Open Access Article
Open Access ArticleNanotechnology meets immunology towards a rapid diagnosis solution: the COVID-19 outbreak challenge†
Santos B. S. A. S.a,
Cunha J. L. R.ag,
Carvalho I. C.b,
Costa J. M. C.a,
Longo B. C.a,
Galinari G. C. F.a,
Diniz P. H. S. M.ac,
Mendes G. M. M. ad,
Fonseca F. G.f,
Abrahão J. S.f,
Mansur A. A. P.b,
Leite M. F.e,
Oréfice R. L.
ad,
Fonseca F. G.f,
Abrahão J. S.f,
Mansur A. A. P.b,
Leite M. F.e,
Oréfice R. L. b,
Lobato Z. I. P.a and
Mansur H. S.‡
b,
Lobato Z. I. P.a and
Mansur H. S.‡
 *b
*b
aUniversidade Federal de Minas Gerais (UFMG), Departamento de Medicina Veterinária Preventiva, Belo Horizonte, MG, Brazil
bUniversidade Federal de Minas Gerais (UFMG), Departamento de Engenharia Metalúrgica e de Materiais, MG, Brazil
cUniversidade Federal de Minas Gerais (UFMG), Colégio Técnico, Belo Horizonte, MG, Brazil
dUniversidade Federal de Minas Gerais (UFMG), Faculdade de Farmácia, Belo Horizonte, MG, Brazil
eUniversidade Federal de Minas Gerais (UFMG), Departamento de Fisiologia e Biofísica, Belo Horizonte, MG, Brazil
fUniversidade Federal de Minas Gerais (UFMG), Instituto de Ciências Biológicas, Belo Horizonte, MG, Brazil
gUniversity of York, York Biomedical Institute, New York, UK
First published on 4th November 2022
Abstract
The current COVID-19 pandemic presents one of the greatest challenges in human history. There is a consensus that the rapid and accurate diagnosis of COVID-19 directly affects procedures to avoid dissemination, promote treatments, and favor the prognosis of infected patients. This interdisciplinary study aims at designing new synthetic peptides inspired by the SARS-CoV-2 spike protein (SARS-CoV-2S) to produce rapid detection tests relying on nanomaterial-based colorimetric properties. Hence, in silico analyses of SARS-CoV-2S were performed using advanced bioinformatic simulation tools and algorithms. Five novel peptide sequences were proposed, and three were selected (P2, J4, and J5) based on their prospective reactivity against positive serum from naturally COVID-19-infected humans. Next, hyperimmune sera against the selected peptides were produced in rabbits. Concurrently, gold nanoparticles (AuNP) were synthesized using a green aqueous method under mild conditions through in situ reduction by trisodium citrate salt. They were extensively characterized by their morphological, physicochemical, and optical properties. The AuNPs demonstrated colloidal chemical stability in aqueous media, with an average size of approximately 29 nm (metallic core), and zeta potential before and after bioconjugation of −43 mV and −31 mV, respectively. Moreover, they presented an intense reddish-bluish color due to the surface plasmon resonance (SPR) effect, with maxima at λ = 525 nm and 536 nm, before and after bioconjugation, respectively, evidencing their applicability as colorimetric biomarkers for antigen–antibody immunoassay detection. To develop a rapid COVID-19 diagnosis test using lateral flow assay (LFA), semi-purified anti-SARS-CoV-2S sera against the three selected peptides were bioconjugated to the AuNPs as the highly optically sensitive agents using a considerably low antibody concentration (0.2 mg mL−1). All tested peptide sequences (P2, J4, and J5) induced antibodies capable of identifying the presence of SARS-CoV-2 virus inactivate suspensions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, or 1
100, or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilutions). For LFA positive test control, an anti-rabbit antibody was used. In summary, this research comprises several contributions and advances to the broad and multidisciplinary field of nanomaterials-based immunodiagnosis tools, encompassing: (a) the novelty of designing and synthesizing new immunogenic peptides inspired by SARS-CoV-2 virus epitopes using in silico bioinformatics; (b) the peptides induced the immune response in rabbit animal model producing hyperimmune serum; (c) the semi-purified hyperimmune serum rendered effective antibodies to detect SARS-CoV-2 virus in cell suspension; (d) colloidal gold nanoparticles were produced and bioconjugated to the antibodies for qualitative colorimetric detection. As the overall result of this study, it was designed, developed, produced, and validated a new simple, rapid, and sensitive LFA diagnostic test for the SARS-CoV-2 virus using a nanotechnology-based qualitative colorimetric assay, which can be envisioned as promising nanoplatforms for detecting other diseases.
1000 dilutions). For LFA positive test control, an anti-rabbit antibody was used. In summary, this research comprises several contributions and advances to the broad and multidisciplinary field of nanomaterials-based immunodiagnosis tools, encompassing: (a) the novelty of designing and synthesizing new immunogenic peptides inspired by SARS-CoV-2 virus epitopes using in silico bioinformatics; (b) the peptides induced the immune response in rabbit animal model producing hyperimmune serum; (c) the semi-purified hyperimmune serum rendered effective antibodies to detect SARS-CoV-2 virus in cell suspension; (d) colloidal gold nanoparticles were produced and bioconjugated to the antibodies for qualitative colorimetric detection. As the overall result of this study, it was designed, developed, produced, and validated a new simple, rapid, and sensitive LFA diagnostic test for the SARS-CoV-2 virus using a nanotechnology-based qualitative colorimetric assay, which can be envisioned as promising nanoplatforms for detecting other diseases.
1 Introduction
The continuous emergence of new pathogenic viruses, such as “Severe Acute Respiratory Syndrome Coronaviruses” (SARS-CoV), the “Middle East Respiratory Syndrome Coronavirus” (MERS-CoV), and more recently, the COVID-19 (“Coronavirus Disease of 2019”) outbreak, have been seriously jeopardizing human health for centuries. In particular, the COVID-19 pandemic has been causing an immense impact on global society1 for more than 2 years, posing unprecedented challenges to humanity.Initially, the novel coronavirus was identified in Wuhan, China, in December 2019 and rapidly overspread around the globe, eliciting extraordinary global measures. The clinical spectrum of COVID-19 ranges from asymptomatic infections and mild upper respiratory tract illnesses to severe viral pneumonia with respiratory failure, multiorgan failure, and death.2 Thus, the World Health Organization declared the SARS-CoV-2 a global pandemic on March 11, 2020.3 To date, two years later, there are already over 267.5 million confirmed COVID-19 cases and over 5.2 million deaths reported globally.4,5 In this scenario, Brazil was considered particularly highly vulnerable to the COVID-19 pandemic because all government levels were not integrated, and they have not promptly adopted strict public health restrictions, which was aggravated by its continental dimension and the enormous socioeconomic asymmetry.6–8
Since the very beginning of the SARS-CoV-2 reported cases, it was almost a consensus among health professionals, public authorities, and society that the strategy to fight the pandemic would require different crucial fronts, including (i) social distancing and sanitary actions combined with rapid and early diagnosis of infected individuals to minimize or ideally to avoid the virus dissemination; (ii) intensive research for developing new vaccines preventing the infection through massive vaccination, and (iii) dramatic search for effective treatments for the infected patients to reduce hospitalization and mortality.9–14
In this view, from the strategic diagnosis front against COVID-19, most health facilities rely on polymerase chain reaction (PCR)-based tests based on the amplification of genetic material (i.e., RNA) to detect the virus. While considered the “gold standard” method for diagnosis and extremely sensitive, PCR poses several challenges, including a relatively long time to get the results, a “cold chain” is usually required to transport the samples from patients to the lab, sophisticated facilities and expensive equipment are needed, and well-trained lab technicians are essential to perform the testing. Thus, simpler and less-expensive tests based on antigen–antibody affinity are highly demanded and have been intensively researched.14–16
Many studies have focused their primary efforts on searching antigenic sites in SARS-CoV-2 for immunogenic responses using bioinformatic tools to design novel methods for virus or antibody detection of infected patients that can also be applied later for developing effective vaccines to prevent new infections. Various advanced bioinformatics tools have remarkably improved the search for immunogenic peptides. The in silico analysis and simulation have been applied to identify novel immunogenic epitopes that could be used to develop novel synthetic antigenic peptides for the development of antibody/antigen-based diagnostic tests for detecting SARS-CoV-2.16 The in silico research has focused on the four main structural proteins of the SARS-CoV-2 virus, namely, spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. These proteins facilitate several pivotal biological functions, such as binding the virus to the host cell receptors, the fusion of virus envelop to the host cell, the fusion of viral RNA inside the host cell, and virus replication. In particular, as the most accepted mechanism, the SARS-CoV-2 virus interacts with the human ACE2 (angiotensin converting enzyme-2) receptor for binding to the cell for priming the S-protein (spike), which is critical for infecting the host cell membrane. Hence, several strategies are being explored to target or block one or more of these structural proteins to avoid infection, including in silico-designed peptides that could be further tested in vitro and in vivo experiments for their potential rapid diagnosis and therapeutic applications. These screened peptides also present potential for functionalizing nanomaterials for detecting the (S) spike protein of the virus through rapid diagnostic tests.17 Bearing in mind the high demand for simple and rapid viral detection methods as an alternative to PCR-based diagnosis, lateral-flow assay (LFA) is probably the most commonly used diagnostic test among all available techniques. LFAs provide a simple and fast diagnostic tool at the point of care (POC) for both clinical professionals and the public through over-the-counter (OTC) devices.14 Essentially, the LFAs assays mainly involve antibody-antigen recognition by specific coupling where the system is assembled onto a nitrocellulose membrane for immunological diagnosis (virus, antigen, antibody, or analytes). The detection relies on specific biomolecule recognition where optically active capture agents, mostly metallic nanoparticles, are conjugated to antigens or antibodies. In particular, gold nanoparticles (AuNPs) conjugated with immunological molecules have been the predominant choice for building LFA assays based on the colorimetric method. In this sense, nanotechnology, amalgamated with biology and medicine, plays a vital role in developing a rapid, simple, accurate, and inexpensive diagnostic tool for detecting virus-related infections based on LFA immunoassay.9,10,14 In brief, the nanotechnology-based colorimetric test relies on the unique optical properties of metal nanoparticles called localized surface plasmon resonance (SPR). SPR has been widely utilized in biomedical applications due to the tunable light absorption and scattering wavelength in the visible region (i.e., color spectrum).
Due to their exceptional optical properties, stability, and biocompatible properties, gold nanoparticles (AuNPs) are the most common optically-active nanomaterials in diagnosing viruses. As a result of the LSPR effect, the aggregation of AuNPs causes a redshift in the absorption peak position, leading to a noticeable visual change (“naked eye”) in the solution color, usually from red to blue.19,20 Thus, the bioconjugation of AuNP with antigenic biomolecules (or antibodies) permits a simple, fast, and reliable virus detection method by developing color in the test line when the coupling with the antibody (or analyte) occurs associated with the virus-infected sample.9,13,14 In this view, the development of simple and rapid LFA-based diagnostic tests for the direct detection of SARS-CoV-2, abbreviating the usual time window necessary for conventional diagnostic methods associated with the potential utilization at the POC, is exceedingly needed as a crucial weapon to fight against the COVID-19 pandemic.1,9,11,15,21–23
Herein, we present all stages of the development of an LFA-based diagnostic immunoassay for directly detecting SARS-CoV-2, encompassing the in silico design of novel synthetic peptides with immunogenic epitopes inspired by the virus spike-protein, the production of high-affinity antibodies through semi-purified hyperimmune sera from rabbits, the synthesis of colloidal gold nanoparticles as optically active nanoprobes, their bioconjugation with the affinity biomolecules, which were all integrated into a nitrocellulose membrane strip, demonstrating effective recognition of COVID-19-infected patient sera.
2 Materials and methods
2.1 Design and selection of in silico peptides
The selection of peptides from the spike protein to be used in SARS-CoV-2 diagnosis was based on three parameters: (i) conservation among SARS-CoV-2 lineages; (ii) high B-cell epitope prediction by bioinformatic tools; and (iii) specificity to SARS-CoV-2.| Lineage | Sequences |
|---|---|
| B.1.1.7 | 300 |
| B.1.351 | 259 |
| B.1.427 | 300 |
| B.1.429 | 300 |
| B.1.525 | 300 |
| B.1.526 | 300 |
| B.1.526 | 300 |
| B.1.617.1 | 113 |
| B.1.617.2 | 78 |
| P.1 | 300 |
| P.2 | 300 |
After the alignment, the number of variant positions, amino acid distribution, and sequence conservation were determined using a script in R-language, which uses the libraries “Biostrings” (https://bioconductor.org/packages/release/bioc/htmL/Biostrings.html) and “seqinr” (https://cran.r-project.org/web/packages/seqinr/index.html). The final output figure was created using the “ggplot2” library (https://ggplot2.tidyverse.org/).
| Program | Weight |
|---|---|
| ABCpred | 1 |
| Bepipred2.0 | 3 |
| NetSurfP | 3 |
| SVMTriP | 2 |
| LBtope | 1 |
| ElliPro-linear | 1 |
| ElliPro-conformational | 2 |
| DiscoTope2.0 | 1 |
2.2 Selection of the designed synthetic peptides using human sera by ELISA assay
The designed peptides were synthesized and evaluated by ELISA (enzyme-linked immunosorbent assay) assay to assess their reactivity against SARS-CoV-2-positive human sera to differentiate between positive and negative samples. These assays were conducted using blood samples collected before the beginning of vaccination campaigns, which were kindly provided by volunteers who tested positive or negative (i.e., negative and positive control sera) for COVID-19 by standard PCR tests (NCT04323527 Ethics Committee approval). All blood samples were collected in tubes containing EDTA (ethylenediaminetetraacetic acid) at the Laboratory of Research in Animal Virology at the Veterinary School of UFMG, Brazil. They were centrifuged for 10 min at 3000 rpm for serum collection, aliquoted into 1.5 mL Eppendorf tubes, and stored at −20 °C for further analyses. The National Institute of Vaccine Science and Technology (CT-Vacinas/UFMG, Brazil) tested and validated these samples using antibodies against the SARS-CoV-2S-protein.The ELISA tests were performed using the synthetic peptides non-covalently immobilized onto polystyrene plates. Then, the positive and negative human sera naturally infected with SARS-CoV-2 were added to the plate. The immunocomplex presence was observed by the immunoenzymatic reaction using an anti-human immunoglobulin antibody (IgG) labeled with peroxidase enzyme (anti-human IgG antibody conjugated with horseradish peroxidase). Orthophenylenediamine (OPD) compound was used as the assay substrate, which was added to the plate after the secondary antibody (production of the compound 2,3-diaminophenazine, DAP, in the presence of hydrogen peroxide). The protocol is described in the sequence.
Briefly, each well of a 96-well flat-bottomed polystyrene microplate (Corning Incorporation, USA) was coated with 2 ng μL−1 of each peptide in bicarbonate-carbonate buffer (0.1 M, pH 9.6) and incubated overnight at 5% CO2 and 37 °C. The plates were blocked with BSA (bovine serum albumin, 5% w/v, in phosphate-buffered saline, PBS, pH 7.4) and kept at 5% CO2 and 37 °C for 1 h. Then, plasma samples diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBST (phosphate buffered saline with Tween 20 at 0.05% v/v) containing 2.5% (w/v) of BSA were added to the plates and incubated for 1 h at 5% CO2 and 37 °C. After four washes with PBST 0.05%, the plates were incubated with the anti-human IgG conjugated with peroxidase (Sigma Aldrich, P8170, diluted 1
100 in PBST (phosphate buffered saline with Tween 20 at 0.05% v/v) containing 2.5% (w/v) of BSA were added to the plates and incubated for 1 h at 5% CO2 and 37 °C. After four washes with PBST 0.05%, the plates were incubated with the anti-human IgG conjugated with peroxidase (Sigma Aldrich, P8170, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 in PBS with 2.5% w/v of BSA) for 1 h at 5% CO2 and 37 °C. The reaction was revealed using OPD substrate (0.5 mg mL−1, Sigma-Aldrich, USA) diluted in phosphate-citrate buffer (0.05 M, pH = 5.0), and the reaction was stopped with 4.0 M H2SO4. The optical density (OD492 nm) was determined using a microplate spectrophotometer (Multiskan GO, Thermo Scientific, USA).
3000 in PBS with 2.5% w/v of BSA) for 1 h at 5% CO2 and 37 °C. The reaction was revealed using OPD substrate (0.5 mg mL−1, Sigma-Aldrich, USA) diluted in phosphate-citrate buffer (0.05 M, pH = 5.0), and the reaction was stopped with 4.0 M H2SO4. The optical density (OD492 nm) was determined using a microplate spectrophotometer (Multiskan GO, Thermo Scientific, USA).
2.3 Hyperimmune serum production in vivo – rabbit animal model
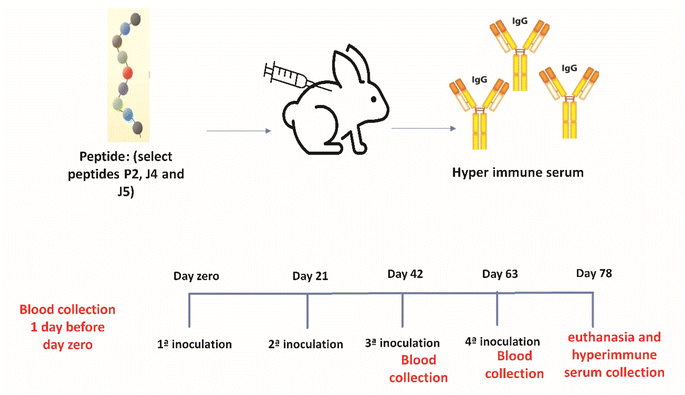
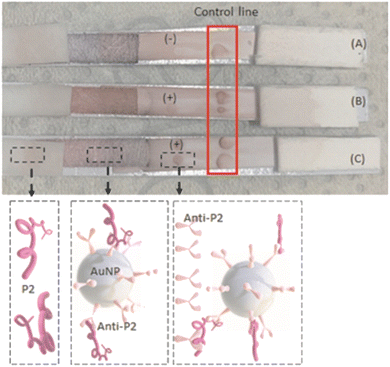
For the production of hyperimmune serum (Fig. 1), two female rabbits (New Zealand rabbits) were used for each of the three novel synthetic peptides (P2, J4, and J5) selected based on ELISA described in the previous section. Four inoculations were performed subcutaneously with a volume of 1 mL of emulsion per animal, divided into two injections of 0.5 mL, at a concentration of 150 μg of peptide per animal, with an interval of 21 days between inoculations. The animals were kept in the vivarium of the Veterinary School of UFMG, with ad libitum feeding and daily monitoring to control the clinical signs and maintain the environment.The first inoculation was performed using a peptide emulsion and Freund's Complete Adjuvant (FCA), and from the second to fourth inoculation, Freund's incomplete adjuvant (FIA) was used.
Partial bleeding was performed through the rabbit's marginal vein after 15 days of the third inoculation to verify the humoral response against the peptides. Total bleeding was performed 15 days after the fourth inoculation by cardiac puncture after deep anesthesia of the animals (Ketamine 80 mg kg−1 and Xylazine 15 mg kg−1).
The reactivity of the hyperimmune serum was evaluated by ELISA as described in Section 2.2, where plates were sensitized with the respective peptide using: (i) the sera of immunized rabbits as a test; (ii) reactive human sera as positive controls; and (iii) pre-inoculations rabbit serum as negative controls.
After confirming the hyperimmune serum reactivity by ELISA, sera were semi-purified as described in the sequence. Rabbits sera were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 with 60 mM acetate buffer (pH 4.0) under magnetic stirring, where the pH was adjusted to 4.5 with tris(hydroxymethyl)aminomethane (1.0 M). Then, caprylic acid droplets were added to the solution (25 μL for each 1 mL of the diluted sample) and kept for 30 min under constant stirring at room temperature. Next, the solution was centrifuged at 1000×g for 30 min. The supernatant was then paper-filtered to remove particles that had not settled by centrifugation. The filtered solution was diluted ten times, and the pH was adjusted to 7.4 (TRIS 1.0 M). In a clean cold chamber, saturated ammonium sulfate solution (v/v) was added by dropping in the solution under stirring for 30 min. The solution was then centrifuged (10
5 with 60 mM acetate buffer (pH 4.0) under magnetic stirring, where the pH was adjusted to 4.5 with tris(hydroxymethyl)aminomethane (1.0 M). Then, caprylic acid droplets were added to the solution (25 μL for each 1 mL of the diluted sample) and kept for 30 min under constant stirring at room temperature. Next, the solution was centrifuged at 1000×g for 30 min. The supernatant was then paper-filtered to remove particles that had not settled by centrifugation. The filtered solution was diluted ten times, and the pH was adjusted to 7.4 (TRIS 1.0 M). In a clean cold chamber, saturated ammonium sulfate solution (v/v) was added by dropping in the solution under stirring for 30 min. The solution was then centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 15 min), the supernatant was discarded, and the pellet was resuspended in PBS 1X (50% of the initial volume). Dialysis was performed 3 times with PBS 1X for each 10 mL of the resuspended pellet. Samples were maintained under stirring for 24 h at 4 °C (PBS solution was changed at least one time during the process). Then, the translucid content was collected, aliquoted, and stored at −20 °C. One of the aliquots was submitted to protein dosage using the Bradford method.32 The semi-purified hyperimmune sera containing the antibodies produced by the rabbits induced by the inoculation of the new peptides were used in all further immunological tests of this study. Regarding their original peptide sequences (P2, J4, and J5), these “anti-peptides” or antibodies were referred to as anti-P2, anti-J4, and anti-J5.
000×g for 15 min), the supernatant was discarded, and the pellet was resuspended in PBS 1X (50% of the initial volume). Dialysis was performed 3 times with PBS 1X for each 10 mL of the resuspended pellet. Samples were maintained under stirring for 24 h at 4 °C (PBS solution was changed at least one time during the process). Then, the translucid content was collected, aliquoted, and stored at −20 °C. One of the aliquots was submitted to protein dosage using the Bradford method.32 The semi-purified hyperimmune sera containing the antibodies produced by the rabbits induced by the inoculation of the new peptides were used in all further immunological tests of this study. Regarding their original peptide sequences (P2, J4, and J5), these “anti-peptides” or antibodies were referred to as anti-P2, anti-J4, and anti-J5.
All experimental procedures, methods, and protocols in this study were conducted in accordance with the institutional ethical review committees on animal use and ethics policy (approved by Project Licence# NCT04323527, Federal University of Minas Gerais, UFMG-Brazil), in compliance with the Brazilian Guidelines by the National Council for the Control of Animal Experimentation.
2.4 Synthesis of gold nanoparticles for LFA-based colorimetry assay
In a three-necked glass flask connected to a condenser/reflux containing 90 mL of DI water, 1 mL of an aqueous auric chloride solution (Au3+, 10 mM) was added and heated to approximately 100 °C (Fig. 2, Step I). After boiling, 1.75 mL of an aqueous sodium citrate solution (34 mM, 1 wt%) was added to the Au3+ solution (Au3+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) citrate = 1
citrate = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) and reacted for 10 min (referred to as stage 1). After stage 1, 1 mL of auric chloride solution and 0.74 mL of citrate solution (Au3+
6) and reacted for 10 min (referred to as stage 1). After stage 1, 1 mL of auric chloride solution and 0.74 mL of citrate solution (Au3+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) citrate = 1
citrate = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
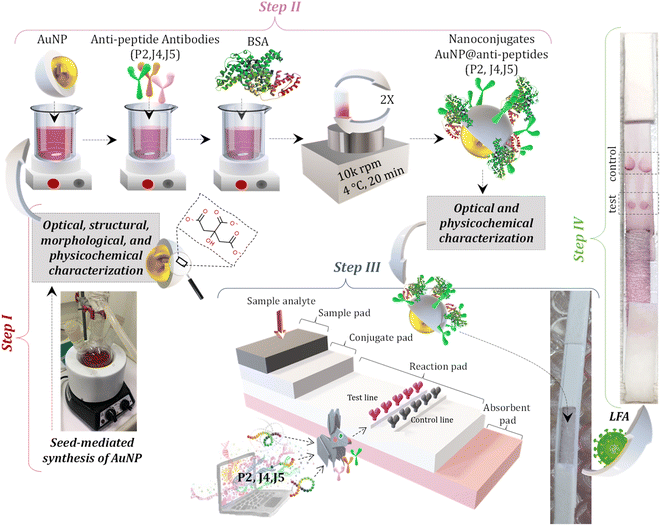
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) were added to the reaction flask to grow Au0 nuclei by in situ reduction. After the addition, the solution was kept under stirring for 30 min at a temperature of 100 °C. This procedure was repeated six times to produce colloidal gold nanoparticles (AuNPs) with an estimated average diameter of approximately ∼30 nm, named BIOCOR_VID19_30. At the end of the reaction, the final volume was completed to 100 mL for standardization purposes. After cooling to room temperature, the resulting AuNP solution was stored in plastic containers (Fig. 2), kept at 4 °C, and protected from light.
2.5) were added to the reaction flask to grow Au0 nuclei by in situ reduction. After the addition, the solution was kept under stirring for 30 min at a temperature of 100 °C. This procedure was repeated six times to produce colloidal gold nanoparticles (AuNPs) with an estimated average diameter of approximately ∼30 nm, named BIOCOR_VID19_30. At the end of the reaction, the final volume was completed to 100 mL for standardization purposes. After cooling to room temperature, the resulting AuNP solution was stored in plastic containers (Fig. 2), kept at 4 °C, and protected from light.
2.5 Conjugation of anti-peptide antibodies to gold nanoparticles optical probes
For the bioconjugation procedures, solutions of purified anti-peptide J4, J5, and P2 (i.e., antibody, hyperimmune sera produced in rabbits see Section 2.3) (pH ∼7.4, 2.27 mg mL−1), named HAb and gold nanoparticles (BIOCOR_VID19_30, pH 6, ∼1 nM) were utilized. Before the conjugation reaction, the pH of the AuNP colloidal solution (BIOCOR_VID19_30) was raised to 8.6 (K2CO3, 25 mM). Then, each HAb solution was added to BIOCOR_VID19_30 to reach a final antibody concentration of 100 μg mL−1 and a total reaction volume of 1 mL. The conjugation by electrostatic coupling between AuNP with antibodies was performed for 1 h at room temperature, protected from light. After that, 100 μL of a 10% w/v BSA solution in borate buffer (2 mM, pH 8.5) was added to the reaction medium, and after a brief stirring for homogenization (∼5 min), the resulting solution was left for one hour at room temperature, protected from light, to stabilize the newly formed nanoconjugates composed by AuNPs coupled to the antibodies. Then, these nanoconjugates were submitted to two centrifugation cycles (10 min, 4 °C, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) for the removal of the supernatant and resuspension in a 1% w/v BSA in borate buffer. At the end of the second cycle, the solution was resuspended in 500 μL of 1% w/v BSA in borate buffer, where the final concentration of the conjugated antibody was 200 μg mL−1, and the concentration of AuNPs (BIOCOR_VID19_30) of ∼1.9 nM. These nanoconjugates (i.e., AuNPs-antibody) were named as follows: (i) BIOCOR_VID19_30-HAb_200J4: anti-J4 antibody conjugated to AuNPs; (ii) BIOCOR_VID19_30-HAb_200J5: anti-J5 antibody conjugated to AuNPs; (iii) BIOCOR_VID19_30-HAb_200P2: anti-P2 antibody conjugated to AuNPs; and (iv) SN: for the reference “blank” sample. Fig. 2 schematically depicts the gold nanoparticle synthesis protocol, conjugation stage, membrane-strip assembly, and LFA.
000 rpm) for the removal of the supernatant and resuspension in a 1% w/v BSA in borate buffer. At the end of the second cycle, the solution was resuspended in 500 μL of 1% w/v BSA in borate buffer, where the final concentration of the conjugated antibody was 200 μg mL−1, and the concentration of AuNPs (BIOCOR_VID19_30) of ∼1.9 nM. These nanoconjugates (i.e., AuNPs-antibody) were named as follows: (i) BIOCOR_VID19_30-HAb_200J4: anti-J4 antibody conjugated to AuNPs; (ii) BIOCOR_VID19_30-HAb_200J5: anti-J5 antibody conjugated to AuNPs; (iii) BIOCOR_VID19_30-HAb_200P2: anti-P2 antibody conjugated to AuNPs; and (iv) SN: for the reference “blank” sample. Fig. 2 schematically depicts the gold nanoparticle synthesis protocol, conjugation stage, membrane-strip assembly, and LFA.
2.6 Characterization of AuNPs and AuNPs@antibody nanoconjugates
The structural and morphological characterizations of AuNPs (BIOCOR_VID19_30) were performed by analyses of high-definition images of transmission electron microscopy (TEM, Tecnai G2-20-FEI, FEI Company, USA) and selected area electron diffraction patterns (SAED) at an accelerating voltage of 200 kV. Energy-dispersive X-ray (EDX) spectra were collected for chemical analysis. The average size and the size distribution of BIOCOR_VID19_30 were obtained from TEM images, measuring at least 300 randomly selected nanoparticles, and using the image processing program (ImageJ, version 1.50, public domain, National Institutes of Health). For evaluating the position of the characteristic peaks of surface plasmon resonance (SPR) of AuNP and AuNP@antibody nanoconjugates (i.e., BIOCOR_VID19_30 and BIOCOR_VID19_30-Hab), their respective colloidal solutions were submitted to ultraviolet-visible absorption spectroscopy (UV-vis). All UV-vis spectra were acquired at 490 < λ < 700 nm using FluoroMax-Plus – CP (Horiba Scientific). To evaluate the hydrodynamic diameter of the colloidal suspensions of nanoconjugates, dynamic light scattering (DLS) analyses were carried out at room temperature (25 ± 2 °C) using the ZetaPlus instrument with a minimum of ten replicates (Brookhaven Instruments Corporation, 35 mW-red diode laser source, at wavelength λ = 660 nm). Zeta potential (ZP) measurements were also carried out (with the same equipment) to evaluate the chemical stability and surface charges of AuNP-based colloidal nanosystems.2.7 Lateral flow assay (LFA) assembly for SARS-CoV-2 immunodiagnostic test
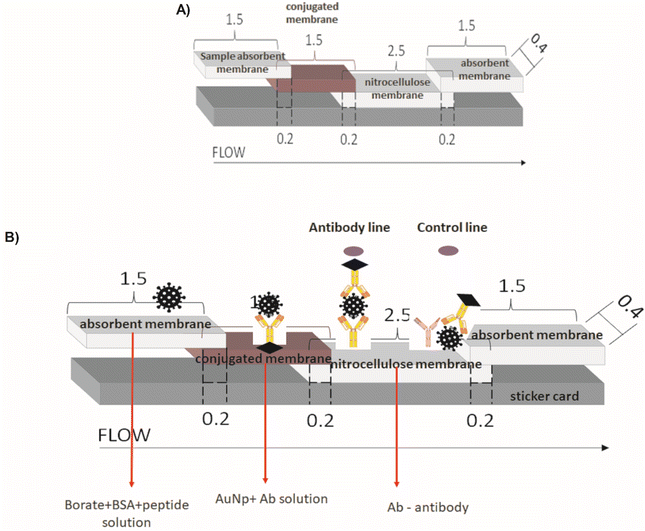
The major parts and components of the LFA strips are presented in detail as follows:Test line: For the test site preparation, two droplets (0.25 μL each) of semi-purified rabbit antibodies (anti-J4, J5, or P2) at 2.27 mg mL−1, or semi-purified serum from non-inoculated rabbit (negative serum control – SN) were poured onto the center of a nitrocellulose membrane (NC, 2.5 cm × 0.4 cm) and allowed to dry for 1 h at room temperature.
Control line: For the control line, two droplets (0.25 μL each) of anti-rabbit IgG (Rabbit Anti-Goat IgG H&L, catalog#AB7085, ABCAM, USA) were poured at the end of a nitrocellulose membrane (NC, 2.5 cm × 0.4 cm), and allowed to dry for 1 h at room temperature.
Membrane Conjugate: For preparing the conjugated pads, 50 μL of each sample solution (BIOCOR_VID19_30-HAb_200J4; BIOCOR_VID19_30-HAb_200J5; BIOCOR_VID19_30-HAb_200P2) was pipetted and slowly poured onto the glass fiber membrane pad (1.5 cm × 0.4 cm), allowing it to dry in an oven at 40 °C for 1 h. The prepared membranes were stored in a zip-lock plastic bag at ambient conditions (18 ± 2 °C and 40 ± 1% humidity).
Membrane Assembly: Membranes were mounted on adhesive cards with an overlap of 0.2 cm between the NC membrane (2.5 cm × 0.4 mm) and absorbent (1.5 cm × 0.4 cm), NC membrane, and conjugated membrane, the conjugate and sample membrane (1.5 cm × 0.4 cm). Fig. 3 summarizes the complete assembly for the LFA test.
2.8 Experimental method of LFA immunoassay
The novel synthetic peptides (i.e., P2, J4, and J5) or their mixture were tested at the concentration of 5 μg mL−1 in a total volume of 150 μL. Thus, each synthetic peptide (P2, J4, and J5) was mixed into 2 wt% BSA in 2 mM borate buffer (pH = 8.5) and used as the sample solution. The viral suspension containing inactivated SARS-CoV-2 in Vero cells was used at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, and 1
100, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilutions. The samples analyzed in the test were prepared to the final volume of 150 μL containing: 5 μL of each sample solution under investigation (i.e., P2, J4, J5; or a mixture of the three peptides; or samples of inactivated SARS-CoV-2 virus) and 145 μL of 2 wt% BSA in 2 mM borate buffer (pH = 8.5).
1000 dilutions. The samples analyzed in the test were prepared to the final volume of 150 μL containing: 5 μL of each sample solution under investigation (i.e., P2, J4, J5; or a mixture of the three peptides; or samples of inactivated SARS-CoV-2 virus) and 145 μL of 2 wt% BSA in 2 mM borate buffer (pH = 8.5).
2.9 Statistical analysis
The statistical analyses were performed using GraphPad Prism 5 software for Windows (GraphPad Software, Inc., La Jolla, CA, USA).3 Results and discussions
3.1 In silico modeling for synthetic peptides design and selection
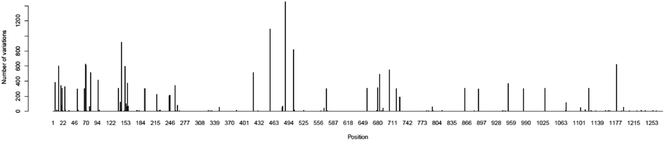
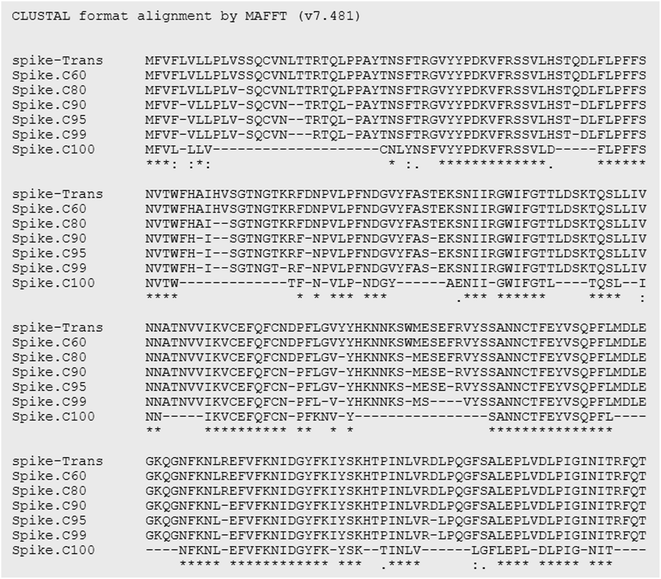
Essentially, peptides are short amino acid sequences that meet increasing interest as candidates for the diagnostic test. The rational design of peptides at protein/protein and protein/membrane interfaces is also raising interest. Overall, computational tools to assist the structural characterization of peptidic sequences and the exploration of peptide–protein interactions are needed.33–35 Peptides are potential targets for infectious disease diagnostics with several advantages. They can be easily synthesized, possess short lengths (usually 10–20 amino acids), and allow the selection of species-specific sequences. Moreover, the broad range of genomes/predicted proteomes of pathogens allows the evaluation of their conservation among strains. They may be used as a target to detect antibodies against a pathogen in patients as an indirect test or can be used to design antibodies that recognize the pathogen proteins as a direct test. In particular, regarding the SARS-CoV-2 virus, the early and rapid detection of infected cases and providing effective therapeutics against the disease are in urgent demand.Along with conventional clinical protocols, these specifically designed peptides can be conjugated to nanomaterials for developing innovative diagnostics and therapeutic alternatives to fight against the SARS-CoV-2 virus (coronavirus disease 2019, COVID-19).9,36 To select potential diagnostic targets in the SARS-CoV-2 spike protein, we evaluated its conservation in 2850 isolates from 11 different viral lineages (Table 1), showing that there are conserved regions interrupted by regions that vary among isolates (Fig. 4). In our approach, we aimed to select regions that were conserved in most isolates, as shown in Fig. 5.
Besides sequence conservation, we evaluated their potential antigenicity using a consensus combination of 6 B-cell epitope predictors based on linear and conformational sequences. A higher weight was given to the widely used programs BepiPred2.0, SVMTrip, ElliPro (conformational epitope prediction), and the NetSurfP program, which estimates the probability of the amino acid to be exposed in the protein structure to be recognized by antibodies. Finally, peptides similar to other Coronavirus, Flu viruses, or repetitive were excluded. We selected 57 peptides that met this criterion and were ranked based on their conservation score and prediction of B cell epitopes. Among them, we pre-selected 5 peptides for soluble synthesis, as described in Table 3. The molecular weight (MW), isoelectric point (Ip), aliphatic index (AI), and charge of peptides were predicted using the ProtParam website (https://web.expasy.org/cgi-bin/protparam/protparam).
| Peptide IDa | Immunogenicityb (%) | Conservationc | MW (Da) | Ip | CPd | CNe | AI |
|---|---|---|---|---|---|---|---|
| a Peptide sequences under Brazilian patent deposit pending.b Percentage of immunogenicity when compared with the highest scoring peptide in the spike protein, based on the combination of the B-cell epitope predictors.c Number of residues that were identical in at least 90% of the evaluated isolates.d Total number of positively charged peptides.e Total number of negatively charged peptides. | |||||||
| P1 | 99.99 | 100 | 2055.27 | 4.68 | 2 | 3 | 70.56 |
| P2 | 95.29 | 83.33 | 2288.57 | 8.44 | 2 | 3 | 37.78 |
| P3 | 92.94 | 77.78 | 2087.28 | 8.76 | 2 | 1 | 43.33 |
| J4 | 86.17 | 94.44 | 2090.28 | 8.15 | 2 | 1 | 81.11 |
| J5 | 98.82 | 77.78 | 1980.22 | 7.94 | 1 | 0 | 65.00 |
3.2 ELISA assay for peptide selection using human sera
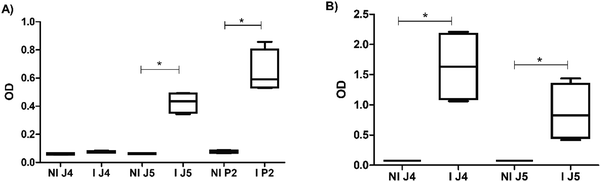
Once the peptides were synthesized, their reactivity to the sera from human patients was evaluated by ELISA as a standard immunoassay. It was observed that peptides P2, J4, and J5 were effective in differentiating between positive and negative human serum samples for SARS-CoV-2S, as shown in Fig. 6. P2 peptide proved less effective for differentiating positive and negative, not effectively recognizing positive samples and thus generating false negatives. These findings demonstrated the successful strategy of this work comprising the design, simulation (in silico), and synthesis of new peptides inspired by antigenic epitopes of SARS-CoV-2S (S-protein), which presented specificity for detecting IgG antibody of SARS-CoV-2-infected human patients. Thus, these promising results have led to the next stage of producing antibodies with the peptides in the rabbit animal model by hyperimmune sera, as reported in the next section.3.3 Hyperimmune serum production
With the three peptides defined in the previous section (i.e., P2, J4, and J5), hyperimmune sera production in rabbits was performed and validated by the ELISA test. Fig. 7 and S1 (ESI†) shows the reactivity of antibodies produced in rabbits inoculated with peptides. Fig. 7(A) indicated that J4 does not produce significant levels of antibodies after the third inoculation when compared with J5. Still, after the fourth inoculation, all three anti-peptide sera showed satisfactory results in ELISA.3.4 Hyperimmune serum semi-purification for immunoglobulin
After confirming the hyperimmune serum reactivity by ELISA, sera were semi-purified (as the methodology in Section 2.3) and aliquoted for use in LFA tests. The initial (total serum) and final (post-purification) dosage of sera can be observed in Table 4. At the end of production and semi-purification of hyperimmune sera, the final concentrations remained satisfactory for use in the LFA test.| Peptide | Initial volume (mL) | Final volume (mL) | Concentration of total protein (mg mL−1) |
|---|---|---|---|
| P2 rabbit 1 | 10 | 7.0 | 2.27 |
| P2 rabbit 2 | 10 | 7.3 | 3.80 |
| J4 rabbit 1 | 10 | 6.8 | 1.20 |
| J4 rabbit 2 | 10 | 7.0 | 1.00 |
| J5 rabbit 1 | 10 | 7.0 | 2.00 |
| J5 rabbit 2 | 10 | 7.4 | 2.12 |
3.5 Characterization of AuNPs and AuNP@antibody bioconjugates
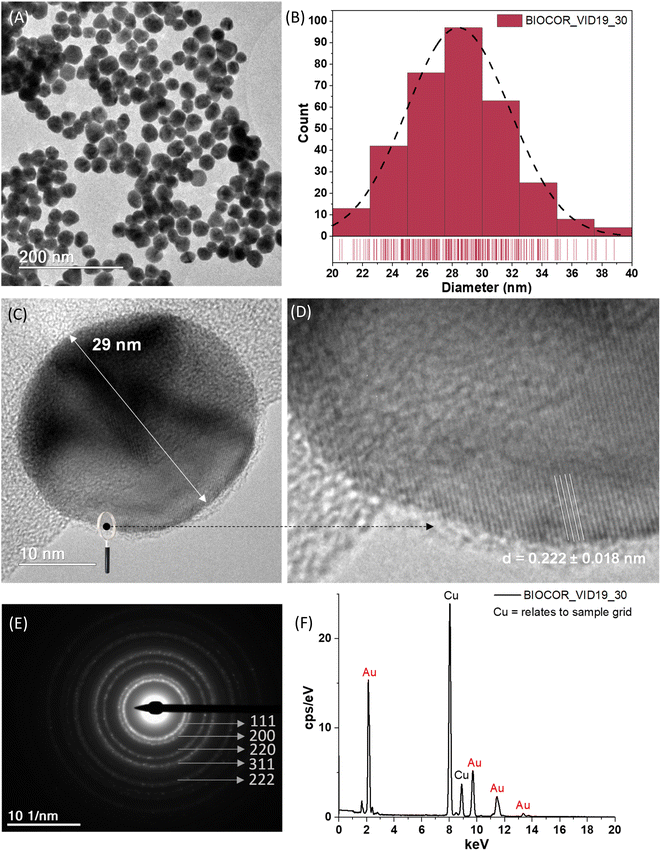
There has been a noticeable increase of novel nanomaterials for immunodiagnosis assays to improve the sensitivity and make them simpler and rapid to be widely accessible at the POC, which was drastically increased by the COVID-19 pandemic. Many nanomaterial-based tools for developing capture agents for virus detection by immunoassays, including LFA, have been developed in the last few years.21,37 Among several alternatives, those involving gold nanoparticles are highlighted due to the ease of synthesis, the wide range of physicochemical and unique optical properties that can be achieved by tuning the size and coupling ligands, and the extraordinary contrast achieved in the testing and control lines. Hence, this study developed a green aqueous process for synthesizing AuNPs through the in situ reduction in TSC and their controlled growth for producing nanoparticles of the expected size, size distribution, and surface chemistry to be applied in LFA tests. The results of the comprehensive characterization of the AuNPs, regarding their morphological features of AuNPs (BIOCOR_VID19_30), are shown in Fig. 8(A–C), including shape, size, and size distribution.In addition, the essential structural aspects, such as d-spacing measurements and crystallographic patterns, were determined by high-resolution images (HRTEM) and SAED, as depicted in Fig. 8(D and E). Also, the chemical analysis of AuNPs assessed by EDS is presented in Fig. 8(F). The step-by-step process for the synthesis using 7 stages of Au3+ and citrate additions cycles resulted in gold nanoparticles with an average size of 28 ± 3 nm (diameter), as evidenced in Fig. 8(A and B). It should be highlighted that based on this seed-mediated synthesis and controlled growth of the AuNP nanocolloids, it was possible to achieve a repeatable procedure to obtain projected sizes with a relatively low dispersity. These findings are significant because one crucial aspect associated with the color of the nanocolloids intrinsically depends on their size, and another because repeatability is vital for allowing the future scale-up to applications in the LFA tests.
In a general view, the morphology of the AuNPs revealed by TEM analyses was chiefly characterized by fairly round-shaped nanostructures. The HRTEM images shown in Fig. 8(C and D) evidenced one typical gold nanoparticle (AuNP, BIOCOR_VID19_30), with well-resolved interplanar spacing (d-spacing = 2.22 ± 0.18 Å), compatible with metallic Au (111) plane. Consistently, SAED analysis, shown in Fig. 8(E), revealed rings mostly related to characteristic reflections of crystalline face-centered-cubic (fcc) gold structures of (111), (200), (220), (311), and (222) (crystallographic database, JCPDS 04-0784). Such a trend confirmed the reduction of Au3+ ions into Au0 by citrate moieties and the formation of metallic gold nanoparticles (AuNPs), endorsed by the EDS spectrum of pure Au atoms (Fig. 8(F)). As expected, due to the extremely low contrast of organic species, compared to gold metallic “core”, citrates and biomolecules were not visible by TEM analysis. That would require special preparation of samples and more sophisticated equipment, which was not the scope of this study. Although previous works applied a layer-by-layer approach, also known as the “seed-mediated process”,20 these findings are relevant to synthesizing gold nanoparticles. The overall characteristics and parameters of the synthesis developed in this study are considered advances in producing uniform gold nanoparticles with tunable size and narrow size distribution through nucleation and controlled growth based on green colloidal chemistry, aiming at building an LFA assembly for immunoassay, which can be expanded to other biomedical applications.
Besides the formation of the AuNPs addressed in the analyses described in previous sections mainly focused on the metallic component, surface chemistry characterization plays a pivotal role in understanding the aqueous colloidal process, their physicochemical properties, and their prospective nanotechnology applications in the biomedical field, such as bioconjugation for immunoassays. Thus, the synthesis of the AuNPs and their conjugation with biomolecules was additionally monitored by changes in zeta potential (ZP, ζ) and hydrodynamic diameter (HD, dynamic light scattering, DLS). In addition, the optical properties were assessed via UV-vis spectroscopy. The UV-vis analyses were performed before and after the conjugation of AuNPs with anti-peptide antibodies for the nanoconjugates (BIOCOR_VID19_30-HAb_200J4; BIOCOR_VID19_30-HAb_200J5; BIOCOR_VID19_30-HAb_200P2), for demonstrating the occurrence of the coupling and interactions which promoted surface changes. To avoid redundancy, as the three synthesized peptides are very similar, and their induced antibodies share a very similar macromolecular structure of immunoglobulins (IgGs), one sample of bioconjugate was selected (BIOCOR_VID19_30-HAb_200X, X = J4, J5 or P2) to represent the results regarding their interactions with the gold nanoparticles (before conjugation, BIOCOR_VID19_30). Fig. 9(A–C) show HD, zeta potential, and UV-vis measurements. Digital images of the AuNPs before and after the coupling with anti-peptide antibodies are depicted in Fig. 9(D).
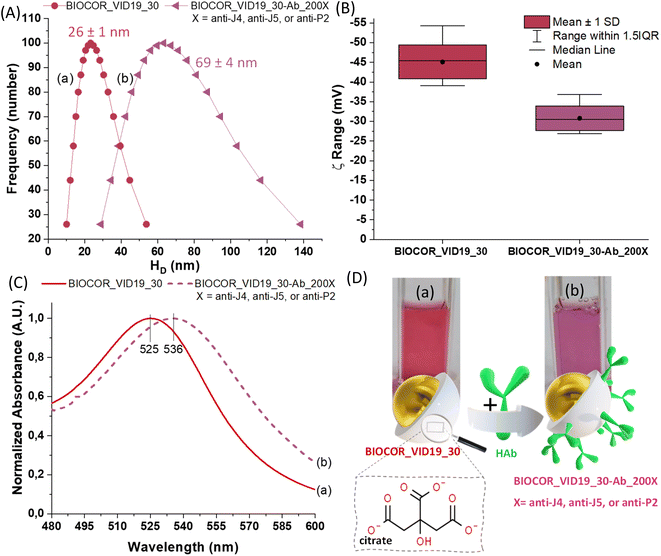 | ||
| Fig. 9 .(A) Size distribution, (B) zeta potential (ξ) distribution, (C) UV-vis, and (D) digital images and schematic illustration of BIOCOR_VID19_30 (a) and BIOCOR_VID19_30-Ab_200X (b). | ||
The hydrodynamic diameter (HD) of the bare AuNPs (BIOCOR_VID19_30) and the AuNP@antibody bioconjugates (BIOCOR_VID19_30-Hab) were analyzed by DLS, and the results are shown in Fig. 9(A). It is worth mentioning that BIOCOR_VID19_30 presented a polydispersity index (PDI) as low as 0.07, indicating a narrow interval of size distribution, characterizing a homogeneous formation of nanoparticles during the process. According to DLS measurements, the coupling reaction caused a significant enlargement of the main HD, from ∼30 nm to ∼70 nm, due to the presence of large-sized antibodies throughout the nanoparticles surface, which were previously covered only with small citrate molecules. These results demonstrated that an effective bioconjugation had taken place where pristine AuNPs were surrounded by anti-peptide antibodies leading to a relative increase in the “colloidal size”.
Before conjugation, ZP measurements of –45 ± 3 mV are consistent with the negatively charged ligand (citrate) at the surface of AuNPs, indicating well-stabilized nanoparticles predominantly due to electrostatic repulsion.18,38 After the conjugation of AuNPs with antibodies, part of the negatively charged citrate moieties was displaced by the large and more neutrally charged biomacromolecules (HAb), resulting in a noticeable shift of the surface charge measurements towards lower ZP (−31 ± 4 mV), as shown in Fig. 9(B).
From the UV-vis spectroscopy analysis, shown in Fig. 10(C), the AuNPs sample (BIOCOR_VID19_30) presents a surface plasma resonance (SPR) peak near 525 nm, which is characteristic of spherical nanogold with an average diameter of ∼30 nm stabilized by citrates,39 and with an extinction coefficient of εSPR = 3.81 × 109 M−1 cm−1. This plasmonic-related absorption, characteristic of AuNPs, is caused by the collective oscillation of the conduction electrons on the AuNP surface caused by the small dimensions of the nanoparticle compared to the excitation wavelength. Thus, considering the well-known SPR effect of metallic nanoparticles intrinsically affected by surface interaction phenomena, changes at the peak position (i.e., wavelength shifts) can be associated with surface chemistry modifications of the nanosystems. Hence, by comparing the curves (a) and (b) shown in Fig. 9(C), the coupling between AuNPs (BIOCOR_VID19_30) and antibodies (i.e., anti-peptides from hyperimmune sera) evidenced a significant SPR “redshift” in the UV-vis spectra of the bioconjugates (BIOCOR_VID19_30-Ab_200X). Such a trend indicates the surface biofunctionalization of the gold nanoparticles (BIOCOR_VID19_30) with the anti-peptides (HAb).18,38
Along with the SPR maxima shifts detected in the UV-vis spectra, qualitative visual changes in sample coloration, from reddish to purple (Fig. 9(D), digital images), endorsed the effective surface functionalization of the AuNPs with antibodies.
Bearing in mind that the AuNP-based bioconjugates were designed and developed for building LFA tests for detecting samples of infected humans by SARS-CoV-2, their relative concentration in the media and the proportion ratio of antibodies to gold nanoparticles are needed for comparison and sensitivity analyses purposes. However, the measurements of these characteristics are very complex, requiring extremely sensitive equipment combined with sophisticated techniques and protocols which are not readily available in most research facilities but also beyond the scope of this study. Nonetheless, in this work, these concentrations and relative ratios were estimated based on several plausible assumptions detailed in the ESI† (section Evaluation of AuNP, antibodies concentration, and AuNP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HAb ratio and Table S1†). In brief, by considering the chemical concentrations of reagents in the reaction during the synthesis of AuNPs and the average size experimentally measured via DLS their concentration can be theoretically calculated. Similarly, the initial antibody concentration assessed by the titration protocols (ELISA) and it was associated with a molecular weight of 150 kDa share high similarities with reasonably regular size and 3D conformation. That would offer a viable alternative to evaluate the relative concentration of AuNPs@antibody in the LFA systems.
HAb ratio and Table S1†). In brief, by considering the chemical concentrations of reagents in the reaction during the synthesis of AuNPs and the average size experimentally measured via DLS their concentration can be theoretically calculated. Similarly, the initial antibody concentration assessed by the titration protocols (ELISA) and it was associated with a molecular weight of 150 kDa share high similarities with reasonably regular size and 3D conformation. That would offer a viable alternative to evaluate the relative concentration of AuNPs@antibody in the LFA systems.
Therefore, the assessment of the AuNP to antibody ratio (BIOCOR_VID19_30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Hab) was performed based on the calculated AuNPs concentration in the medium (0.61 μM) and the theoretical HAb concentration in the medium (100 μg mL−1, 0.67 μM), considering a 100% coupling yield. Intending a more concentrated solution of nanoconjugates for colorimetric purposes in the LFA tests, the conjugated solution had the volume reduced to half of the original. Thus, for the AuNP@anti-peptides nanoconjugates (BIOCOR_VID19_30-Ab_200X; X = P2, J4, and J5), the final concentration of AuNPs = 1.22 μM, and the theoretical estimated concentration of anti-peptide (HAb) ∼200 μg mL−1 (1.32 μM), resulting in a molar ratio of 1
Hab) was performed based on the calculated AuNPs concentration in the medium (0.61 μM) and the theoretical HAb concentration in the medium (100 μg mL−1, 0.67 μM), considering a 100% coupling yield. Intending a more concentrated solution of nanoconjugates for colorimetric purposes in the LFA tests, the conjugated solution had the volume reduced to half of the original. Thus, for the AuNP@anti-peptides nanoconjugates (BIOCOR_VID19_30-Ab_200X; X = P2, J4, and J5), the final concentration of AuNPs = 1.22 μM, and the theoretical estimated concentration of anti-peptide (HAb) ∼200 μg mL−1 (1.32 μM), resulting in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.1 (AuNP
1.1 (AuNP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) anti-peptide).
anti-peptide).
Notably, within the statistical analysis variation, the HD values obtained by DLS (26 ± 1 nm) for gold nanocolloids (AuNPs, BIOCOR_VID19_30) well matched those of the inorganic core obtained by TEM (28 ± 3 nm), which is reasonable considering citrates are very small organic moieties as surface ligands. This trend is relevant to demonstrate the accuracy of the results obtained by different techniques and validate the nanoparticle size values used to estimate both nanoparticle concentration and reaction yield.
3.6 Lateral flow assay (LFA) for validation for diagnosis of SARS-CoV-2
After designing, synthesizing, characterizing, and validating each component, the final goal of this study was to build an LFA test for potential use as one simple and rapid diagnosis colorimetric assay for the detection of SARS-CoV-2-infected samples from human patients at the point-of-care (POC). In brief, the LFA test was assembled for performing the immunoassays using the novel-designed synthetic peptides (P2, J4, and J5) and conjugated with gold nanoparticles (BIOCOR_VID19_30-HAb_200J4; BIOCOR_VID19_30-HAb_200J5; BIOCOR_VID19_30-HAb_200P2).As a proof of concept and for analyzing possible cross-reactions, the synthetic peptides (P2, J4, and J5) were used as the samples in the LFA. Also, each antibody (i.e., anti-peptide from hyperimmune serum) was used in the bioconjugate solution as well as to label the LFA running membrane (end terminal pad). Fig. 10 illustrates the performance of the assembled LFA test using P2 and anti-P2-based samples as the affinity-binding biomolecules. Two peptide concentrations (P2) were tested (0.3 mg mL−1 (B), and 0.8 mg mL−1 (C)), but they did not directly correlate to the apparent visual intensity at the “test line”. LFA is essentially a qualitative assay; therefore, it depends on several other factors, including the precision of the dispensing tool (syringe, pipette, or automatic dispenser), the solution's concentration, the medium's viscosity, and the time elapsed after initiating the assay, etc.
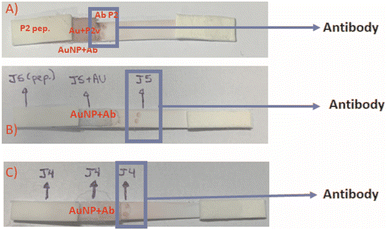
Similarly, the effective bioaffinity between each synthetic peptide with its respective antibody (produced through hyperimmune sera) after bioconjugation with AuNPs nanoprobes was also tested when applied in a lateral flow membrane assay. Fig. 11 shows LFA-based results using peptides P2, J4, and J5 as samples (sample-pad), the colloidal solution of AuNPs conjugated with the corresponding anti-peptides (i.e., AuNPs@anti-P2, AuNPs@anti-J4, AuNPs@anti-J5), and the final positive colorimetric responses.
All the LFA results associated with the validation of the three synthetic peptides, their antibodies, and the AuNP nanoconjugates for application in SARS-CoV-2 diagnosis are summarized in Tables 5 and 6.
| Samples | AuNp solution + Ab conjugate | Hyperimmune serum – Ab | Result |
|---|---|---|---|
| Peptide P2 | AuNp + anti-P2 | Ab P2+J5 | Reacted |
| Peptide P2 | AuNp + anti-P2 | Ab J4 | Reacted |
| Peptide P2 | AuNp + anti-P2 | Ab J5 | Reacted |
| Peptide P2 | AuNp + anti-P2 + anti-J4 | Ab P2 | Reacted |
| Peptide P2 | AuNp + anti-P2 + anti-J5 | Ab P2 | Reacted |
| Peptide P2 | AuNp + P2 + anti-J4 + anti-J5 | Ab P2 | Reacted |
| Peptide P2 | AuNp + anti-J4 + anti-J5 | Ab P2 | Reacted |
| Peptide J4 | AuNp + anti-J4 | Ab J4 | Reacted |
| Peptide J4 | AuNp + anti-J4 | Ab negative + serum | Not reacted |
| Peptide J4 | AuNp + anti-J4 | Ab J5 | Reacted |
| Peptide J4 | AuNp + anti-J4 | Ab P2 | Reacted |
| Peptide J4 | AuNp + anti-P2 + anti-J4 | Ab J4 | Not reacted |
| Peptide J4 | AuNp + anti-P2 + anti-J5 | Ab J4 | Not reacted |
| Peptide J4 | AuNp + anti-P2 + anti-J4 + anti-J5 | Ab J4 | Reacted |
| Peptide J4 | AuNp + anti-J4 + anti-J5 | Ab J4 | Reacted |
| Peptide J5 | AuNp + anti-J5 | Ab J5 | Reacted |
| Peptide J5 | AuNp + anti-J5 | Ab J4 | Not reacted |
| Peptide J5 | AuNp + anti-J5 | Ab P2+J5 | Reacted |
| Peptide J5 | AuNp + anti-J5 | Ab P2 | Reacted |
| Peptide J5 | AuNp + anti-J5 | Ab negative + serum | Not reacted |
| Peptide J5 | AuNp + anti-P2 + anti-J4 | Ab J5 | Reacted |
| Peptide J5 | AuNp + anti-P2 + anti-J5 | Ab J5 | Reacted |
| Peptide J5 | AuNp + anti-P2 + anti-J4 + anti-J5 | Ab J5 | Reacted |
| Peptide J5 | AuNp + anti-J4 + anti-J5 | Ab J5 | Reacted |
| Samples | AuNp solution + Ab conjugate | Hyperimmune serum – Ab | Result |
|---|---|---|---|
| Inactivated SARS-CoV-2 pure | AuNp + anti-P2 | Ab P2 | Reacted |
| Inactivated SARS-CoV-2 pure | AuNp + anti-J4 | Ab J4 | Reacted |
| Inactivated SARS-CoV-2 pure | AuNp + anti-J5 | Ab J5 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
AuNp + anti-P2 | Ab P2 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
AuNp + anti-J4 | Ab J4 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
AuNp + anti-J5 | Ab J5 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
AuNp + anti-P2 | Ab P2 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
AuNp + anti-J4 | Ab J4 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
AuNp + anti-J5 | Ab J5 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
AuNp + anti-P2 | Ab P2 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
AuNp + anti-J4 | Ab J4 | Reacted |
Inactivated SARS-CoV-2 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
AuNp + anti-J5 | Ab J5 | Reacted |
As the rapid response is one of the vital requirements for the diagnosis of COVID19, it should be emphasized that all LFA results presented in this study were obtained in less than 10 min from the initial contact of the sample pad, regardless of the peptide or anti-peptide applied in the test. These results are comparable (Table S2 ESI†) to or even shorter testing times than other POC tests for SARS detection.14,21 As evidenced in Tables 5, 6 and Fig. S2–S4 (ESI†), all three designed synthetic peptides were capable of recognizing and coupling their respective anti-peptide (antibody). These peptides in the running samples (peptides diluted in borate buffer) were capable of binding onto antibodies present in the conjugated solution (i.e., AuNP + antibodies). In the sequence, during the membrane fluid flow, they effectively bound to the antibody present in the “test line”, producing a visually detectable reddish-purple colorful signal (Fig. 10 and 11), which is caused by the accumulation of Au-based nanoconjugates. Thus, these results validated the LFA immunoassay as the primary goal of this study.
As the negative control, sera from rabbits collected before the hyperimmune procedure (i.e., prior to the inoculation of peptides) were applied as they lacked anti-peptide antibodies. The non-appearance of a reddish signal in the test line characterized the absence of non-specific responses in the tests. Notably, the developed LFA systems did not return false-positive results, as shown in Fig. 11(A), even using HAb concentrations in the test lines as high as [HAb] = 2.27 mg mL−1. These findings are critical when developing reliable immunoassays because false-positive results can lead to incorrect actions by health professionals considering “a healthy” individual as an “infected patient”.
Combined peptide solutions (mixture of peptides) were also used to conjugate AuNPs for producing and detecting agents and serving as capture agents in the test spots. This approach aimed at observing if the LFA test's response could be enhanced. As shown in Table 5, the obtained responses did not differ from those acquired using single peptide conjugated solutions, demonstrating that there is no boost of response for more than one peptide in the conjugated solution or the test spot labeling. This simplifies the LFA test and also lowers its costs.
Regarding the cross-reactivity aspect, the P2 peptide presented a more significant cross-reactivity with the other peptides (J4 and J5), as already observed in the previous section based on ELISA standard assay (Fig. S1 ESI†). As all studied peptides were designed from conserved and immunogenic parts of a single protein, the spike-protein from SARS-CoV-2 (SARS-CoV-2S), such a trend was expected. These peptides share a significant correlation among their sequences, thus leading to antigen/antibody cross-recognition between analogous or similar parts.
Once the first set of LFA tests with the peptides and their antibodies were validated, the conclusive stage of evaluation using actual inactivated SARS-CoV-2S samples was performed, using pure and in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilutions; 1
10 dilutions; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 1
100 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 (Table 6 and Fig. S4 ESI†). As shown in Fig. 12, for the LFA tests, the effective specific binding between antigen–antibody has occurred, producing color at the “test line” and, more importantly, validating the new LFA-based diagnosis of this work. The “control line”, where a commercial anti-rabbit antibody was used, reacted adequately, producing color and confirming the LFA test.
1000 (Table 6 and Fig. S4 ESI†). As shown in Fig. 12, for the LFA tests, the effective specific binding between antigen–antibody has occurred, producing color at the “test line” and, more importantly, validating the new LFA-based diagnosis of this work. The “control line”, where a commercial anti-rabbit antibody was used, reacted adequately, producing color and confirming the LFA test.
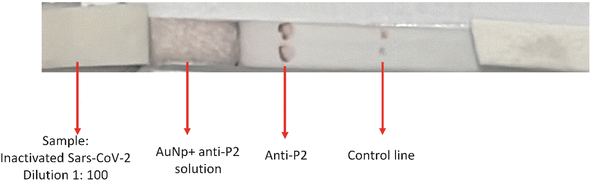 | ||
Fig. 12 .LFA using inactivated SARS-CoV-2 dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, reacting with AuNP + anti-P2 solution and anti-P2. Control line with commercial anti-rabbit antibody. 100, reacting with AuNP + anti-P2 solution and anti-P2. Control line with commercial anti-rabbit antibody. | ||
Thus, based on these results, it is worth mentioning that, compared to other similar assays reported in the literature for COVID-19 rapid diagnosis,12 our newly developed LFA test presents a reliable rapid qualitative color reading detection response within 10 minutes after applying the sample, which endows applicability at the point-of-care by individuals in general and health professionals. Although beyond the scope of this research, it can be envisioned that further studies will need to address other parameters, including the limit of detection, sensitivity, and accuracy for the specificity of detection of COVID-19, where other viruses that could potentially cause cross-reactions, would be discarded (e.g., influenza, dengue, etc.).
4 Conclusions
In this study, it can be summarized that novel SARS-CoV-2-inspired synthetic peptides were in silico designed, modeled, and synthesized. The 3 most suitable peptide sequences were selected based on a set of biochemical and immunological assays to be used as antigenic agents to induce immunogenic responses in vivo using a rabbit animal model for producing hyperimmune serum. The hyperimmune sera were semi-purified and validated for preliminary antibody-antigenic recognition with the respective synthetic peptides (P2, J4, J5, and anti-P2, anti-J4, and anti-J5, respectively). In addition, based on a nanotechnology approach, gold nanoparticles (AuNPs) were synthesized through a green process, producing uniformly spherical nanoparticles with an average size of 29 nm and surface potential of - 45 ± 3 mV. The AuNPs presented an optical absorption maximum at approximately 525 nm, suitable to be used as colorimetric nanoprobes in immunoassays. Thus, the immunogenic synthetic peptide sequences and antibodies (P2, J4, J5, and anti-P2, anti-J4, and anti-J5) were bioconjugated with the AuNPs. They were comprehensively characterized before and after conjugation with the biomolecules (peptides or anti-peptides), demonstrating successful coupling of the nanoassemblies. These AuNP-labeled nanoconjugates were used for producing colorimetric LFA assays based on the detection of the 3 peptides and the inactivated SARS-CoV-2 virus. The results demonstrated that the LFA test effectively detected SARS-CoV-2 virus-infected samples based on the novel in silico-designed synthetic peptides and bioconjugates developed through nanotechnology tools, producing innovative, rapid, and simple immunoassays for potential application at the point-of-care (POC) by individuals in general and health professionals. Therefore, it can be envisioned that the successful strategy adopted in this research, integrating several professionals from different realms of knowledge, spanning from nanotechnology to immunology, encompassing chemistry, materials science, engineering, bioinformatics, and medicine, could be applied to face challenges for the rapid diagnosis of pathogenic-related diseases.Author contributions
Beatriz Senra Alvares da Silva Santos: conceptualization, methodology, visualization, investigation, formal analysis, writing – original draft, writing – review & editing. João Luis Reis Cunha: conceptualization, methodology, visualization, investigation, formal analysis, writing – original draft. Isadora Cota Carvalho: conceptualization, methodology, visualization, investigation, formal analysis, writing – original draft, writing – review & editing. Julia Machado Caetano Costa: methodology, visualization, investigation, formal analysis. Grazielle Cossenzo Florentino Galinari: methodology, visualization, investigation, formal analysis. Bárbara Chrispin Longo: methodology, visualization, investigation, formal analysis. Paulo Henrique Souza Marazzi Diniz: methodology, visualization, investigation, formal analysis. Gabriel Moreira de Mello Mendes: methodology, visualization, investigation, formal analysis. Jônatas Santos Abrahão: production and inactivation of SARS – Cov-2, methodology, visualization, investigation, formal analysis. Flávio Guimarães da Fonseca: peptide validation ELISA, methodology, visualization, investigation, formal analysis. Alexandra Ancelmo Piscitelli Mansur: conceptualization, methodology, funding acquisition, writing – review & editing, resources. Maria de Fátima Leite: supervision, conceptualization, methodology, visualization, validation, funding acquisition, writing – original draft, writing – review & editing, resources. Herman Sander Mansur: supervision, conceptualization, methodology, visualization, validation, funding acquisition, writing – original draft, writing – review & editing, resources, project administration. Zélia Inês Portela Lobato: supervision, conceptualization, methodology, visualization, validation, funding acquisition, writing – original draft, writing – review & editing, resources, project administration. Rodrigo Lambert Orefice: supervision, conceptualization, methodology, visualization, validation, funding acquisition, writing – original draft, writing – review & editing, resources, project administration.Conflicts of interest
The authors confirm no competing interests to declare regarding the publication of this article.Acknowledgements
This work was financially funded by the Brazilian Government Research Agencies: CAPES (Programa Estratégico Emergencial de Combate a Surtos, Endemias, Epidemias e Pandemias – 0683/2020; PROINFRA-2010–2014; PROEX-2010-2021; PNPD-2014-2020); CNPq (PQ1A-303893/2018-4; PDS-103138/2020-0; PIBIC-2017-18-21; UNIVERSAL-421312/2018-1); FAPEMIG (UNIVERSAL-APQ-00291-18; PROBIC-2018); and FINEP (CTINFRA/PROINFRA 2008/2010/2011/2018; SOS/Equipamentos/2018 - 01.19.0032.00). PRPQ/PRPG-UFMG, Special Funding for Open-access publications. The authors would like to express their gratitude to the staff of the Center of Nanoscience, Nanotechnology, and Innovation-CeNano2I/CEMUCASI/UFMG for spectroscopy and of the Microscopy Center/UFMG for performing the HR-TEM analysis. We dedicate this research to all patients, doctors, health professionals, and scientists that tirelessly fought this everyday battle against COVID19 with diligence and courage.References
- N. H. Anh, et al, Gold nanoparticle-based optical nanosensors for food and health safety monitoring: recent advances and future perspectives, RSC Adv., 2022, 12, 10950–10988 RSC.
- L. Mulder, et al, A Comparative Study of Nine SARS-CoV-2 IgG Lateral Flow Assays Using Both Post-Infection and Post-Vaccination Samples, J. Clin. Med., 2022, 11, 2100 CrossRef CAS PubMed.
- D. Cucinotta and M. Vanelli, WHO declares COVID-19 a pandemic, Acta Biomedica, 2020, 91, 157–160 Search PubMed.
- WHO, World Health Organization, Weekly epidemiological update on COVID-19, 2021, vol. 69, available at: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19–-7-december-2021 Search PubMed.
- B. Bruijns, L. Folkertsma and R. Tiggelaar, FDA authorized molecular point-of-care SARS-CoV-2 tests: A critical review on principles, systems and clinical performances, Biosens. Bioelectron.: X, 2022, 11, 100158 CAS.
- I. J. B. Do Nascimento, et al, Hospitalization, mortality and public healthcare expenditure in Brazil during the COVID-19 crisis: vulnerabilities in the spotlight, Sao Paulo Med. J., 2022, 140, 290–296 CrossRef PubMed.
- Lancet, Editorial COVID-19 : learning from experience, Lancet, 2020, 395, 1011 CrossRef.
- M. M. Santos, et al, Brazilian Older People Hospitalized by COVID-19: Characteristics and Prognostic Factors in a Retrospective Cohort Study, 2021 Search PubMed.
- M. A. Derakhshan, A. Amani and R. Faridi-Majidi, State-of-the-Art of Nanodiagnostics and Nanotherapeutics against SARS-CoV-2, ACS Appl. Mater. Interfaces, 2021, 13, 14816–14843 CrossRef CAS PubMed.
- C. Weiss, et al, Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic, ACS Nano, 2020, 14, 6383–6406 CrossRef CAS PubMed.
- H. Che, et al, A rapid water bath PCR combined with lateral flow assay for the simultaneous detection of SARS-CoV-2 and influenza B virus, RSC Adv., 2022, 12, 3437–3444 RSC.
- U. Amara, et al, Insight into prognostics, diagnostics, and management strategies for SARS CoV-2, RSC Adv., 2022, 12, 8059–8094 RSC.
- R. Medhi, et al, Nanoparticle-Based Strategies to Combat COVID-19, ACS Appl. Nano Mater., 2020, 3, 8557–8580 CrossRef CAS.
- A. Pramanik, et al, Bioconjugated Nanomaterial for Targeted Diagnosis of SARS-CoV-2, Acc. Mater. Res., 2022, 3, 134–148 CrossRef CAS.
- C. M. Bachman, et al, Clinical validation of an open-access SARSCOV-2 antigen detection lateral flow assay, compared to commercially available assays, PLoS ONE, 2021, 16, 1–12 CrossRef PubMed.
- K. Cortés-Sarabia, et al, Utility of in silico-identified-peptides in spike-S1 domain and nucleocapsid of SARS-CoV-2 for antibody detection in COVID-19 patients and antibody production, Sci. Rep., 2022, 12, 15057 CrossRef PubMed.
- Y. Badhe, R. Gupta and B. Rai, In silico design of peptides with binding to the receptor binding domain (RBD) of the SARS-CoV-2 and their utility in bio-sensor development for SARS-CoV-2 detection, RSC Adv., 2021, 11, 3816–3826 RSC.
- A. J. Caires, et al, Gold nanoparticle-carboxymethyl cellulose nanocolloids for detection of human immunodeficiency virus type-1 (HIV-1) using laser light scattering immunoassay, Colloids Surf., B, 2019, 177, 377–388 CrossRef CAS PubMed.
- A. J. Caires, et al, A carboxymethylcellulose-mediated aqueous colloidal process for building plasmonic–excitonic supramolecular nanoarchitectures based on gold nanoparticles/ZnS quantum emitters for cancer theranostics, Green Chem., 2021, 23, 8260–8279 RSC.
- N. G. Bastús, J. Comenge and V. Puntes, Kinetically controlled seeded growth synthesis of citrate-stabilized gold nanoparticles of up to 200 nm: Size focusing versus Ostwald ripening, Langmuir, 2011, 27, 11098–11105 CrossRef PubMed.
- J. Wang, et al, Gold nanoparticles in virus detection: Recent advances and potential considerations for SARS-CoV-2 testing development, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, 1–30 Search PubMed.
- J. Heskin, et al, Evaluating the performance characteristics of five lateral flow assays for the detection of the SARS – CoV – 2 nucleocapsid antigen, Sci. Rep., 2022, 1–6 Search PubMed.
- M. Harun-Ur-Rashid, et al, Rapid diagnosis of COVID-19 via nano-biosensor-implemented biomedical utilization: a systematic review, RSC Adv., 2022, 12, 9445–9465 RSC.
- K. Katoh and D. M. Standley, MAFFT multiple sequence alignment software version 7: Improvements in performance and usability, Mol. Biol. Evol., 2013, 30, 772–780 CrossRef CAS PubMed.
- G. Sanchez, Las instituciones de ciencia y tecnología en los procesos de aprendizaje de la producción agroalimentaria en Argentina, in El sistema argentino de innovación: instituciones, empresas y redes: el desafío de la creación y apropiación de conocimiento, ed. D. Suárez, Universidad Nacional de General Sarmiento, Los Polvorines, 1a edn, 2013, pp. 285–310 Search PubMed.
- M. C. Jespersen, et al, BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes, Nucleic Acids Res., 2017, 45, W24–W29 CrossRef CAS PubMed.
- H. Singh, H. R. Ansari and G. P. S. Raghava, Improved Method for Linear B-Cell Epitope Prediction Using Antigen's Primary Sequence, PLoS ONE, 2013, 8, 1–8 CrossRef.
- S. Sinha, R. K. Grewal and S. Roy, Modeling phage–bacteria dynamics Methods Mol. Biol., 2020, 2131, 309–327 Search PubMed.
- J. Kringelum, et al, Reliable B Cell Epitope Predictions: Impacts of Method Development and Improved Benchmarking, PLoS Comput. Biol., 2012, 8, e1002829 CrossRef CAS PubMed.
- J. Ponomarenko, et al, ElliPro: A new structure-based tool for the prediction of antibody epitopes, BMC Bioinf., 2008, 9, 1–8 CrossRef PubMed.
- S. F. Altschul, et al, Basic local alignment search tool, J. Mol. Biol., 1990, 215, 403–410 CrossRef CAS PubMed.
- N. J. Kruger, The Bradford Method for Protein Quantitation, The Protein Protocols Handbook, 1996, vol. 32, p. 15–20 Search PubMed.
- W. K. B. Chan, et al, In silico analysis of SARS-CoV-2 proteins as targets for clinically available drugs, Sci. Rep., 2022, 12, 1–12 CrossRef PubMed.
- A. Lamiable, et al, PEP-FOLD3: faster denovo structure prediction for linear peptides in solution and in complex, Nucleic Acids Res., 2016, 44, W449–W454 CrossRef CAS PubMed.
- S. Rajpoot, et al, In silico design of a Novel Tridecapeptide targeting spike Protein of SARS-CoV-2 Variants of Concern, Int. J. Pept. Res. Ther., 2022, 28, 1–15 CrossRef.
- H. Can, et al, In silico discovery of antigenic proteins and epitopes of SARS-CoV-2 for the development of a vaccine or a diagnostic approach for COVID-19, Sci. Rep., 2020, 10, 1–16 CrossRef PubMed.
- P. Sadeghi, et al, Lateral flow assays (LFA) as an alternative medical diagnosis method for detection of virus species: The intertwine of nanotechnology with sensing strategies, TrAC, Trends Anal. Chem., 2021, 145, 116460 CrossRef CAS PubMed.
- S. G. Hwang, et al, Rapid and simple detection of Tamiflu-resistant influenza virus: Development of oseltamivir derivative-based lateral flow biosensor for point-of-care (POC) diagnostics, Sci. Rep., 2018, 8, 1–11 Search PubMed.
- Daruich De Souza, B. Ribeiro Nogueira and M. E. C. M. Rostelato, Review of the methodologies used in the synthesis gold nanoparticles by chemical reduction, J. Alloys Compd., 2019, 798, 714–740 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra05096j |
| ‡ Present address: Federal University of Minas Gerais, Av. Antônio Carlos, 6627 – Escola de Engenharia, Bloco 2 – Sala 2233, 31.270–901, Belo Horizonte/MG, Brazil., E-mail: hmansur@demet.ufmg.br; Fax: +55-31-34091843; Tel: +55-31-34091843. |
| This journal is © The Royal Society of Chemistry 2022 |