, 6, 5549–5561 RSC.
- N. Seedher and S. Bhatia, AAPS PharmSciTech, 2003, 4, 36–44 CrossRef PubMed.
- X. Chen, X. Ling, L. Zhao, F. Xiong, G. Hollett, Y. Kang, A. Barrett and J. Wu, ACS Appl. Mater. Interfaces, 2018, 10, 33976–33985 CrossRef CAS PubMed.
- Y. Liu, Q. Chen, P. Sun, Y. Li, Z. Yang and T. Xu, Mater. Today Energy, 2021, 20, 100634 CrossRef CAS.
- H. Peng, Q. Yu, S. Wang, J. Kim, A. E. Rowan, A. K. Nanjundan, Y. Yamauchi and J. Yu, Adv. Sci., 2019, 6, 1900431 CrossRef PubMed.
- S. Lee, D. Jeong, C. Kim, C. Lee, H. Kang, H. Y. Woo and B. J. Kim, ACS Nano, 2020, 14, 14493–14527 CrossRef PubMed.
- E.-S. Ha, S.-K. Lee, D. H. Choi, S. H. Jeong, S.-J. Hwang and M.-S. Kimet, J. Pharm. Invest., 2020, 50, 231–250 CrossRef CAS.
- I. Ullah, M. K. Baloch, S. Niaz, A. Sultan and I. Ullah, J. Solution Chem., 2019, 48, 1603–1616 CrossRef CAS.
- Y. J. Ng, H. R. Lim, K. S. Khoo, K. W. Chew, D. J. C. Chane, M. Bilalf, H. S. H. Munawarohg and P. L. Show, Environ. Res., 2022, 212, 113126 CrossRef CAS PubMed.
- P. Johnson, A. Trybala, V. Starov and V. J. Pinfield, Adv. Colloid Interface Sci., 2021, 288, 102340 CrossRef CAS PubMed.
- E. A. Ocherednyuk, R. I. Garipova, I. M. Bogdanov, B. Kh. Gafiatullin, E. D. Sultanova, D. A. Mironova, A. G. Daminova, V. G. Evtugyn, V. A. Burilov, S. E. Solovieva and I. S. Antipin, Colloids Surf., A, 2022, 648, 129236 CrossRef CAS.
- D. H. Schwarz, A. Engelke and G. Wenz, Int. J. Pharm., 2017, 531, 559–567 CrossRef CAS PubMed.
- T. Yumoto, S. Satake, S. Hino, K. Sugikawa, R. Kawasaki and A. Ikeda, Org. Biomol. Chem., 2020, 18, 6702–6709 RSC.
- K. Yamana, R. Kawasaki, K. Sugikawa and A. Ikeda, ACS Appl. Bio Mater., 2020, 3, 3217–3225 CrossRef CAS PubMed.
- Y. Goto, S. Hino, K. Sugikawa, R. Kawasaki and A. Ikeda, Asian J. Org. Chem., 2020, 9, 1589–1596 CrossRef CAS.
- A. Ikeda, K. Hayashi, T. Konishi and J. Kikuchi, Chem. Commun., 2004, 1334–1335 RSC.
- R. Kawasaki, K. Yamana, R. Shimada, K. Sugikawa and A. Ikeda, ACS Omega, 2021, 6, 3209–3217 CrossRef CAS PubMed.
- S. Hino, R. Funada, K. Sugikawa, K. Koumoto, T. Suzuki, T. Nagasakic and A. Ikeda, Photochem. Photobiol. Sci., 2019, 18, 2854–2858 CrossRef CAS PubMed.
- A. Ikeda, T. Iizuka, N. Maekubo, K. Nobusawa, K. Sugikawa, K. Koumoto, T. Suzuki, T. Nagasaki and M. Akiyama, Chem.–Asian J., 2017, 12, 1069–1074 CrossRef CAS.
- R. Omokawa, R. Kawasaki, K. Sugikawa, T. Nishimura, T. Nakaya and A. Ikeda, ACS Appl. Polym. Mater., 2021, 3, 3708–3713 CrossRef CAS.
- C. Kojima, T. Suehiro, K. Watanabe, M. Ogawa, A. Fukuhara, E. Nishisaka, A. Harada, K. Kono, T. Inui and Y. Magata, Acta Biomater., 2013, 9, 5673–5680 CrossRef CAS.
- K. Lin, D. Zhang, M. H. Macedo, W. Cui, B. Sarmento and G. Shen, Adv. Funct. Mater., 2019, 29, 1804943 CrossRef.
- K. Matsumoto, A. Kawamura and T. Miyata, Chem. Lett., 2015, 44, 1284–1286 CrossRef CAS.
- L.-L. Wang, J.-T. Chen, L.-F. Wang, S. Wu, G.-z. Zhang, H.-Q. Yu, X.-d. Ye and Q.-S. Shi, Sci. Rep., 2017, 7, 12787 CrossRef PubMed.
- Q. Xu, C. He, C. Xiao and X. Chen, Macromol. Biosci., 2016, 16, 635–646 CrossRef CAS.
- S. Roberts, T. S. Harmon, J. L. Schaal, V. Miao, K. Li, A. Hunt, Y. Wen, T. G. Oas, J. H. Collier, R. V. Pappu and A. Chilkoti, Nat. Mat., 2018, 17, 1154–1163 CrossRef CAS PubMed.
- Y. Shen, X. Fu, W. Fu and Z. Li, Chem. Soc. Rev., 2015, 44, 612–622 RSC.
- R. A. Quirk, W. C. Chan, M. C. Davies, S. J. B. Tendler and K. M. Shakesheff, Biomaterials, 2001, 22, 865–872 CrossRef CAS PubMed.
- A. Béduer, C. Vieu, F. Arnauduc, J.-C. Sol, I. Loubinoux and L. Vaysse, Biomaterials, 2012, 33, 504–514 CrossRef PubMed.
- Y. F. Makableh, R. Vasan, S. Lee and O. M. Manasreh, Appl. Phys. Lett., 2013, 102, 051904 CrossRef.
- A. R. Bhat, V. U. Irorere, T. Bartlett, D. Hill, G. Kedia, M. R. Morris, D. Charalampopoulos and I. Radecka, AMB Express, 2013, 3, 36 CrossRef PubMed.
- L. Wang, D. Hu, X. Kong, J. Liu, X. Li, K. Zhou, H. Zhao and C. Zhou, Chem. Eng. J., 2018, 346, 38–49 CrossRef CAS.
- R. Kawasaki, S. Kawamura, S. Hino, K. Yamana and A. Ikeda, Mater. Adv., 2022, 3, 467–473 RSC.
- T. Nishimura, Y. Hatatani, M. Ando, Y. Sasaki and K. Akiyoshi, Chem. Sci., 2022, 13, 5243–5251 RSC.
- L. Shi, F. Carn, F. Boué and E. Buhler, Phys. Rev. E, 2016, 94, 032504 CrossRef PubMed.
- Z. H. Loh, A. K. Samanta and P. W. S. Heng, Asian J. Pharm., 2015, 10, 255–274 Search PubMed.
- V. Sanna, A. M. Roggio, N. Pala, S. Marceddu, G. Lubinu, A. Mariani and M. Sechi, Int. J. Biol. Macromol., 2015, 72, 531–536 CrossRef CAS PubMed.
- P. P. d. Santos, S. H. Flôres, A. d. O. Rios and R. C. Chisté, Trends Food Sci. Technol., 2016, 53, 23–33 CrossRef.
- T. Feczkó, O. Varga, M. Kovács, T. Vidóczy and B. Voncina, J. Photochem. Photobiol., A, 2011, 222, 293–298 CrossRef.
- X. Li, S. Kolemen, J. Yoon and E. U. Akkaya, Adv. Funct. Mater., 2017, 27, 1604053 CrossRef.
- Y. Herdiana, N. Wathoni, S. Shamsuddin, I. M. Joni and M. Muchtaridi, Polymers, 2021, 13, 1717 CrossRef CAS PubMed.
- M. Shang, X. Sun, L. Guo, D. Shi, P. Liang, D. Meng, X. Zhou, X. Liu, Y. Zhao and J. Li, Int. J. Nanomed., 2020, 15, 537–552 CrossRef CAS PubMed.
- J. Sobczyński, H. H. Tønnesen and S. Kristensen, Pharmazie, 2013, 68, 100–109 Search PubMed.
- T. Entradas, S. Waldron and M. Volk, J. Photochem. Photobiol., B, 2020, 204, 11787 CrossRef PubMed.
- X. Wang, Q. Tu, B. Zhao, Y. An, J.-C. Wang, W. Liu, M.-S. Yuan, S. M. Ahmed, J. Xu, R. Liu, Y. Zhang and J. Wang, Biomaterials, 2013, 34, 1155–1169 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of the experimental procedure and analytical data. See DOI: https://doi.org/10.1039/d2ra06054j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |