Open Access Article
Open Access ArticleEnhancing the compatibility of BioCaRGOS silica sol-gel technology with ctDNA extraction and droplet digital PCR (ddPCR) analysis†
Chinmay S. Potnis a,
Rajat Chauhanb,
Theodore S. Kalbfleischb,
Evan Alexanderc,
Lindsay Eichholdb,
Meenakshi Bansald,
Craig A. Grapperhausa,
Robert S. Keyntone,
Mark W. Linder*c and
Gautam Gupta
a,
Rajat Chauhanb,
Theodore S. Kalbfleischb,
Evan Alexanderc,
Lindsay Eichholdb,
Meenakshi Bansald,
Craig A. Grapperhausa,
Robert S. Keyntone,
Mark W. Linder*c and
Gautam Gupta *b
*b
aDepartment of Chemistry, University of Louisville, Louisville, 40292, Kentucky, USA
bDepartment of Chemical Engineering, University of Louisville, Louisville, 40292, Kentucky, USA. E-mail: gautam.gupta@louisville.edu
cDepartment of Pathology and Laboratory Medicine, University of Louisville, Louisville, 40292, Kentucky, USA. E-mail: mark.linder@louisville.edu
dDepartment of Chemistry, Thomas More University, Crestview Hills, KY 41017, USA
eWilliam States Lee College of Engineering, University of North Carolina at Charlotte, 28223, USA
First published on 13th October 2022
Abstract
Previously, our group had demonstrated long term stabilization of protein biomarkers using BioCaRGOS, a silica sol-gel technology. Herein, we describe workflow modifications to allow for extraction of cell free DNA (cfDNA) from primary samples containing working concentrations of BioCaRGOS, as well as the compatibility of BioCaRGOS with droplet digital PCR (ddPCR) analysis for pancreatic cancer biomarkers i.e., KRAS circulating tumor DNA (ctDNA). Preliminary attempts to extract ctDNA from BioCaRGOS containing samples demonstrated interference in the extraction of primary samples and the interference with ddPCR analysis when BioCaRGOS was directly introduced to stabilize sample extracts. In our modified technique, we have minimized the interference caused by methanol with ddPCR by complete removal of methanol from the activated BioCaRGOS formulation prior to addition to the biospecimen or ctDNA extract. Interference of the silica matrix present in BioCaRGOS with ctDNA extraction was eliminated through the introduction of invert filtration of the sample prior to extraction. These modifications to the workflow of BioCaRGOS containing samples allow for use of BioCaRGOS for stabilization of trace quantities of nucleic acid biomarkers such as plasma ctDNA, while retaining the capability to extract the biomarker and quantify based on ddPCR.
Introduction
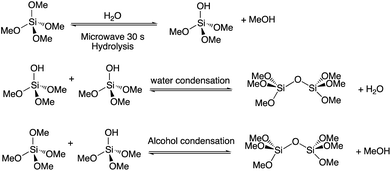
Entrapment of biomolecules such as nucleic acids, proteins and other sensitive enzymes into inorganic silica matrices has gained significant attention over the last two decades.1–4 The basic concept of the sol-gel process involves hydrolysis and polymerization of inorganic metal alkoxide monomers such as tetramethyl orthosilicate (TMOS) or tetraethyl orthosilicate (TEOS).5 Hydrolysis of these alkoxides in the presence of water leads to the formation of orthosilicic acid (SiOH4) which then spontaneously forms a three-dimensional –Si–O–Si– polymer network.5,6 The BioCaRGOS (“Capture and Release Gels for Optimized Storage of biospecimens”), technology previously described by our group involves hydrolyzing the silica precursor TMOS, in a standard benchtop microwave in just 30 seconds as demonstrated in Scheme 1. We have previously reported, long term stabilization of nucleic acids (DNA, miRNA-21) and proteins such as hemoglobin using BioCaRGOS.7–9Circulating tumor DNA (ctDNA) in plasma has been demonstrated as a promising predictive and prognostic biomarker for a variety of cancers.10,11 ctDNA represents a small fraction of the total cell-free DNA (cfDNA) released from cells and can be found in various body fluids including plasma, and urine samples.12 KRAS mutation is found in nearly 30% of human cancers and is the most prevalent mutation form that drives the most prevalent type of pancreatic cancer.13,14 Due to the extremely low abundance of ctDNA (<0.1% mutant allele fraction of total cfDNA), current analytical protocols involve extraction and concentration of cfDNA from the biospecimen of interest followed by measurement of the ctDNA mutant allele fraction using ultrasensitive technologies such as droplet digital PCR (ddPCR).15,16 This prompted us to investigate the compatibility of the novel BioCaRGOS silica sol-gel technology with the extraction and downstream analysis of plasma mutant KRAS ctDNA.
Herein, we report the compatibility of BioCaRGOS with a routinely employed plasma ctDNA extraction technology and measurement of mutant KRAS ctDNA levels by ddPCR extracted from a semi-synthetic plasma reference sample. We have demonstrated an efficient way of minimizing the interference of methanol, generated as a by-product during the hydrolysis of TMOS, with ddPCR measurements and of removing silica from the BioCaRGOS prior to extraction of cfDNA to eliminate interference with plasma ctDNA extraction.
Experimental
Methods
TMOS and sodium chloride were purchased from Sigma Aldrich (Saint Louis, MO, USA). Nuclease free water was purchased from New England BioLabs (MA, USA). The 96 well plates were purchased from Thermo Fisher Scientific (MA, USA). Tris EDTA buffer (10 mM Tris–HCl, 1 MM EDTA) pH 7.4 and RNase Free water was purchased from Bioworld (Dubline, OH, USA). Methanol elimination was performed on Buchi R-124 Rotary Vap System (Delaware, US). Raman spectra were acquired on Reva Educational Raman platform (Hellma, Plainview, NY). The KRAS circulating tumor DNA (ctDNA) based reference materials were purchased from Seracare (Milford, MA, USA). QIAamp circulating nucleic acid kit was purchased from Qiagen (Hilden, Germany). Human KRAS G12D ctDNA was measured using the BioRad KRASG12D ddPCR assay reagent with ddPCR Supermix for probes per manufacturers recommendations followed by quantification of KRAS G12D utilizing the QX200 Droplet Digital PCR (ddPCR) system (BioRad laboratories, Hercules, CA, USA).BioCaRGOS synthesis
A 10.0 v/v% TMOS stock-solution was prepared in doubly de-ionized water and transferred to a 40.0 mL glass vial, screw capped and hydrolyzed via microwave for thirty seconds. Post-microwave, the solution was then subjected to rotary evaporation for 30 minutes at a controlled pressure (25 mbar) and temperature (45 °C) to remove the methanol by-product of the polymerization reaction. This methanol free TMOS stock solution was allowed to cool to room temperature. After the equilibrium was reached, doubly de-ionized water was added to CaRGOS, to formulate final concentrations of 0, 0.5, 1.0, 1.5 and 2 (v/v%). Then, 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, and 0.15 M NaCl was added to formulate BioCaRGOS.Evaluation of CaRGOS using Raman spectroscopy
Raman spectroscopy was performed on BioCaRGOS with buffer [5 v/v% TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl, 3.0 mL] using a Reva Educational Raman platform (Hellma, Plainview, NY, USA). The laser power of 450.0 mW and current 959.0 mA were optimized to analyze the samples. The laser temperatures [diode 1/4 30 °C; case 1/4 24.4 °C] and spectrometer temperature [23.1 °C] were optimized for the collection of Raman spectra.Circulating cell free (cfDNA) nucleic acid extraction
Cell free DNA (cfDNA) reference material consisting of a synthetic plasma matrix containing ∼0.01 ng μL−1 G12 D in the presence or absence of BioCARGOS [0.5 v/v%; 10 mM Tris–HCl (pH 7.5), 0.15 M NaCl] were formulated in 15.0 mL centrifuge tubes and stored for a desired amount of time. The extraction was carried out using a nucleic acid extraction kit [QIAamp, Qiagen (Hilden, Germany)] according to the manufacturer's instruction. The extracted DNA was then stored in the AVE buffer, provided by manufacturer, and used for the downstream ddPCR analysis.Circulating tumor (ctDNA) analysis using seracare samples
The measurement was performed using the Seraseq ctDNA reference material v2 (SeraCare), which is a full-process plasma-like material supporting the assessment of the entire workflow from extraction through the analysis. It includes 40 clinically relevant mutations across 28 genes all at the same allele frequency. It includes single well characterized GM24385 human genomic DNA as background wild-type material. This specific ctDNA mutation mix v2 product has an allele frequency of 2%.Invert syringe filtration
Reference material with and without BioCaRGOS [0.5 v/v% TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl] was subjected to invert syringe filtration using a PES filter membrane pore size of 0.45 μm. The process was completed in 5 minutes and then subjected to nucleic acid extraction protocol for cfDNA.Droplet digital PCR (ddPCR)
Droplet digital PCR was performed using the QX200™ Droplet Digital™ PCR System from BioRad (Hercules, CA) according to the manufacturer's instructions. These samples contained cfDNA sample, 2× ddPCR supermix, and 20× primer probe mix in a final reaction volume of 20 μL. Droplets were generated using the Droplet Generator (DG) with 70 μL DG oil per well with a DG8 cartridge and cartridge holder, 20 μL PCR reaction mix, and DG8 gasket. Droplets were dispensed into the 96-well PCR plate by pipetting 40 μL from the DG8 cartridge into each well. The PCR plate was then heat-sealed with a foil seal and the sealed plate was placed in the PCR thermocycler. The S1000 thermal cycler was used for optimal annealing with a 10 minute activation (95 C), 40 cycles, denaturation (15 s, 95 °C; 60 s, 60 °C), followed by one cycle (98 °C, 10 minutes). The data analysis on the results (n = 3) is performed using QuantaSoft Software, version 1.7, Regulatory Edition, after carrying out PCR experiments on the QX200 system. After the reaction, the droplets were read using the Droplet Reader, and QuantaSoft software converted the data into concentrations using Poisson distribution statistical analysis.Methanol elimination
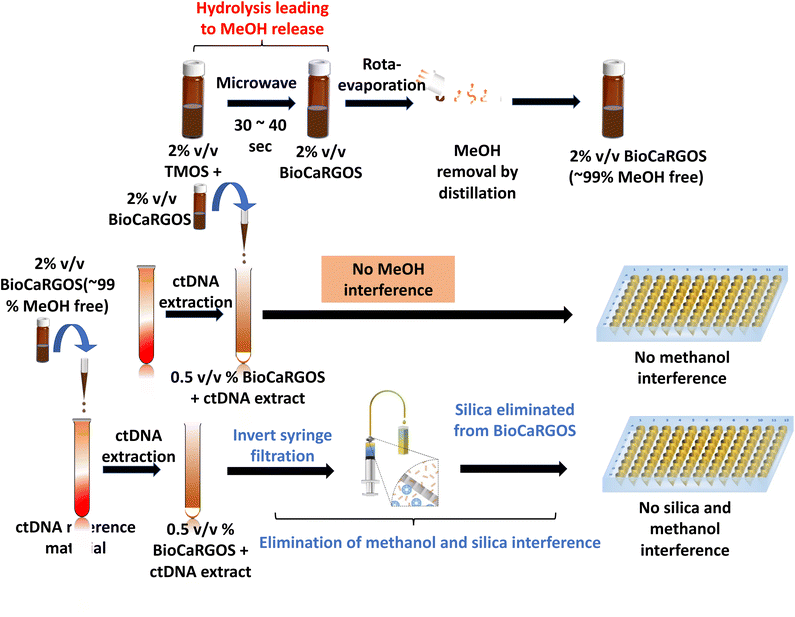
Various rotary evaporation parameters were employed to optimize the methanol removal: temperature (45, 55 and 70 °C), pressure (10, 20, 25, 30 and 40 mbar) and time (10, 15, 20, 25 and 30 minutes). As a result, the most suitable set of parameters were optimized as [temp (45 °C), pressure (25 mbar) and time (30 min)], which was repeatedly observed to demonstrate ∼99% methanol elimination from the hydrolyzed TMOS solution.Results and discussions
BioCaRGOS compatibility with ddPCR
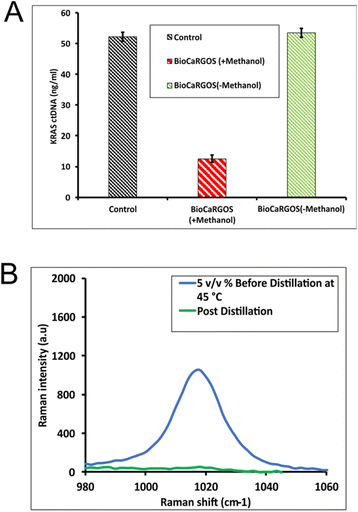
To investigate the compatibility of BioCaRGOS with ctDNA in downstream analysis, we first carried out the extraction of ctDNA (∼0.01 ng μl−1 KRAS G12 D concentration) from the QIAamp cfDNA extraction kit as per the manufacturer's protocol. The extracted KRAS ctDNA was spiked into BioCaRGOS [0.5 v/v% TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl] and then analyzed by ddPCR. Upon analysis, we observed low quantities of ctDNA obtained in BioCaRGOS samples as compared to control samples [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl] with only 10–15% KRAS ctDNA recovery [i.e., 15.65 ± 2.19 ng ml−1; 4475.15 ± 591.89 copies per ml] from BioCaRGOS (+Silica/Methanol) formulations, against their control group [52.15 ± 1.05 ng ml−1; 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 864.20 ± 299.37 copies per ml] (Fig. 1a). We attributed this low number of copies of KRAS ctDNA to interference of the significant amount of methanol (by-product from TMOS hydrolysis) with the extracted KRAS ctDNA. We further validated this by externally adding 98% MeOH as a negative control interference [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl] to study the effect of methanol which showed significant reduction in number of copies per mL (ESI Fig. 5†).
864.20 ± 299.37 copies per ml] (Fig. 1a). We attributed this low number of copies of KRAS ctDNA to interference of the significant amount of methanol (by-product from TMOS hydrolysis) with the extracted KRAS ctDNA. We further validated this by externally adding 98% MeOH as a negative control interference [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl] to study the effect of methanol which showed significant reduction in number of copies per mL (ESI Fig. 5†).
Raman spectroscopy was used to confirm methanol as the interfering agent as it allows for facile monitoring of reaction progress during BioCaRGOS preparation. The peak positions of the silica precursor (Si(OCH3)4), intermediates [(Si(OCH3)3OH, Si(OCH3)2(OH)2), Si(OCH3)(OH)3)], silicic acid (Si(OH)4 and methanol (CH3OH) are expected at 640–650 cm−1, 673–725 cm−1, 750–780 cm−1 and 1020 cm−1, respectively.17 Experimentally, we observe a peak at 646 cm−1 for 5 v/v% TMOS/water solution prior to hydrolysis indicating the presence of intact Si(OCH3)4. After ∼30 s microwave exposure, the Si(OCH3)4 peak at 646 cm−1 is absent and new peaks attributed to Si(OH)4 and methanol are observed at 750 cm−1 and 1020 cm−1, respectively. Notably, peaks for partially hydrolyzed TMOS species at 673 cm−1, 697 cm−1, and 725 cm−1 are not observed indicating completed hydrolysis of Si(OCH3)4.17 Next, the methanol by-product was removed from the hydrolyzed TMOS solution by distillation via rotary evaporation. The methanol content was monitored at different time points [0, 15, 20, 25 and 30 minutes] of the rotary evaporation by Raman spectroscopy (ESI Fig. 2–4†). The optimization process of methanol removal is described in detail in ESI. Fig. 1b shows the high methanol content in 5 v/v% TMOS pre-distillation and after nearly ∼99% methanol is eliminated post-distillation. It was observed that rotary evaporation at 45 °C, 55 °C and 70 °C did not show any significant difference in terms of elimination efficiency and therefore 45 °C was chosen as the optimum temperature for distillation of MeOH (ESI Fig. 2–4†).
After the complete removal of methanol, the hydrolyzed TMOS solution was buffered [0.5 v/v% TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl], the extracted KRAS ctDNA was added, and then analyzed by ddPCR. The BioCaRGOS (-methanol) formulations [0.5 v/v% TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl, KRAS ctDNA (post-extraction)] showed analogous KRAS ctDNA levels [50.15 ± 1.01 ng ml−1; 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859.20 ± 294.12 copies per ml] against controls [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl, KRAS ctDNA (post-extraction)] [ 52.15 ± 1.05 ng ml−1; 14
859.20 ± 294.12 copies per ml] against controls [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl, KRAS ctDNA (post-extraction)] [ 52.15 ± 1.05 ng ml−1; 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 864.20 ± 299.37 copies per ml] (Fig. 1b). The results confirm that methanol was responsible for the interference observed in the methanol containing BioCaRGOS samples.
864.20 ± 299.37 copies per ml] (Fig. 1b). The results confirm that methanol was responsible for the interference observed in the methanol containing BioCaRGOS samples.
BioCaRGOS compatibility with DNA extraction
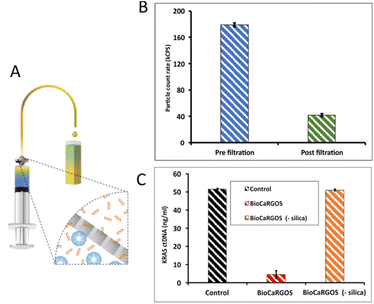
Our next goal was to investigate the compatibility of BioCaRGOS with a complex semi-synthetic plasma matrix requiring ctDNA extraction. To investigate the impact of colloidal silica present in the BioCaRGOS on the extraction of ctDNA, a BioCaRGOS stock solution [ 0.5 v/v%TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl] was added directly to the plasma reference materials. The resulting formulations with BioCaRGOS (pre-extraction) were incubated for an hour, along with their control samples, prior to extraction and ddPCR characterization. Fig. 2 demonstrates significant interference attributed to colloidal silica with the extraction process and extracted ctDNA, demonstrating only 5–9% KRAS ctDNA recovery [i.e., 4.50 ± 2.21 ng ml−1; 1268.15 ± 591.89 copies per ml] from BioCaRGOS (-methanol) formulations, against their control group [52.15 ± 1.05 ng ml−1; 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 864.20 ± 299.37 copies per ml]. Additionally, adsorption of the sample in the presence of BioCARGOS to the QIAamp Mini membrane took 3–4 hours as compared to control samples which were adsorbed in 5 min.
864.20 ± 299.37 copies per ml]. Additionally, adsorption of the sample in the presence of BioCARGOS to the QIAamp Mini membrane took 3–4 hours as compared to control samples which were adsorbed in 5 min.
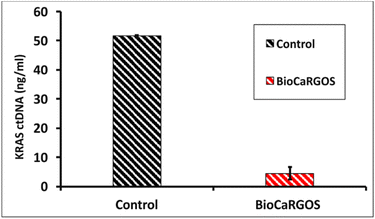 | ||
| Fig. 2 ddPCR characterization of KRAS ctDNA concentrations (ng ml−1) extracted from seracare reference material containing BioCARGOS (+silica), prior to the ctDNA extractions, against controls. | ||
Elimination of silica from BioCaRGOS to minimize the interference with ctDNA extraction
Silica gels have demonstrated a great promise in preserving the structural and functional integrity of proteins and nucleic acids against storing, shipping, and mechanical variations, via encapsulating a wide range of biospecimens. Herein, we demonstrate a technique called as invert syringe filtration (ISF) where a syringe is inverted and a filter membrane of pore size 0.45 μm is attached to the syringe to separate the silica from BioCaRGOS (Fig. 3a). Different pore sizes of filter membrane such as 0.1, 0.2, and 0.22 μm were investigated to optimize the removal of silica from BioCaRGOS. The data suggested that 0.45 μm worked most efficiently in removing silica. We confirmed the silica elimination by dynamic light scattering (DLS), which showed a significant decrease in the kCPS count rate after invert syringe filtration. The kCPS is a function of size and concentration of scattering particles. In nanoscale particle measurements, kCPS can be used to determine the concentration of sample present.18 Fig. 3b shows kCPS count rate of BioCaRGOS [0.5 v/v% TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl] pre and post inverted syringe filtration (ISF). We employed this ISF technique to circumvent silica interference prior to biospecimen analysis in downstream process. Although the hydrodynamic size measurements have demonstrated similar colloidal sizes for pre and post filtered samples, a significant reduction in the colloidal silica count rate (kCPS) was observed in the filtered samples. Therefore, we have demonstrated the effect of inverted syringe filtration on TMOS formulations [0.5, v/v% TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl]. During the hydrodynamic size measurements for the colloidal particles (2–1000) nm, the silica-colloids count rate for pre-filtered samples was observed in 160–175 kCPS range and in 40–50 kCPS range for the post filtered BioCaRGOS formulation [0.5 v/v% TMOS; 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 0.15 M NaCl] demonstrating 75–80% reduction in colloidal count rate in the filtered samples (Fig. 3b).Hence, to eliminate the silica interference during the ctDNA extraction process, we performed inverted syringe filtration on the seracare reference materials to remove silica from the BioCaRGOS solution containing [TMOS 0.5 v/v%; 10 mM Tris–HCl (pH 7.5)], 1 mM EDTA, 0.15 M NaCl, 5 ml seracare reference material]. After inverted syringe filtration, the BioCaRGOS (-methanol) formulations [ (0.5 v/v% TMOS; cfDNA (post-extraction)] indicated no significant difference in ctDNA measurements between control and inverted syringe filtered BioCaRGOS treated reference material KRAS ctDNA levels [51.04 ± 0.6 ng ml−1; 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 466 ± 181.8 copies per ml] against controls [ 51.59 ± 0.4 ng ml−1; 15
466 ± 181.8 copies per ml] against controls [ 51.59 ± 0.4 ng ml−1; 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 633 ± 121.21 copies per ml] (Fig. 3c). I we obtained nearly 96% recovery after removal of silica and methanol from BioCaRGOS solutions demonstrating a very facile and efficient method for release of ctDNA from the silica matrix with comparable integrity to that of ctDNA present in control (buffer) solutions as illustrated in Fig. 4.
633 ± 121.21 copies per ml] (Fig. 3c). I we obtained nearly 96% recovery after removal of silica and methanol from BioCaRGOS solutions demonstrating a very facile and efficient method for release of ctDNA from the silica matrix with comparable integrity to that of ctDNA present in control (buffer) solutions as illustrated in Fig. 4.
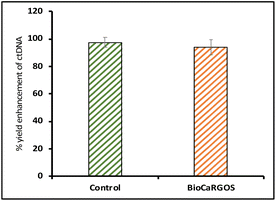 | ||
| Fig. 4 Percentage yield enhancement of ctDNA in BioCaRGOS samples demonstrating ∼96% release from silica matrix as compared to control samples. | ||
Conclusion
In summary, we have demonstrated that BioCaRGOS is highly compatible with sensitive analytical techniques. The mutant KRAS ctDNA was selected as the biomarker of interest as it is found in extremely low concentrations in clinical samples. The main goal of this article was to test the compatibility of BioCaRGOS with the extraction of ctDNA from semi-synthetic plasma reference material. Preliminary data suggested that the methanol by-product from BioCaRGOS synthesis interfered with ddPCR measurements. This interference was eliminated by incorporating distillation via rotary evaporation to remove methanol prior to addition of biospecimen. We further demonstrated that BioCaRGOS interfered with the extraction process of ctDNA by causing slow filtration through extraction columns and low recovery of ctDNA was observed by ddPCR analysis. We minimized this interference by incorporating an inverted filtration technique that improved the flow through rate as well as the yield of ctDNA recovery. These modifications to the workflow of BioCaRGOS containing samples allow for use of BioCaRGOS for stabilization of trace quantities of nucleic acid biomarkers such as plasma ctDNA while retaining the capability of recovery of the biomarker and PCR-based quantification. Fig. 5 illustrates complete workflow of synthesis of BioCaRGOS and using it in the ctDNA extraction process. Moreover, we have developed a highly downstream compatible sol-gel based encapsulation matrix called BioCaRGOS. Thus, we have tailored BioCaRGOS for not only long-term stabilization of biospecimens but improved the compatibility of BioCaRGOS with the downstream applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors acknowledge financial support from National Cancer Institute (NCI) exploratory/development grant [ID: grant number: 1R21CA251042-01].References
- J. Livage, T. Coradin and C. Roux, J. Condens. Matter Phys., 2001, 33, R673–R691 CrossRef.
- A. C. Pierre, Biocatal. Biotransform., 2004, 3, 145–170 CrossRef.
- J. Tu, A. L. Boyle, H. Friedrich, P. H. H. Bomans, J. Bussmann, N. A. J. M. Sommerdijk, W. Jiskoot and A. Kros, ACS Appl. Mater. Interfaces, 2016, 47, 32211–32219 CrossRef PubMed.
- F. Tang, L. Li and D. Chen, Adv. Mater., 2012, 12, 1504–1534 CrossRef PubMed.
- R. B. Bhatia, C. J. Brinker, A. K. Gupta and A. K. Singh, Chem. Mater., 2000, 8, 2434–2441 CrossRef.
- N. A. Montoya, R. E. Roth, E. K. Funk, P. Gao, D. R. Corbin and M. B. Shiflett, Microporous Mesoporous Mater., 2022, 111750 CrossRef CAS.
- C. W. Narvaez Villarrubia, K. C. Tumas, R. Chauhan, T. MacDonald, A. M. Dattelbaum, K. Omberg and G. Gupta, Emergent Mater., 2021, 307–314 Search PubMed.
- J. Boylan, R. Chauhan, K. Koneru, M. Bansal, T. Kalbfleisch, C. S. Potnis, K. Hartline, R. S. Keynton and G. Gupta, RSC Adv., 2021, 22, 13034–13039 RSC.
- R. Chauhan, T. S. Kalbfleisch, C. S. Potnis, M. Bansal, M. W. Linder, R. S. Keynton and G. Gupta, RSC Adv., 2021, 50, 31505–31510 RSC.
- Y. Ma and S. Gamagedara, Biomarkers Med., 2015, 9, 845–850 CrossRef CAS PubMed.
- A. McGuigan, P. Kelly, R. C. Turkington, C. Jones, H. G. Coleman and R. S. McCain, World J. Gastroenterol., 2018, 43, 4846–4861 CrossRef PubMed.
- D. Fernandez-Garcia, A. Hills, K. Page, R. K. Hastings, B. Toghill, K. S. Goddard, C. Ion, O. Ogle, A. R. Boydell, K. Gleason, M. Rutherford, A. Lim, D. S. Guttery, R. C. Coombes and J. A. Shaw, Breast Cancer Res., 2019, 1, 149 CrossRef PubMed.
- Z. Mao, H. Xiao, P. Shen, Y. Yang, J. Xue, Y. Yang, Y. Shang, L. Zhang, X. Li, Y. Zhang, Y. Du, C.-C. Chen, R.-T. Guo and Y. Zhang, Cell Discovery, 2022, 1, 5 CrossRef PubMed.
- S. R. Hingorani, E. F. Petricoin, A. Maitra, V. Rajapakse, C. King, M. A. Jacobetz, S. Ross, T. P. Conrads, T. D. Veenstra, B. A. Hitt, Y. Kawaguchi, D. Johann, L. A. Liotta, H. C. Crawford, M. E. Putt, T. Jacks, C. V. E. Wright, R. H. Hruban, A. M. Lowy and D. A. Tuveson, Cancer Cell, 2003, 6, 437–450 CrossRef.
- G. Johansson, D. Andersson, S. Filges, J. Li, A. Muth, T. E. Godfrey and A. Ståhlberg, Biomol. Detect. Quantif., 2019, 100078 CrossRef CAS PubMed.
- A. R. Thierry, S. El Messaoudi, P. B. Gahan, P. Anker and M. Stroun, Cancer Metastasis Rev., 2016, 3, 347–376 CrossRef PubMed.
- T. Woignier, C. Fernandez-Lorenzo, J. L. Sauvajol, J. F. Schmit, J. Phalippou and R. Sempere, J. Sol-Gel Sci. Technol., 1995, 3, 167–172 CrossRef.
- J. Smeraldi, R. Ganesh, J. Safarik and D. Rosso, J. Environ. Monit., 2012, 1, 79–84 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra05862f |
| This journal is © The Royal Society of Chemistry 2022 |