Open Access Article
Open Access ArticleEnhanced and specific epoxidation activity of P450 BM3 mutants for the production of high value terpene derivatives
Danilo Correddu a,
Sabrina Helmy Alya,
Giovanna Di Nardoa,
Gianluca Catuccia,
Cristina Prandi
a,
Sabrina Helmy Alya,
Giovanna Di Nardoa,
Gianluca Catuccia,
Cristina Prandi b,
Marco Blangetti
b,
Marco Blangetti b,
Chiara Bellomo
b,
Chiara Bellomo b,
Elisabetta Bonometti
b,
Elisabetta Bonometti b,
Guido Viscardi
b,
Guido Viscardi b and
Gianfranco Gilardi
b and
Gianfranco Gilardi *a
*a
aDepartment of Life Sciences and Systems Biology, University of Torino, Via Accademia Albertina 13, 10123, Torino, Italy. E-mail: gianfranco.gilardi@unito.it
bDepartment of Chemistry, University of Torino, Via P. Giuria 7, 10125, Torino, Italy
First published on 28th November 2022
Abstract
Terpenes are natural molecules of valuable interest for different industrial applications. Cytochromes P450 enzymes can functionalize terpenoids to form high value oxidized derivatives in a green and sustainable manner, representing a valid alternative to chemical catalysis. In this work, an enhanced and specific epoxidation activity of cytochrome P450 BM3 mutants was found for the terpenes geraniol and linalool. This is the first report showing the epoxidation of linalool by P450 BM3 and its mutant A2 (Asp251Gly/Gln307His) with the formation of valuable oxide derivatives, highlighting the relevance of this enzymes for industrial applications.
Introduction
Terpenes are a large group of natural molecules mostly present in plants and fungi. These compounds and their derivatives find applications in many industrial fields and are particularly useful for human healthcare.1,2 The functionalization of terpenes using microorganisms or enzymes represents a sustainable and alternative approach compared to the chemical routes.3 Cytochromes P450 are a family of enzymes that are employed in biotechnology thanks to their ability to perform challenging reactions for the modification and valorization of a large variety of molecules.4–6 Indeed, P450s from plants, fungi and bacteria are known to take part on the pathways of synthesis or modification of terpenes, and therefore are considered of extreme importance for applications on biotransformation processes. Among bacterial P450s, the cytochrome P450 BM3 from Bacillus megaterium and laboratory-evolved mutants have been shown to functionalize terpenoids such as ionone,7–10 limonene,11–13 farnesol14–16 and geraniol,16,17 in in vitro and in vivo catalysis.Previously, a library of P450 BM3 mutants was obtained by a directed evolution approach in our laboratories.18,19 Different mutants with an increased activity towards the native substrate lauric acid were identified via a high-throughput screening method allowing to select, produce and characterize BM3 mutants with an augmented activity and substrate scope.20 The aim of this work was to investigate the activity of P450 BM3 wild type and mutants towards the terpenes geraniol and linalool.
Results and discussion
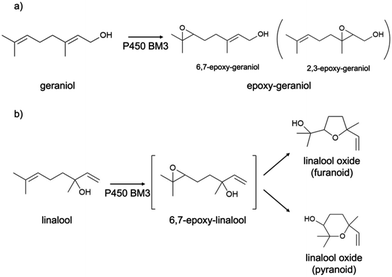
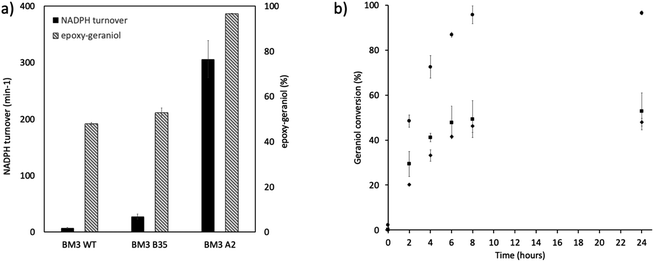
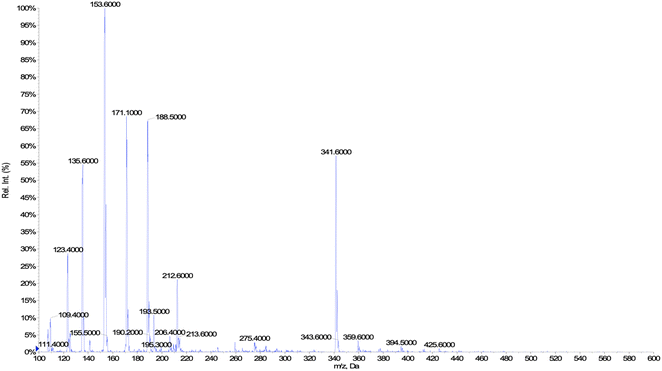
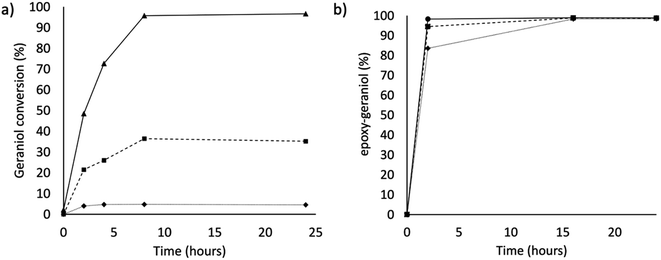
In this work, the P450 BM3 mutants B35 (His138Tyr) and A2 (Asp251Gly/Gln307His) were initially used to evaluate the functionalization of the terpene geraniol, that is known to be preferably oxidised to 6,7-epoxy-geraniol by P450 BM3 (Scheme 1a).16,17 Firstly, the oxidation rate of NADPH for BM3 WT and the mutants B35 and A2 in the presence of geraniol was measured spectrophotometrically following the reduction of absorbance at 340 nm. The NADPH oxidation rate of BM3 WT was 7.1 ± 1.2 min−1 while the mutants showed dramatic increase with the B35 and A2 having respectively a turnover of 27.5 ± 4.7 min−1 and 305.8 ± 32.7 min−1 (Fig. 1a).These values fall in the range of reported catalytic activity towards these compounds for previously characterized BM3 mutants.16,17 However, since it is known that the level of uncoupling in the mutant A2 is higher compared to WT,21 the activity of the enzymes was further investigated by analysing the reaction products by HPLC. The reactions catalysed by P450 mutants B35 and A2 yield a product with the same retention time as BM3 WT suggesting that the mutants B35 and A2 maintained an unaltered chemoselectivity. This is consistent with the location of the mutated residues that are not in the active site of the enzyme. As it was previously shown, the augmented catalytic activity of the BM3 A2 mutant is a consequence of structural rearrangements of the enzyme, which gains flexibility and also causes the rotation of a key residue (Phe87) in the active site.21 The reaction product was further investigated by HPLC-MS. The analysis revealed the specific production of an epoxide derivative of geraniol, which has a molecular mass of 170.25. The ESI spectrum of Fig. 2 shows a pattern of adduct ions that can be related to ionised forms of epoxy-geraniol, with the corresponding molecular masses. Previously, other P450 BM3 mutants have been shown to preferably oxidize the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond in position 6, 7. However the chemoselectivity was not always maintained. In fact, a second epoxide and two hydroxylated products of geraniol were detected.16,17 In our experiments, the higher activity of A2 towards geraniol resulted in a dramatic increase of product over time (Fig. 1a). Substrate conversion was calculated at different time points in a reaction mixture containing 1 μM enzyme, 1 mM geraniol and 10 mM NADPH at 30 °C (Fig. 1b). After 24 hours, BM3 WT converted the 48% of geraniol and similar percentage of conversion were obtained with the mutant B35 (52.8%), indicating that the mutation His138Tyr increases the uncoupling level rather than product formation in this mutant. However, the A2 mutant showed a nearly total substrate conversion (96.6%) already after 8 hours, which is higher compared to the ones of previously reported BM3 mutants reaching up to the 80%.17 The activity of the A2 enzyme towards geraniol was then evaluated increasing the concentrations of the substrate from 1 mM to 3 and 6 mM, keeping the enzyme concentration fixed at 1 μM. The percentage of conversion decreased when the concentration of substrate increased indicating a possible substrate/product inhibition (Fig. 3a). In order to avoid the expensive use of the NADPH cofactor, the catalytic activity of BM3 WT and mutants towards geraniol was then evaluated in whole-cell biotransformation experiments using a cofactor regeneration system (Fig. 3b).22,23 For this experiment, the starting concentration of geraniol used was 1 mM, which was the only one that did not lead to substrate inhibition. In this experiment, BM3 WT and B35 could totally convert geraniol to epoxy-geraniol only after 16 hours reaction, while the reaction of BM3 A2 was complete after 2 hours. Thus, due to the high expression levels of A2 in Escherichia coli (20 mg g−1 of wet cell paste)18 the use of whole-cells significantly decreases the time for an almost complete substrate conversion. BM3 A2 resulted being the best enzyme in terms of activity and can maintain the chemoselectivity, thus showing great potential for exploitation in a biotechnological process. Given that the mutant BM3 A2 was the best enzyme for the conversion of terpenes, the attention focused on expanding the substrate scope to a terpene of a shorter carbon chain, linalool, for which the activity of BM3 or mutants has never been shown. Linalool is present in high quantity in essential oils of plants such as basil and lavender.24 As a fragrance, it is used in nearly all the cosmetic products such as perfumes, shampoos and soaps.25,26 Furthermore, it finds medical applications for its beneficial effects such as anti-carcinogenic.27 Previously, the cytochrome P450lin (CYP111) from Pseudomonas incognita showed the ability to convert linalool into 8-hydroxy-linalool as the first step of its degradation pathway.28 Nevertheless, the most interesting transformation of linalool is the one that leads to the production of highly value forms of linalool oxides. These compounds possess potent pharmaceutical effects as anxiolytic,29 antinociceptive and anticonvulsant.30 Linalool oxides are known to be formed via a cyclization reaction starting from the molecule 6,7-epoxy-linalool, which is formed enzymatically.31,32 Linalool epoxidation takes place in plants and fungi via an enzymatic reaction that most probably involves non-identified P450s. For this reason, several works reported biotransformation processes involving whole microorganisms resulting in high conversion yields.33,34 However, the enzymes that are involved in the biosynthesis pathway have never been identified.
C bond in position 6, 7. However the chemoselectivity was not always maintained. In fact, a second epoxide and two hydroxylated products of geraniol were detected.16,17 In our experiments, the higher activity of A2 towards geraniol resulted in a dramatic increase of product over time (Fig. 1a). Substrate conversion was calculated at different time points in a reaction mixture containing 1 μM enzyme, 1 mM geraniol and 10 mM NADPH at 30 °C (Fig. 1b). After 24 hours, BM3 WT converted the 48% of geraniol and similar percentage of conversion were obtained with the mutant B35 (52.8%), indicating that the mutation His138Tyr increases the uncoupling level rather than product formation in this mutant. However, the A2 mutant showed a nearly total substrate conversion (96.6%) already after 8 hours, which is higher compared to the ones of previously reported BM3 mutants reaching up to the 80%.17 The activity of the A2 enzyme towards geraniol was then evaluated increasing the concentrations of the substrate from 1 mM to 3 and 6 mM, keeping the enzyme concentration fixed at 1 μM. The percentage of conversion decreased when the concentration of substrate increased indicating a possible substrate/product inhibition (Fig. 3a). In order to avoid the expensive use of the NADPH cofactor, the catalytic activity of BM3 WT and mutants towards geraniol was then evaluated in whole-cell biotransformation experiments using a cofactor regeneration system (Fig. 3b).22,23 For this experiment, the starting concentration of geraniol used was 1 mM, which was the only one that did not lead to substrate inhibition. In this experiment, BM3 WT and B35 could totally convert geraniol to epoxy-geraniol only after 16 hours reaction, while the reaction of BM3 A2 was complete after 2 hours. Thus, due to the high expression levels of A2 in Escherichia coli (20 mg g−1 of wet cell paste)18 the use of whole-cells significantly decreases the time for an almost complete substrate conversion. BM3 A2 resulted being the best enzyme in terms of activity and can maintain the chemoselectivity, thus showing great potential for exploitation in a biotechnological process. Given that the mutant BM3 A2 was the best enzyme for the conversion of terpenes, the attention focused on expanding the substrate scope to a terpene of a shorter carbon chain, linalool, for which the activity of BM3 or mutants has never been shown. Linalool is present in high quantity in essential oils of plants such as basil and lavender.24 As a fragrance, it is used in nearly all the cosmetic products such as perfumes, shampoos and soaps.25,26 Furthermore, it finds medical applications for its beneficial effects such as anti-carcinogenic.27 Previously, the cytochrome P450lin (CYP111) from Pseudomonas incognita showed the ability to convert linalool into 8-hydroxy-linalool as the first step of its degradation pathway.28 Nevertheless, the most interesting transformation of linalool is the one that leads to the production of highly value forms of linalool oxides. These compounds possess potent pharmaceutical effects as anxiolytic,29 antinociceptive and anticonvulsant.30 Linalool oxides are known to be formed via a cyclization reaction starting from the molecule 6,7-epoxy-linalool, which is formed enzymatically.31,32 Linalool epoxidation takes place in plants and fungi via an enzymatic reaction that most probably involves non-identified P450s. For this reason, several works reported biotransformation processes involving whole microorganisms resulting in high conversion yields.33,34 However, the enzymes that are involved in the biosynthesis pathway have never been identified.
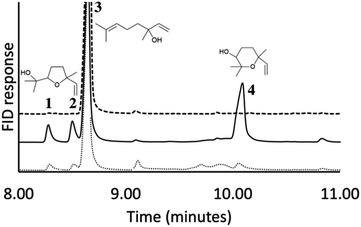
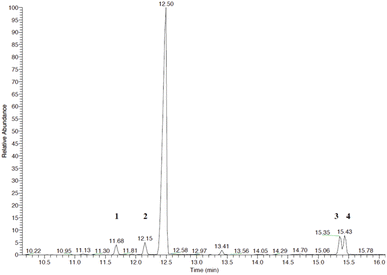
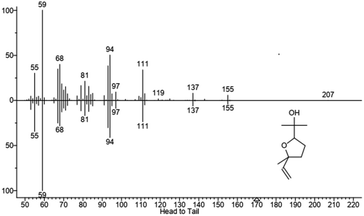
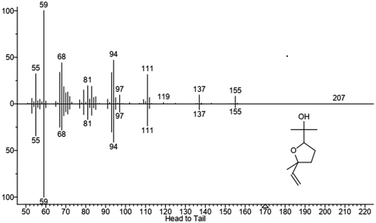
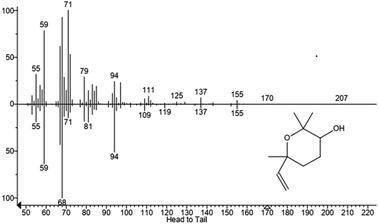
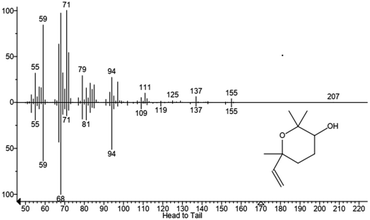
We hypothesized that the production of epoxy-linalool may be catalysed by enzymes that can oxidize other terpenes like geraniol. For this reason, the activity of P450 BM3 WT and the mutant A2 was investigated in in vitro reactions using purified enzymes and were analysed by gas chromatography (Fig. 4). GC-MS analysis revealed the formation of four different linalool derivatives, which were identified as linalool oxides in the form of cis- and trans-pyranoid and furanoid (Fig. 5–9), according to a previous characterization of the same compounds.35
These are formed spontaneously from 6,7-epoxy-linalool, which represents the only P450 enzymatic product (Scheme 1b). However, 6,7-epoxy-linalool cannot be detected probably due to the fast cyclization reaction. Nevertheless, the identification of diastereomers of linalool oxides was possible, as reported in previous characterization.35
In order to exclude the formation of epoxy-linalool by auto-oxidation, control samples were prepared without enzymes and that did not show any formation of product(s) (Fig. 4). As for other non-native substrates, also for linalool the percentage of conversion by BM3 WT was very low (3%). This increased when the reaction was carried out by the mutant BM3 A2, reaching up to 40% substrate conversion. Furthermore, also in this case the specificity of the reaction was confirmed by the absence of other hydroxylated linalool products from the GC-MS analysis.
Conclusions
Overall, this work highlights the high potential of P450 BM3 WT and laboratory evolved BM3 mutants to produce high value derivatives of terpenes. Indeed, the enhanced catalytic activity of the BM3 mutant A2 (Asp251Gly/Gln307His), can be exploited in whole cell biotransformation processes to maximise the yield of production without losing reaction selectivity. Moreover, for the first time the specific epoxidation of linalool, leading to the formation of linalool oxides is reported for the bacterial P450 BM3. Such an activity is largely increased in A2, a BM3 mutant that not only has an increased activity for terpenes compared to WT, but also shows high chemoselectivity. This feature is crucial for the selective production of valuable compounds using a biocatalytic approach.Experimental
Materials
All chemicals were purchased from Sigma-Aldrich, Thermo Fisher Scientific and Bio-Rad unless otherwise stated.Expression and purification
Escherichia coli supercompetent cells DH5α and BL21(DE3) were purchased from Thermo Fisher Scientific. Cells were routinely grown in Luria–Bertani (LB) medium supplemented with antibiotic at 37 °C. Wild type P450 BM3 and mutants were expressed in E. coli BL21(DE3) cells transformed with the IPTG-inducible pT7Bm3Z vector and purified as previously described.18,36,37 Proteins were stored in 50 mM 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid HEPES pH 7.4 at −20 °C.NADPH consumption assays
The rate of NADPH oxidation was measured in a reaction mix containing incubating the enzymes (1 μM), substrate up to saturating concentrations in 30 mM phosphate buffer pH 7.4 at 20 °C. After pre-incubation, 150 μM NADPH was added to the mix and the oxidation was followed spectrophotometrically at 340 nm (ε340 = 6.22 mM−1 cm−1). Fittings of the data were performed by using Sigma Plot.Product conversion
Reactions mixtures containing P450 enzyme (BM3 WT, B35 or A2) (1 μM), NADPH (10 mM) and the substrate geraniol (1 mM) were incubated up to 24 hours at 30 °C. Reactions were inactivated by heat at 90 °C for 5 minutes. Control reactions were carried out in absence of NADPH. HPLC analysis was performed using an Agilent Series 1200 equipped with a LiChrosphere® 100 RP-18 column applying a gradient of 0.1% formic acid/acetonitrile (from 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 to 30
20 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70).
70).
Reaction mixtures containing P450 enzyme (BM3 WT or A2) (10 μM), NADPH (10 mM) and linalool (1–10 mM) were incubated for 5 hours at 30 °C. Reactions were stopped by adding the same volume of MTBE and centrifuged for 3 minutes at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. The organic phase was dried with MgSO4, and the products were dissolved in 20 μL of MTBE and used for analysis. GC-FID measurements were performed using an Agilent 7890A Gas Chromatograph equipped with an Agilent HP-5 capillary column (30 m, 0.320 mm, 0.25 μm). Helium as a carrier gas was used with a flow of 1 mL min−1. Oven temperature was kept at 60 °C for 1 minute, then 10 °C min−1 to 280 °C and held for 2 minutes. FID detector temperature was set to 300 °C.
000 rpm. The organic phase was dried with MgSO4, and the products were dissolved in 20 μL of MTBE and used for analysis. GC-FID measurements were performed using an Agilent 7890A Gas Chromatograph equipped with an Agilent HP-5 capillary column (30 m, 0.320 mm, 0.25 μm). Helium as a carrier gas was used with a flow of 1 mL min−1. Oven temperature was kept at 60 °C for 1 minute, then 10 °C min−1 to 280 °C and held for 2 minutes. FID detector temperature was set to 300 °C.
Products identification
Geraniol product determination was carried using a QTRAP3200 by Sciex interfaced via ESI TurbolonSpray source to a Shimadzu Nexera HPLC. The elution was carried out with a mixture of formic acid 0.05% and acetonitrile applying a gradient (from 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 to 20
20 to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80). The separation was performed with a Kinetex C18 (Phenomenex) 2.6 μm, 100 × 4.6 mm, 100 Å column at 40 °C, with a flow rate of 0.6 mL min−1 and an injection volume of 10 μL. The MS parameters (positive mode) were optimized by direct infusion on the mass spectrometer to maximize the signal intensities: ion spray +3500 V, 300 °C, declustering potential +20 V, entrance potential +10 V. Gas: curtain 20, GS1 40, GS2 30. Scan: 100–1000 m/z. UV-Vis detection carried out with a DAD in the range of 190–800 nm.
80). The separation was performed with a Kinetex C18 (Phenomenex) 2.6 μm, 100 × 4.6 mm, 100 Å column at 40 °C, with a flow rate of 0.6 mL min−1 and an injection volume of 10 μL. The MS parameters (positive mode) were optimized by direct infusion on the mass spectrometer to maximize the signal intensities: ion spray +3500 V, 300 °C, declustering potential +20 V, entrance potential +10 V. Gas: curtain 20, GS1 40, GS2 30. Scan: 100–1000 m/z. UV-Vis detection carried out with a DAD in the range of 190–800 nm.
Linalool products determination was carried out using a Thermo Finnigan Trace GC-MS Plus spectrometer equipped with an Agilent HP-5 ms capillary column (30 m, 0.320 mm, 0.25 μm) using ramp of 30 °C min−1 from 40 to 250 °C in order to separate products 3 and 4 (see Fig. 5).
Whole cell biotransformation
For P450 BM3-mediated whole-cell catalysis, cell pellet was resuspended in 5 mL of 30 mM phosphate buffer (pH 7.4) to a final OD600 nm of 10. The reaction was conducted by mixing geraniol (1 mM) and NADP+ (0.3 mM), trisodium citrate (50 mM), MgCl2, (10 mM) to the cell suspension to sustain the catalysis. Control reaction was performed with pellet obtained from non-induced cultures, where the operon was repressed by adding 2% of glucose. Reaction mixtures were incubated at 30 °C with 200 rpm shaking. Samples of 200 μL were collected and mixed with 0.25% v/v acetonitrile and centrifuged for 5 minutes at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 rpm. Supernatant was used for HPLC analysis.
300 rpm. Supernatant was used for HPLC analysis.
Author contributions
Danilo Correddu: conceptualization, investigation, data curation, formal analysis, visualization, writing – original draft, review and editing. Sabrina Helmy Aly: investigation, writing – review. Giovanna Di Nardo: conceptualization, resources, supervision, writing – review and editing. Gianluca Catucci: investigation, writing – review. Cristina Prandi: conceptualization, resources, writing – review and editing. Marco Blangetti: investigation, writing – review. Chiara Bellomo: investigation, writing – review. Elisabetta Bonometti: investigation. Guido Viscardi: conceptualization, resources, writing – review and editing. Gianfranco Gilardi: conceptualization, resources, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a CRT grant 2017 IDSIME: 2017-1949.References
- R. Paduch, M. Kandefer-Szerszeń, M. Trytek and J. Fiedurek, Arch. Immunol. Ther. Exp., 2007, 55, 315 CrossRef CAS
.
- V. Dell'Oste, F. Spyrakis and C. Prandi, Molecules, 2021, 26, 4579 CrossRef
.
- S. Janocha, D. Schmitz and R. Bernhardt, in Biotechnology of Isoprenoids, ed. J. Schrader and J. Bohlmann, Springer International Publishing, Cham, 2015, pp. 215–250 Search PubMed
.
- V. B. Urlacher and M. Girhard, Trends Biotechnol., 2019, 37, 882–897 CrossRef CAS PubMed
.
- D. Correddu, G. Di Nardo and G. Gilardi, Trends Biotechnol., 2021, 39, 1184–1207 CrossRef CAS
.
- G. Di Nardo and G. Gilardi, Trends Biochem. Sci., 2020, 45, 511–525 CrossRef CAS
.
- S. Eiben, L. Kaysser, S. Maurer, K. Kühnel, V. B. Urlacher and R. D. Schmid, J. Biotechnol., 2006, 124, 662–669 CrossRef CAS PubMed
.
- S. C. Maurer, H. Schulze, R. D. Schmid and V. Urlacher, Adv. Synth. Catal., 2003, 345, 802–810 CrossRef CAS
.
- V. B. Urlacher, A. Makhsumkhanov and R. D. Schmid, Appl. Microbiol. Biotechnol., 2006, 70, 53–59 CrossRef CAS
.
- D. Appel, S. Lutz-Wahl, P. Fischer, U. Schwaneberg and R. D. Schmid, J. Biotechnol., 2001, 88, 167–171 CrossRef CAS PubMed
.
- R. Fasan, Y. T. Meharenna, C. D. Snow, T. L. Poulos and F. H. Arnold, J. Mol. Biol., 2008, 383, 1069–1080 CrossRef CAS PubMed
.
- A. Seifert, M. Antonovici, B. Hauer and J. Pleiss, ChemBioChem, 2011, 12, 1346–1351 CrossRef CAS
.
- A. Seifert, S. Vomund, K. Grohmann, S. Kriening, V. B. Urlacher, S. Laschat and J. Pleiss, ChemBioChem, 2009, 10, 853–861 CrossRef CAS
.
- C.-K. J. Chen, T. Kh. Shokhireva, R. E. Berry, H. Zhang and F. A. Walker, J. Biol. Inorg Chem., 2008, 13, 813–824 CrossRef CAS
.
- M. B. Murataliev, L. N. Trinh, L. V. Moser, R. B. Bates, R. Feyereisen and F. A. Walker, Biochemistry, 2004, 43, 1771–1780 CrossRef CAS
.
- C.-K. J. Chen, R. E. Berry, T. Kh. Shokhireva, M. B. Murataliev, H. Zhang and F. A. Walker, J. Biol. Inorg Chem., 2010, 15, 159–174 CrossRef CAS PubMed
.
- Y. Watanabe, S. Laschat, M. Budde, O. Affolter, Y. Shimada and V. B. Urlacher, Tetrahedron, 2007, 63, 9413–9422 CrossRef CAS
.
- G. E. Tsotsou, A. Sideri, A. Goyal, G. Di Nardo and G. Gilardi, Chem.–Eur. J., 2012, 18, 3582–3588 CrossRef CAS PubMed
.
- G. Di Nardo and G. Gilardi, Int. J. Mol. Sci., 2012, 13, 15901–15924 CrossRef CAS PubMed
.
- G. E. Tsotsou, A. E. G. Cass and G. Gilardi, Biosens. Bioelectron., 2002, 17, 119–131 CrossRef CAS PubMed
.
- G. Di Nardo, V. Dell'Angelo, G. Catucci, S. J. Sadeghi and G. Gilardi, Arch. Biochem. Biophys., 2016, 602, 106–115 CrossRef CAS PubMed
.
- G. Catucci, G. Gilardi and S. J. Sadeghi, Microb. Cell Fact., 2020, 19, 74 CrossRef CAS PubMed
.
- S.
P. Hanlon, A. Camattari, S. Abad, A. Glieder, M. Kittelmann, S. Lütz, B. Wirz and M. Winkler, Chem. Commun., 2012, 48, 6001 RSC
.
- J. S. Dambolena, M. P. Zunino, A. G. López, H. R. Rubinstein, J. A. Zygadlo, J. W. Mwangi, G. N. Thoithi, I. O. Kibwage, J. M. Mwalukumbi and S. T. Kariuki, Innovative Food Sci. Emerging Technol., 2010, 11, 410–414 CrossRef CAS
.
- A. C. Aprotosoaie, M. Hăncianu, I.-I. Costache and A. Miron, Flavour Fragrance J., 2014, 29, 193–219 CrossRef CAS
.
- C. S. Letizia, J. Cocchiara, J. Lalko and A. M. Api, Food Chem. Toxicol., 2003, 41, 943–964 CrossRef CAS PubMed
.
- M.-Y. Chang, D.-E. Shieh, C.-C. Chen, C.-S. Yeh and H.-P. Dong, Int. J. Mol. Sci., 2015, 16, 28169–28179 CrossRef CAS
.
- J. D. Ropp, I. C. Gunsalus and S. G. Sligar, J. Bacteriol., 1993, 175, 6028–6037 CrossRef CAS PubMed
.
- F. N. Souto-Maior, F. L. de Carvalho, L. C. S. L. de Morais, S. M. Netto, D. P. de Sousa and R. N. de Almeida, Pharmacol., Biochem. Behav., 2011, 100, 259–263 CrossRef CAS
.
- F. N. Souto-Maior, D. V. da Fonsêca, P. R. R. Salgado, L. de O. Monte, D. P. de Sousa and R. N. de Almeida, Pharm. Biol., 2017, 55, 63–67 CrossRef CAS
.
- F. Luan, A. Mosandl, M. Gubesch and M. Wüst, J. Chromatogr. A, 2006, 1112, 369–374 CrossRef CAS PubMed
.
- R. J. W. Meesters, M. Duisken and J. Hollender, Xenobiotica, 2007, 37, 604–617 CrossRef CAS
.
- A. Sales, L. F. Afonso, J. A. Americo, M. de Freitas Rebelo, G. M. Pastore and J. L. Bicas, Biotechnol. Lett., 2018, 40, 561–567 CrossRef CAS PubMed
.
- M.-A. Mirata, M. Wüst, A. Mosandl and J. Schrader, J. Agric. Food Chem., 2008, 56, 3287–3296 CrossRef CAS PubMed
.
- D. Wang, K. Ando, K. Morita, K. Kubota and A. Kobayashi, Biosci., Biotechnol., Biochem., 1994, 58, 2050–2053 CrossRef CAS
.
- A. Sideri, A. Goyal, G. Di Nardo, G. E. Tsotsou and G. Gilardi, J. Inorg. Biochem., 2013, 120, 1–7 CrossRef CAS
.
- H. Y. Li, K. Darwish and T. L. Poulos, J. Biol. Chem., 1991, 266, 11909–11914 CrossRef CAS
.
| This journal is © The Royal Society of Chemistry 2022 |