Open Access Article
Open Access ArticleHighly sensitive benzothiazole-based chemosensors for detection and bioimaging of peroxynitrite in living cells†
Yaqiong
Kong
 a,
Rong
Wu
a,
Xiaodong
Wang
a,
Guoxu
Qin
a,
Fengyi
Wu
a,
Rong
Wu
a,
Xiaodong
Wang
a,
Guoxu
Qin
a,
Fengyi
Wu
 a,
Chunyu
Wang
a,
Chunyu
Wang
 ac,
Minmin
Chen
a,
Nannan
Wang
*a,
Qian
Wang
*b and
Duojun
Cao
*a
ac,
Minmin
Chen
a,
Nannan
Wang
*a,
Qian
Wang
*b and
Duojun
Cao
*a
aEngineering Technology Center of Department of Education of Anhui Province, Institute of Novel Functional Materials and Fine Chemicals, College of Chemistry and Materials Engineering, Chaohu University, Chaohu 238024, PR China. E-mail: 053076@chu.edu.cn; nnw1990@126.com
bDepartment of Radiation Oncology, China-Japan Union Hospital of Jilin University, Changchun 130000, PR China. E-mail: wangqian1991@jlu.edu.cn
cSchool of Information Science and Technology, University of Science and Technology of China, Hefei, Anhui 230026, China
First published on 30th September 2022
Abstract
It is well accepted that peroxynitrite (ONOO−) plays a crucial role in various physiological and pathological processes. Thus, the detection and imaging of ONOO− in vitro and in vivo with high selectivity and sensitivity is of great significance. Here we report two simple benzothiazole-based fluorescent chemosensors, BS1 and BS2. Under physiological pH, both probes could quickly sense ONOO− with a remarkable “turn-on” fluorescence signal at 430 nm. The limit of detection (LOD) of BS1 and BS2 toward ONOO− was 12.8 nM and 25.2 nM, respectively, much lower than the reported values. Experimental results indicated that BS1 with a diphenyl phosphonate unit presented higher selectivity for ONOO− than BS2. Furthermore, based on the advantages of lower cytotoxicity and pH-stabilities of BS1, probe BS1 was successfully employed to detect and image ONOO− in HepG2 cells. More importantly, we used BS1 to successfully showcase drug-induced hepatotoxicity via imaging ONOO− upregulated by acetaminophen (APAP), and also evaluated the remediation effect of GSH. All the results illustrated that the fluorescent probe BS1 has great potential for the detection of ONOO− and to further uncover the roles of ONOO− during the drug-induced liver injury (DILI) process.
1. Introduction
The liver is a very important organ in the human body, which has the function of regulating metabolism and drug detoxification.1 Drug-induced liver injury (DILI) has always been common in clinical patients and could cause acute liver failure, which generally endangers human health and even life.2 According to statistics, the annual incidence of DILI is approximately one in 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people.3,4 Consequently, the development of an accurate and reliable method for early diagnosis of drug-induced hepatotoxicity is necessary and beneficial. However, due to the complexity of the pathophysiological mechanisms, the early diagnosis of DILI by the Roussel Uclaf causality assessment method (RUCAM) involved some drawbacks, such as relatively cumbersome operations and delayed diagnosis, and has rarely obtained satisfactory predicted results.5 Generally, some reports revealed that drug metabolism could cause liver injury with the generation of reactive species including reactive oxygen species (ROS) and reactive nitrogen species (RNS).6,7 Therefore, ROS and RNS could be used as biomarkers for the early diagnosis of drug-related hepatotoxicity.8,9
000 people.3,4 Consequently, the development of an accurate and reliable method for early diagnosis of drug-induced hepatotoxicity is necessary and beneficial. However, due to the complexity of the pathophysiological mechanisms, the early diagnosis of DILI by the Roussel Uclaf causality assessment method (RUCAM) involved some drawbacks, such as relatively cumbersome operations and delayed diagnosis, and has rarely obtained satisfactory predicted results.5 Generally, some reports revealed that drug metabolism could cause liver injury with the generation of reactive species including reactive oxygen species (ROS) and reactive nitrogen species (RNS).6,7 Therefore, ROS and RNS could be used as biomarkers for the early diagnosis of drug-related hepatotoxicity.8,9
Peroxynitrite (ONOO−), a highly reactive oxygen and nitrogen species (RONS), originates from the chemical transformation between nitric oxide (˙NO) and superoxide anions (O2˙−).10–14 It has been universally known that ONOO− is not only a kind of biological endogenous oxidant but an efficient nitration agent, which could easily nitrify or oxidize amino acids like tyrosine and consume biothiols, and consequently do irreversible damage to proteins, nucleic acid and lipids.15–19 Therefore, the level change of ONOO− in the organism could been considered as an important pathogenic indicator for early diagnosis of DILI.17,20–24 Despite of the general recognition in the formation of ONOO− and its importance in physiological and pathological processes, it has still many difficulties in tracking and monitoring directly in situ. The reasons lie in dynamic changes of ONOO− concentration and short half-life time (∼10 ms) in vivo and in vitro as well as mutual competition and inter-conversion among endogenous biomolecules, which make selective detection of ONOO− in intricate physiological environment more challenging.11,25–27 Thus, given the significant role and specialty of ONOO−, it is in urgent need of a reliable and effective analytical method in detecting and visualizing ONOO− in practical biological systems.
Fortunately, owing to the real-time visualization, instantaneous response, convenience and noninvasive nature, fluorescence-based techniques makes accurate and efficient determination and imaging of ONOO− possible.28–32 Recently, much efforts have been devoted to exploiting and designing novel fluorescent ONOO− probes with excellent selectivity and sensitivity. From the recent reviews of fluorescent chemosensors for ONOO− detection, it could be concluded that these fluorescent ONOO− probes were developed based on the strong oxidation, nitration, and nucleophilicity of ONOO−, which usually incorporate some particular reactive moieties into different chromophores, such as alkene,33,34 N-aminophenol,35,36 active ketone,37–39 arylboronic esters,40–43 organoselenium/organotellurium44,45 and diphenyl phosphonate unit.46,47 Taking advantage of these reactive sites, the probes would release their masked fluorescence upon attacked by ONOO−, which makes them promising biosensors for ONOO− detection and imaging. However, there are also many disadvantages of sensors reported already, like poor biocompatibility, photobleaching, low sensitivity, obvious interference from other RONS, making them unsuitable for the determination of ONOO− in real bio-samples. Herein, in face of the arduous problems, it is still of great significance to exploit exquisite fluorescent ONOO− probes and uncover the biofunctions of ONOO− in living systems.
Inspired by the previous works, two simple benzothiazole-based fluorescent probes, BS1 and BS2, for the detection and imaging of ONOO− in vitro and vivo were designed for that benzothiazole probes share the merits of high quantum yield and large Stokes shift.48 And the two probes were easily obtained based on the condensation reaction of 2-aminothiophenol and aromatic aldehydes in the presence of catalytic amount of lanthanum(III) nitrate hexahydrate in a considerable yield under mild conditions.49 The difference of the probes in structure is that BS1 possesses a reactive electron withdrawing diphenyl phosphonate unit while BS2 bears a pinacol boronic ester group for monitoring ONOO−. Both probes showed excellent sensitivity and rapid response toward ONOO− in comparison with the previously reported ONOO− probes (Table S1†). However, it's worth noting that the probe BS1 with diphenyl phosphonate unit showed better selectivity than BS2 bearing a pinacol boronic ester group. Thus, BS1 was then employed to monitor the fluctuation of ONOO− level regulated by antipyretic acetaminophen (APAP) drug via fluorescence imaging. Encouragingly, with the help of probe BS1, the experiments demonstrated that APAP-induced hepatotoxicity was accompanied with the up-regulation of ONOO−. Moreover, the use of GSH (ONOO− scavenger) could alleviate drug-induced damage, which might open the door for the nontoxic metabolism of APAP. This study would provide a potential approach for the early diagnosis and therapy of DILI.
2. Experimental
Materials and instruments, synthesis and characterization of BS1 and BS2, general procedure for spectroscopic studies, cell culture and fluorescent imaging studies were listed in the ESI.†3. Results and discussions
3.1 Synthesis of probes BS1 and BS2
The reasonable route for preparing sensors BS1 and BS2 were presented in Scheme 1. Structurally, the two simple probes BS1 and BS2 share same benzothiazole moiety and were prepared in a similar approach where La(NO3)3·6H2O served as an efficient catalyst. The concrete synthesis information was outlined in ESI.† The structures of probes BS1, BS2 and intermediates involved were exactly characterized through NMR and HR-MS spectroscopy (Fig. S1–S7†).3.2 The optical response of BS1 and BS2 toward ONOO−
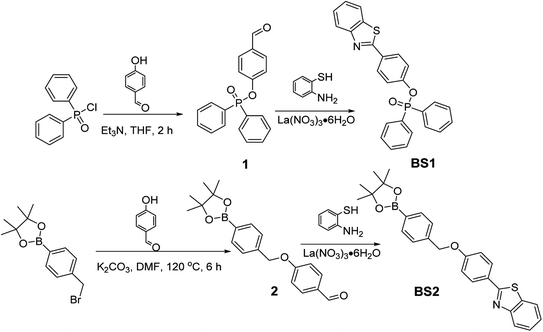
The response of BS1 and BS2 toward ONOO− were evaluated in the DMSO/PBS solution (10 µM, 25% DMSO in PBS, pH = 7.4). As shown in Fig. 1a and S14a,† the free BS1 and BS2 solutions showed their maximum absorption peaks centered approximately at 305 nm and 300 nm, respectively, which could be assigned to the characteristic absorption of benzothiazole unit.50 However, the absorption spectra exhibited apparent bathochromic shift (356 nm) when the above solutions were treated with ONOO−. Furthermore, in the absence of ONOO−, both of the solutions almost had no emission at 430 nm while significant fluorescence enhancement with remarkable color changing from colorless to bright blue were observed after the addition of ONOO− (insets of Fig. 1a and S14a†), which were due to the ONOO−-promoted deprotection of diphenyl phosphonate group and hydrolysis of benzeneboronic ester.43,47 Meanwhile, relative fluorescent quantum yield(ΦF) of BS1 and BS2 varied from 0.05% to 48.9% and from 0.03% to 42.7%, respectively, where the ethanolic solution of anthracene (ΦF = 27%) was used as a ref. 51. The detailed photophysical properties of BS1 and BS2 were listed in the Table S2.†The concentration-dependent fluorescence titrations of BS1 and BS2 toward ONOO− were also investigated in the DMSO/PBS solution at ambient temperature. As seen from Fig. 1b and S14b,† upon the excitation at 365 nm, both of emission intensities gradually rose with the increasing concentration of ONOO−. After addition of 6 µM ONOO− into the solution of BS1, the emission signaling achieved a plateau with a nearly 850-fold fluorescent enhancement (Fig. S13a†). The similar enhancement was observed for BS2 when the concentration of ONOO− reached 10 µM (Fig. S15a†). What is noteworthy is that there were excellent linear relationships (R2BS1 = 0.9989, R2BS2 = 0.9980) between ONOO− concentration and emission intensities of probes (Fig. S13b and S15b†). And the limit of detection (LOD) were calculated to be 12.8 nM and 25.2 nM, respectively. Additionally, time-dependent fluorescence changes of BS1 and BS2 were carried out as well. The probes had a rapid response toward ONOO− and reached their equilibrium states within 300 s (Fig. S18†). And according to the formula Ln[(Fmax − Ft)/Fmax] = −k't,52,53 their pseudo-first-order rate constants were determined as 1.49 × 10−2 s−1 (Fig. S19a†) and 1.23 × 10−2 s−1 (Fig. S19b†), respectively. Altogether, the above performances illustrated that the probes presented highly sensitive and rapid response to ONOO−.
With regard to practical applications of a good chemosensor, its characteristic pH-dependent feature must be taken carefully account. The fluorescence response of BS1 and BS2 with and without ONOO− under different pH conditions were investigated and the results were presented in Fig. S21.† Both solutions of free BS1 and BS2 kept stable and displayed nearly no emission signaling at a wide pH values varying from 2–10. However, under the same conditions, the fluorescence intensities of the solutions enhanced immediately and remarkably upon addition of ONOO−, illustrating the two probes were sufficiently feasible to sense ONOO− under normal physiological environment.
3.3 Specificity of BS1 and BS2 toward ONOO−
As is widely acknowledged that high selectivity is of great significance for an excellent chemosensor.32,53 Herein, the specific recognition of BS1 and BS2 toward ONOO− were exactly examined in the presence of various bio-species, including cations, reactive oxygen and nitrogen species (RONS) and reactive sulfur species (RSS), etc., which may influence the response efficiency of the probes for ONOO−. As shown in the Fig. 1c, it could be markedly observed that the probe BS1 presented superior response for ONOO− with notable fluorescence enhancement over other representative analytes, suggesting ONOO− triggered the cleavage of diphenyl phosphonate group of BS1.54,55 Moreover, BS2 exhibited a similar pattern as BS1 toward ONOO− (Fig. S14c†). But, it was worth noting that H2O2 and NaClO could also turn on the fluorescence of BS2 to some extent owing to hydrolysis of benzeneboronic ester induced by the two species.56,57 Thus, it could be concluded that selectivity of probe BS1 toward ONOO− with diphenyl phosphonate unit was superior to BS2 bearing a pinacol boronic ester group. Afterwards, to ensure that BS1 or BS2 is suitable to sense ONOO− without any anti-interference of other biological substrates, the competition tests were implemented. As clearly seen from the Figs. S16 and 17,† upon introduction of ONOO− into the solutions of two probes which pretreated with corresponding competing species, the fluorescence signaling rose dramatically, indicating both probes could resist the interference of other biologically relevant species.3.4 Investigation of the sensing mechanism
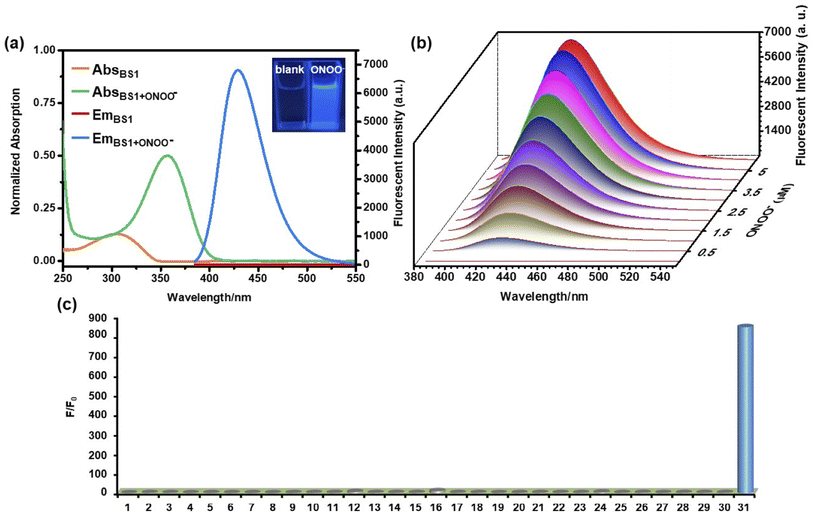
As mentioned above, the mechanism of BS1 and BS2 sensing ONOO− were based on the cleavage of diphenyl phosphonate group and hydrolysis of benzeneboronic ester, respectively. As a result, both BS1 and BS2 transformed to the same phenolic hydroxyl structure BS-OH. Herein, to verify our proposed reaction mechanism, high resolution mass spectrometry analysis was firstly carried out. From Figs. S6 and 7,† BS1 and BS2 exhibited obvious peaks at m/z = 428.0898 and 444.1824, corresponding to [BS1 + H+] and [BS2 + H+], respectively. After interacting with ONOO−, a new peak of BS1 at m/z = 228.0486 was found and assigned to the compound [BS-OH + H+] (Fig. S8†). For BS2, the peak of [BS-OH + H+] at m/z = 228.0485 was also observed (Fig. S9†). Besides, the 1H NMR of the reaction product for BS1 was recorded in Fig. S10.† It was obviously found that the peak at 10.24 ppm appeared after reaction which was assigned to the phenolic proton. And also, the reaction of BS1 with ONOO− was analyzed via HPLC. As shown in Fig. S11,† the probe BS1, BS1 with ONOO− and BS-OH displayed a single peak with the retention time at 2.50 min, 1.09 min and 1.06 min, respectively. The peak of the reaction of BS1 with ONOO− at 1.09 min in Fig. S11(b)† was in accordance with the peak of BS-OH at 1.06 min. For BS2, the FTIR analysis of BS2 with ONOO− was collected in Fig. S12.† The FTIR spectrum of the product was identical to the spectrum based on the previous works58,59 All the results of spectral analysis were consistent with the proposed sensing mechanism showed in Scheme 2. What's more, according to the absorption spectra analysis (Fig. 1a and S14a†), the obvious redshift of maximum peaks from 305 to 356 nm and fluorescence enhancement at 430 nm of BS1 or BS2 demonstrated that an intramolecular charge transfer (ICT) process happened via intra-molecular p–π conjugation from hydroxyl to benzothiazole unit. To further elucidate the ICT process, a density functional theory (DFT) calculation was exploited based on the Gaussian 16 program with the B3LYP/6-31G(d, p) method. From Fig. S20,† the frontier molecular orbital energies of optimized structures of BS1, BS2 and BS-OH were calculated. Taking BS1 as an example, in comparison with the π-electrons distribution of BS1 and BS-OH, it could be clearly found that the BS-OH was in form of phenolate, a latent donor, which activated the ICT process from hydroxyl to benzothiazole unit. Therefore, a new push–pull conjugated system formed. And the orbital energy gaps of BS1, BS2 and BS-OH were calculated to be 4.36 eV, 4.32 eV and 4.29 eV, respectively. Thus, the above experimental and theoretical results rationalized the sensing mechanism.3.5 Visualizing exogenous ONOO− in living cells
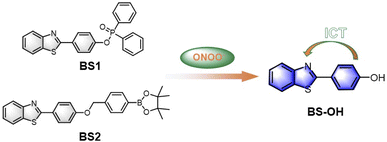
Encouraged by the predominant vitro sensing performances of the probes, we further attempted to investigated the biosensing and bioimaging of ONOO− in living cells. Considering that probe BS1 has much better selectivity and sensitivity, BS1 was chosen to monitor ONOO− level in biological systems. Prior to fluorescence imaging, the cytotoxicity of BS1 was identified by the CCK-8 assay in living HepG2 cells. From Fig. S22,† it can be seen that the cell viability was as high as 90% after incubation with BS1 at a concentration of 100 µM for 24 h. Moreover, when the cells continued to be incubated with BS1 for 48 h, an increased cell proliferation was observed (Fig. S23†). The above results confirmed the low cytotoxicity of BS1 to HepG2 cells.Subsequently, the ability of BS1 to monitor and image exogenous ONOO− was evaluated in living HepG2 cells. Herein, 3-morpholinosydnon imine hydrochloride (SIN-1) was utilized as ONOO− donor. The results were displayed in Fig. 2. In the control group, negligible fluorescence signal was detected after the HepG2 cells were incubated with BS1. Contrastingly, the cells which were pretreated with SIN-1 and then treated with BS1 emitted a remarkable enhancement of intracellular fluorescence intensity in the blue channel. Furthermore, the intact cells in the bright field images implied the good viability of the HepG2 cells during the entire process of the experiment. Consequently, the data convinced that BS1 possesses the ability of cell membrane permeability and ONOO− detection, which is promising to be a bioimaging agent for exploring ONOO− fluctuation in the physiological conditions.
3.6 APAP-induced hepatotoxicity and remediation effect of GSH
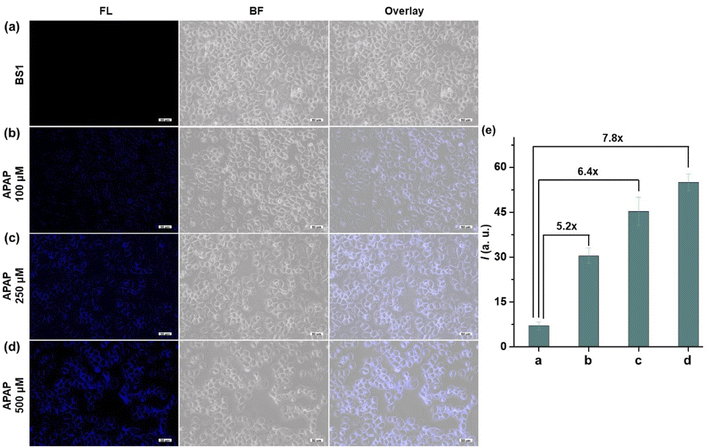
It is well-known that APAP was a common medicine to treat pain and fever. However, overdose of APAP could cause hepatotoxicity relating to excessive oxidative stress, which might be possible to induce a burst of ROS and RNS including ONOO− in the organism. Therefore, we built the dose-dependent APAP-induce injury model to confirm whether BS1 could visualize DILI in HepG2 cells. As presented in Fig. 3, an apparent fluorescence signal in HepG2 cells was progressively enhanced by increasing the concentration of APAP (0, 100, 250, 500 µM). The obtained results not only proved that APAP-induce liver injury had a definite dose-dependent relationship, but also verified that BS1 could be a favorable tool to reveal the upregulated ONOO− by excessive oxidative stress under APAP administration. Furthermore, as a common hepatoprotective medicine, glutathione (GSH) is an antioxidant which serves as an ONOO− scavenger in the organism. In order to validate whether GSH could alleviate the liver damage caused by APAP, the remediation experiment of GSH was performed. As shown in Fig. S24,† the group of cells treated with APAP and BS1 exhibited an obvious blue fluorescence intensity. Meanwhile, the other group of cells were pretreated with GSH, followed by APAP, and then incubated with BS1. As expected, the blue fluorescence signal significantly attenuated, which could be attributed to the cell injury remediation effect via GSH depleting the major of endogenous ONOO−. The results indicated that GSH might be an effective alternative antidote for APAP-induced liver damage. In total, BS1 possesses the ability to be a suitable tool to visualize the diagnosis of diseases and evaluate the remediation effect of hepatoprotective drugs for DILI.4. Conclusion
In summary, two benzothiazole-based fluorescent probes (BS1 and BS2) were designed and constructed for detecting ONOO−. At a physiological pH, both probes could quickly and sensitively detect ONOO− with remarkable “turn-on” fluorescence signal based on the cleavage of diphenyl phosphonate and hydrolysis of boronic pinacol ester. And nearly 850-fold signal enhancement was also found after BS1 reacting with ONOO−, and its LOD value was calculated to be as low as 12.8 nM with an excellent linear relationship. For BS2, the values were 695-fold and 25.2 nM, respectively. Compared with BS2, BS1 bearing diphenyl phosphonate unit exhibited better selectivity and was successfully employ to detect and image ONOO− in HepG2 cells. Significantly, experiments also proved that BS1 could monitor upregulated ONOO− levels after APAP-induced hepatotoxicity, and a remediation effect of GSH was also evaluated. These results suggested that the fluorescent probe BS1 could be a new promising biomarker to reveal the roles of ONOO− during the DILI process.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by the Natural Science Foundation of Chaohu University [Grant no. XLY-202002, XLY-202212, XLY-202211], the Discipline Construction Quality Improvement Project [Grant no. kj21fdzy03]. Y. Kong would like to thank Chaohu University for the Start-Up grant [Grant no. KYQD-202006]. X. Wang would like to thank the Key Research and Development Program of Anhui Province [2022a05020019]. Q. Wang would like to thank the National Nature Science Foundation of China [Grant no. 82003208] and Science and Technology Project of Jilin Province Education Department [Grant no. JJKH20221069KJ]. M. Chen would like to thank the Natural Science Foundation of Anhui Province (Grant no. 2108085QC147). The authors thank the Shiyanjia lab (http://www.shiyanjia.com) for NMR and HRMS measurements.References
- A. Srivastava, J. L. Maggs, D. J. Antoine, D. P. Williams, D. A. Smith and B. K. Park, Handb. Exp. Pharmacol., 2010, vol. 196, pp. 165–194 Search PubMed.
- L. Yuan and N. Kaplowitz, Clin. Liver Dis., 2013, 17, 507–518 CrossRef PubMed.
- M. Holt and C. Ju, Handb. Exp. Pharmacol., 2010, 196, 3–27 CrossRef CAS PubMed.
- J. L. Walgren, M. D. Mitchell and D. C. Thompson, Crit. Rev. Toxicol., 2002, 35, 325–361 CrossRef PubMed.
- D. J. Antoine, D. P. Williams and B. K. Park, Expert Opin. Drug Metab. Toxicol., 2008, 4, 1415–1427 CrossRef CAS PubMed.
- S. Russmann, G. A. Kullak-Ublick and I. Grattagliano, Curr. Med. Chem., 2009, 16, 3041–3053 CrossRef CAS PubMed.
- D. Pessayre, A. Mansouri, D. Haouzi and B. Fromenty, Cell Biol. Toxicol., 1999, 15, 367–373 CrossRef CAS PubMed.
- A. J. Shuhendler, K. Pu, L. Cui, J. P. Uetrecht and J. Rao, Nat. Biotechnol., 2014, 32, 373–380 CrossRef CAS PubMed.
- H. Jaeschke, G. J. Gores, A. I. Cederbaum, J. A. Hinson, D. Pessayre and J. J. Lemasters, Toxicol. Sci., 2002, 65, 166–176 CrossRef CAS PubMed.
- C. Prolo, N. Rios, L. Piacenza, M. N. Alvarez and R. Radi, Free Radical Bio. Med., 2018, 128, 59–68 CrossRef CAS PubMed.
- Z. Mao, J. Xiong, P. Wang, J. An, F. Zhang, Z. Liu and J. S. Kim, Coordin. Chem. Rev., 2022, 454, 214356 CrossRef CAS.
- L. Wang, J. Shao, B. Cheng, X. Li and J. Ma, J. Iran. Chem. Soc., 2019, 16, 437–447 CrossRef CAS.
- R. Radi, J. S. Beckman, K. M. Bush and B. A. Freeman, J. Biol. Chem., 2015, 290, 30726–30727 CrossRef PubMed.
- G. Ferrer-Sueta, N. Campolo, M. Trujillo, S. Bartesaghi, S. Carballal, N. Romero, B. Alvarez and R. Radi, Chem. Rev., 2018, 118, 1338–1408 CrossRef CAS PubMed.
- P. L. Pacher, J. S. Beckman and L. Liaudet, Physiol. Rev., 2007, 87, 315–424 CrossRef CAS PubMed.
- Z. X. Chen and S. Pervaiz, Front. Biosci., 2009, 14, 4809–4814 CAS.
- C. Szabo, H. Ischiropoulos and R. Radi, Nat. Rev. Drug discov., 2007, 6, 662–680 CrossRef CAS PubMed.
- V. Shafirovich, A. Dourandin, W. Huang and N. E. Geacintov, J. Biol. Chem., 2001, 276, 24621–24626 CrossRef CAS PubMed.
- G. Ferrer-Sueta and R. Radi, ACS Chem. Biol., 2009, 4, 161–177 CrossRef CAS PubMed.
- C. Cover, A. Mansouri, T. R. Knight, M. L. Bajt, J. J. Lemasters, D. Pessayre and H. Jaeschke, J. Pharmacol. Exp. Ther., 2005, 315, 879–887 CrossRef CAS PubMed.
- J. Li, S. Peng, Z. Li, F. Zhao, X. Han, J. Liu, W. Cao and Y. Ye, Talanta, 2022, 238, 123007 CrossRef CAS PubMed.
- S. Qin, H. Lu, J. Zhang, X. Ji, N. Wang, J. Liu, W. Zhao and J. Wang, Dyes Pigm., 2022, 203, 110345 CrossRef CAS.
- S. Feng, Z. Zheng, S. Gong and G. Feng, Sens. Actuators B Chem., 2022, 361, 131751 CrossRef CAS.
- Y. Deng and G. Feng, Anal. Chem., 2020, 92, 14667–14675 CrossRef CAS PubMed.
- J. Kim, J. Park, H. Lee, Y. Choi and Y. Kim, Chem. Commun., 2014, 50, 9353–9356 RSC.
- H. Masumoto, R. Kissner, W. H. Koppenol and H. Sies, FEBS Lett., 1996, 398, 179–182 CrossRef CAS PubMed.
- C. Ducrocq, B. Blanchard, B. Pignatelli and H. Ohshima, Cell. Mol. Life Sci., 1999, 55, 1068–1077 CrossRef CAS PubMed.
- H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620–2640 CrossRef CAS.
- C. Wu and D. T. Chiu, Angew. Chem., Int. Ed., 2013, 52, 3086–3109 CrossRef CAS PubMed.
- L. Wang, M. S. Frei, A. Salim and K. Johnsson, J. Am. Chem. Soc., 2019, 141, 2770–2781 CrossRef CAS.
- D. Cao, Z. Liu, P. Verwilst, S. Koo, P. Jangjili, J. S. Kim and W. Lin, Chem. Rev., 2019, 119, 10403–10519 CrossRef CAS.
- Y. Kong, X. Wan, Z. Liu, F. Chen, F. Wu, G. Qin, D. Cao and Y. Cui, Sens. Actuators B Chem., 2022, 350, 130852 CrossRef CAS.
- Z. Li, S. H. Yan, C. Chen, Z. R. Geng, J. Y. Chang, C. X. Chen, B. H. Huang and Z. L. Wang, Biosens. Bioelectron., 2017, 90, 75–82 CrossRef CAS PubMed.
- L. Wu, J. Liu, X. Tian, R. R. Groleau, S. D. Bull, P. Li, B. Tang and T. D. James, Chem. Sci., 2021, 12, 3921–3928 RSC.
- X. Li, R. R. Tao, L. J. Hong, J. Cheng, Q. Jiang, Y. M. Lu, M. H. Liao, W. F. Ye, N. N. Lu, F. Han, Y. Z. Hu and Y. H. Liu, J. Am. Chem. Soc., 2015, 137, 12296–12303 CrossRef CAS PubMed.
- Y. Lei, W. Ren, C. K. Wang, R. R. Tao, H. J. Xiang, L. L. Feng, Y. P. Gao, Q. Jiang, X. Li, Y. Hu and F. Han, Theranostics, 2019, 9, 5672–5680 CrossRef CAS PubMed.
- D. Cheng, W. Xu, L. Yuan and X. B. Zhang, Anal. Chem., 2017, 89, 7693–7700 CrossRef CAS PubMed.
- W. Wang, J. Xiong, X. Song, Z. Wang, F. Zhang and Z. Mao, Anal. Chem., 2020, 92, 13305–13312 CrossRef CAS PubMed.
- J. Zhang, X. Zhen, J. Zeng and K. Pu, Anal. Chem., 2018, 90, 9301–9307 CrossRef CAS PubMed.
- J. S. Hu, C. Shao, X. Wang, X. Di, X. Xue, Z. Su, J. Zhao, H. L. Zhu, H. K. Liu and Y. Qian, Adv. Sci., 2019, 6, 1900341 CrossRef PubMed.
- D. Li, S. Wang, Z. Lei, C. Sun, A. M. El-Toni, M. S. Alhoshan, Y. Fan and F. Zhang, Anal. Chem., 2019, 91, 4771–4779 CrossRef CAS PubMed.
- H. H. Han, A. C. Sedgwick, Y. Shang, N. Li, T. Liu, B. H. Li, K. Yu, Y. Zang, J. T. Brewster, M. L. Odyniec, M. Weber, S. D. Bull, J. Li, J. L. Sessler, T. D. James, X. P. He and H. Tian, Chem. Sci., 2019, 11, 1107–1113 RSC.
- Z. Wang, W. Wang, P. Wang, X. Song, Z. Mao and Z. Liu, Anal. Chem., 2021, 93, 3035–3041 CrossRef CAS.
- F. Yu, P. Li, G. Li, G. Zhao, T. Chu and K. Han, J. Am. Chem. Soc., 2011, 133, 11030–11033 CrossRef CAS PubMed.
- F. Yu, P. Li, B. Wang and K. Han, J. Am. Chem. Soc., 2013, 135, 7674–7680 CrossRef CAS PubMed.
- X. Luo, Z. Cheng, R. Wang and F. Yu, Anal. Chem., 2021, 93, 2490–2499 CrossRef CAS.
- R. Yuan, Y. Ma, J. Du, F. Meng, J. Guo, M. Hong, Q. Yue, X. Li and C. Li, Anal. Methods, 2019, 11, 1522–1529 RSC.
- F. Yan, J. Sun, Y. Zang, Z. Sun, H. Zhang and X. Wang, J. Iran. Chem. Soc., 2020, 17, 3179–3203 CrossRef CAS.
- K. A. Shaikh and U. N. Chaudhar, Org. Commun., 2017, 10, 288–297 CrossRef CAS.
- H. Ju, D. J. Chang, S. Kim, H. Ryu, E. Lee, I. H. Park, J. H. Jung, M. Ikeda, Y. Habata and S. S. Lee, Inorg. Chem., 2016, 55, 7448–7456 CrossRef CAS PubMed.
- W. H. Melhuish, J. Phys. Chem. B, 1960, 65, 229–235 CrossRef.
- L. Ji, C. Yang, H. Li, N. Yang, Y. Fu, L. Yang, Q. Wang and G. He, Luminescence, 2021, 36, 4–10 CrossRef CAS.
- Y. Kong, M. Wang, W. Lu, L. Li, J. Li, M. Chen, Q. Wang, G. Qin and D. Cao, Anal. Bioanal. Chem., 2022, 414, 2009–2019 CrossRef CAS PubMed.
- Y. Wu, A. Shi, Y. Li, H. Zeng, X. Chen, J. Wu and X. Fan, Analyst, 2018, 143, 5512–5519 RSC.
- S. V. Mulay, Y. Kim, K. J. Lee, T. Yudhistira, H. S. Park and D. G. Churchill, New J. Chem., 2017, 41, 11934–11940 RSC.
- H. Xiao, P. Li, X. Hu, X. Shi, W. Zhang and B. Tang, Chem. Sci., 2016, 7, 6153–6159 RSC.
- E. W. Miller and C. J. Chang, Curr. Opin. Chem. Biol., 2007, 11, 620–625 CrossRef CAS PubMed.
- K. N. Patil, R. K. Bhat, C. D. Bhenki and V. B. Helavi, Chem. Date Collect., 2019, 24, 100307 CrossRef CAS.
- D. P. Araujo, V. S. S. Morais, A. Fatima and L. V. Modolo, RSC Adv., 2015, 5, 28814–28821 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra04549d |
| This journal is © The Royal Society of Chemistry 2022 |