Open Access Article
Open Access ArticleN-Doped graphene quantum dots (N-GQDs) as fluorescent probes for detection of UV induced DNA damage
Gulshan Jabeenab,
Muhammad Hassan Ahmadc,
Muhammad Aslamb,
Sara Riazc,
Akhtar Hayata and
Mian Hasnain Nawaz *a
*a
aInterdisciplinary Research Centre in Biomedical Materials (IRCBM), COMSATS University Islamabad, Lahore Campus, 54000, Pakistan. E-mail: mhnawaz@cuilahore.edu.pk
bDepartment of Chemistry, Division of Science and Technology, University of Education, Lahore, 54000, Pakistan
cDepartment of Chemistry, COMSATS University Islamabad, Lahore Campus, 54000, Pakistan
First published on 11th August 2022
Abstract
UV induced DNA damage can lead to the development of skin cancer, skin aging and cell death. In this study, we fabricated a fluorescence-based biosensor that can be applied to the detection of DNA damage caused by UV radiation with the help of nitrogen doped graphene quantum dots (N-GQDs) as the probe material. In this paper, N-GQDs with good fluorescence efficiency have been synthesized by the hydrothermal method and were used as a fluorescent probe for the detection of UV damaged DNA. The fluorescence intensity of N-GQDs was quenched by the static quenching of UV damaged DNA through the formation of a N-GQD/UV damaged DNA complex. Moreover, the effects of different values of pH, NaCl and glucose on analytical performances of the sensor were also studied. Thus, using a fluorescence based approach, we demonstrated a quite simple, rapid, and inexpensive biosensor for the detection of DNA damage caused by UV radiation.
1 Introduction
DNA damage refers to chemical or physical changes to DNA in cells, which can affect the interpretation and transmission of genetic information.1 DNA damage is classified into four types: single-base alteration (depurination, deamination and alkylation), double-base alterations (pyrimidine dimers), strand breaks (single and double strand breaks) and cross-linking of DNA.2 All these types of damage are caused by various endogenous and exogenous agents such as reactive oxygen species, free radicals, infrared radiations, ultraviolet (UV) radiations, X-rays, crosslinking and alkylating agents.3 UV radiation is a leading exogenous agent to the integrity of DNA,4 which is classified into three groups on the basis of wavelength range: UV-C (190–290 nm), UV-B (290–320 nm) and UV-A (320–400 nm).5 Fortunately, all organisms have DNA damage response (DDR) mechanism1,6 that detects and repairs the damage in DNA by using certain repair pathways like mismatch repair (MMR), homologous recombination (HR), non-homologous end joining (NHEJ), nucleotide excision repair (NER) and base excision repair (BER).3,7 Nucleotide excision repair (NER) is the only system responsible for removal of the most dangerous UV induced photo-lesions in DNA of human. If there are any defects in NER system, as in Xeroderma Pigmentosum disease (XP), Cockayne, trichothiodystrophy (TTD), cerebro-oculofacio-skeletal (COFS), and UV sensitive syndrome (UVsS),8,9 it can't detect and repair UV induced photo-lesions in DNA.In case of defects in NER system, enzyme linked immunosorbent assay (ELISA), low cytometry, radioimmunoassay (RIA), alkaline gel electrophoresis by T4 endonuclease V, mass spectrometry, near-infrared spectroscopy, atomic force microscopy (AFM) and fluorescent sensor have been used for detection of photo-dimmers in DNA.10–14 Among these methods, fluorescent sensor has gained great interest due to rapid analysis, low cost, highest sensitivity, good selectivity, relatively simple equipment and simple operation which overcome the disadvantages of other techniques such as tedious, time consuming and multistep process, and special instrumentation. To date, carbon dots have been used for the detection of UV induced DNA damage.
The fluorescent probes, graphene quantum dots (GQDs), have received more attention in sensing applications due to their low cytotoxicity, high aqueous dispersibility, high photostability, good biocompatibility,15 unique electronic properties, low preparation cost and fluorescence properties.16 Moreover, they can be easily doped with hetero-atoms like nitrogen, boron, and phosphorous, in order to improve their quantum yield and fluorescence efficiency. In this context, nitrogen (N) doping is accounted to be highly useful in changing the intrinsic properties of GQDs.17,18 Nitrogen doped graphene quantum dots (N-GQDs) are broadly used in fluorescence sensors to detect metal ions, small organic molecules, non-metal ions and biological macromolecules.19 Several protocols, including electrochemical,20 chemical vapor deposition,21 hydrothermal,22–24 oxidative ultra-sonication25 and microwave-assisted hydrothermal methods26 have been used for the synthesis of N-GQDs. Among many synthesis methods of N-GQDs, the hydrothermal synthesis is the most popular, simple and low cost.
Herein, highly fluorescent N-GQDs were prepared by hydrothermal method and were characterized by FT-IR, XRD, UV-Vis and fluorescence spectrophotometer to explore their characteristics. The intercalative and electrostatic interactions were established between ds-DNA and N-GQDs, which played a significant role in the detection of UV damaged ds-DNA. The effects of pH, glucose, NaCl on stability of fluorescence biosensor were further explored.
2 Experimental section
2.1 Materials
Citric acid monohydrate was purchased from Fisher Scientific. Urea and hydrochloric acid were purchased from BDH. Glucose, sodium hydroxide and ds-DNA were purchased from Sigma-Aldrich. Sodium chloride was purchased from Daejung.2.2 Instruments
The UV-Vis absorption spectroscopy was performed on a PerkinElmer Lambda 25 UV-Vis spectrophotometer. The X-ray powder diffraction (XRD) spectra were recorded using X-ray diffractometer. The Fourier transform infrared (FTIR) spectra were recorded on a Thermo Fisher Scientific Nicolet 6700 spectrometer. The photoluminescence (PL) spectroscopy was carried out on a Carry Eclipse Fluorescence spectrophotometer.2.3 Synthesis of N-GQDs
N-Doped GQDs were synthesized by hydrothermal heating of citric acid and urea as a carbon and nitrogen sources, respectively, following a reported method with minor modifications.27 Typically, 7.5 mmol of citric acid (1.56 g) and 30 mmol of urea (1.8 g) were dissolved in 40 mL of de-ionized water, and stirred for 5 min. The mixture was then transferred into a 50 mL Teflon lined stainless autoclave and was placed in oven for 11 h at 150 °C. The final product was collected by adding excess of ethanol into the solution and centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The green precipitates were dried in drying oven at 80 °C for 3 h.
000 rpm for 15 min. The green precipitates were dried in drying oven at 80 °C for 3 h.
2.4 Damage to DNA by UV in the presence of N-GQDs
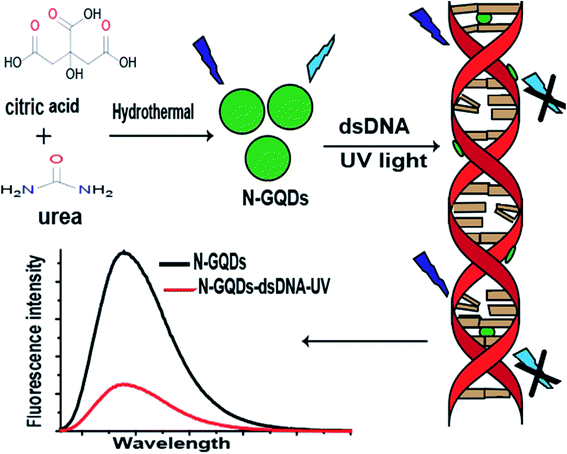
4 mL of ds-DNA solution (20 μg mL−1) was added into 4 mL of as-prepared N-GQDs solution (30-fold diluted with de-ionized water) and placed in UV box equipped with a UV lamps of 6 W. The fluorescence emission spectra of the solution were recorded after different intervals of UV irradiation. The synthesis and application of N-GQDs is illustrated in Fig. 1.3 Results and discussion
3.1 Characterization of N-GQDs
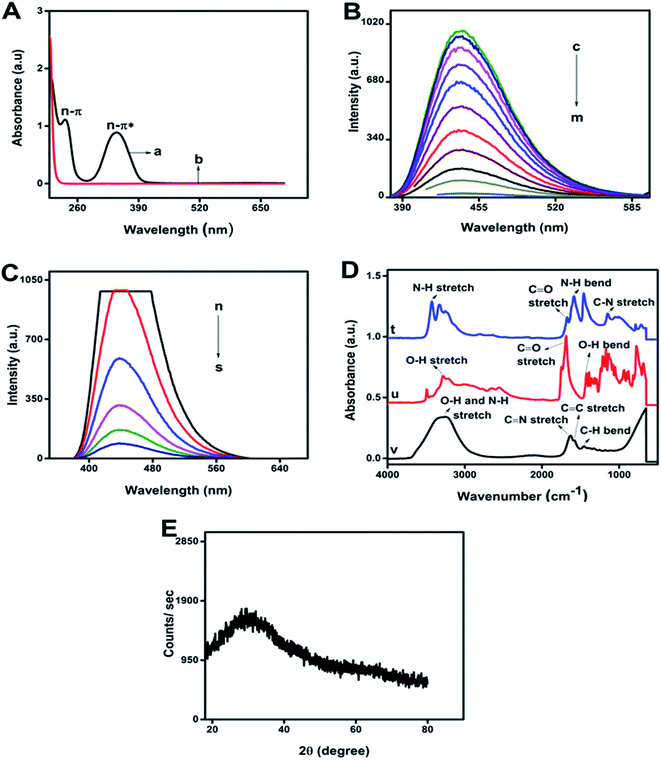
To explore the optical properties of N-GQDs, UV-visible (UV-Vis) absorption spectroscopy was carried out. The absorption spectra of N-GQDs and urea are shown in Fig. 2A. In the absorption spectra of N-GQDs, two absorption peaks were observed, centered at about 234 and 345 nm. The peak at 234 nm indicated the transition of π–π* of aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C domain while at 345 nm showed the transition of n–π* of C
C domain while at 345 nm showed the transition of n–π* of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O.19
O.19
The FL spectra at different excitation wavelength is shown in Fig. 2B. The N-GQDs exhibited an excitation-independent FL behavior. At different excitation wavelengths, only change in peak intensity was observed, with no shift in peak position. By increasing excitation wavelengths, the FL intensity of the peaks was first increased and then decreased. At 340 nm excitation, the maximum FL intensity was observed. Effect of dilution on fluorescence intensity of N-GQDs was observed at an excitation wavelength of 350 nm by diluting the samples with de-ionized water. The FL spectra of N-GQDs at different dilutions can be evidenced in Fig. 2C. It can be seen that the FL intensity of N-GQDs is decreased by increasing their dilution. The structure of the N-GQDs was further characterized using FTIR spectra. The FTIR spectra of N-GQDs aqueous solution, citric acid and urea are shown in Fig. 2D. The broad absorption band in N-GQDs spectra at around 3600–3000 cm−1 indicated hydrogen bond confirming the existence of OH and NH groups in N-GQDs. The stretching vibrations of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C were observed in N-GQDs spectra at 1630 cm−1 and 1570 cm−1, respectively. The strong absorption band was affected by the conjugation of C
C were observed in N-GQDs spectra at 1630 cm−1 and 1570 cm−1, respectively. The strong absorption band was affected by the conjugation of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond with other double bond structures. The peak at around 1478–1636 cm−1 indicated the bending vibration of C–H bond. These observations revealed that the N-GQDs possess amine, and hydroxyl functional groups on their surfaces, which facilitates the homogenous and stable dispersion of N-GQDs in water. The XRD spectra of the N-GQDs (Fig. 2E) revealed that the diffraction peaks of graphitic structure appeared in the range of 27–29 2θ value. The smaller size of the N-GQDs resulted in peak broadening as can be evidenced in the spectra.28
C bond with other double bond structures. The peak at around 1478–1636 cm−1 indicated the bending vibration of C–H bond. These observations revealed that the N-GQDs possess amine, and hydroxyl functional groups on their surfaces, which facilitates the homogenous and stable dispersion of N-GQDs in water. The XRD spectra of the N-GQDs (Fig. 2E) revealed that the diffraction peaks of graphitic structure appeared in the range of 27–29 2θ value. The smaller size of the N-GQDs resulted in peak broadening as can be evidenced in the spectra.28
3.2 Detection of UV induced DNA damage by N-GQDs
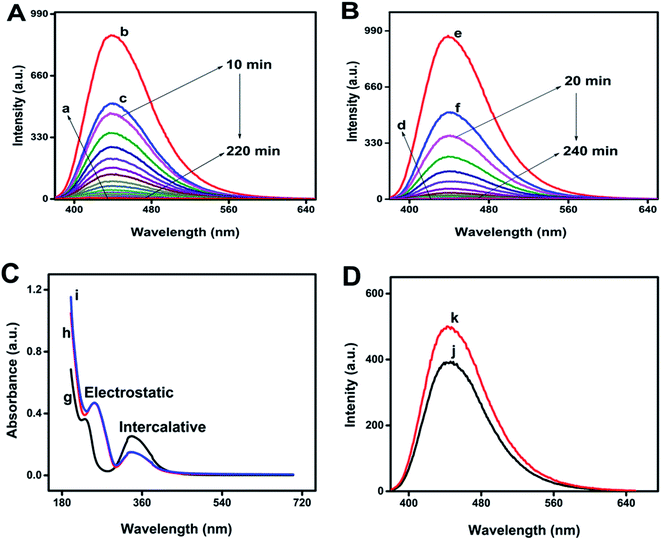
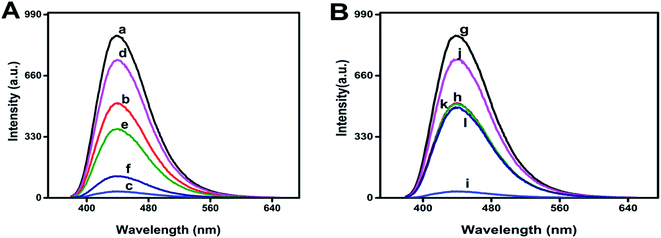
The UV damaged DNA leads to the development of skin cancer and skin aging.9 As can be evidenced from Fig. 3A and B, the fluorescence intensity of N-GQDs is more quenched by UV damaged DNA as compare to un-damaged DNA, which indicates presence of photo-lesions in UV damaged DNA that are main cause of skin cancer. Such photolesions and individual protein bases of DNA are more prone to surround the N-GQDs which eventually results in the quenching of fluorescence properties of these quantum dots.Fluorescence quenching of any fluorescent probes can involve dynamic or static quenching, or their combined quenching mechanism.29 As can be evidenced from Fig. 3A and B, UV damaged ds-DNA quenches N-GQDs emission intensity through static quenching mechanism. The static quenching mechanism between N-GQDs and UV damaged DNA is explained by Fig. 3C and D. As can be seen in Fig. 3C, after interaction with ds-DNA and UV damaged DNA, the absorption spectra of N-GQDs were changed. These changes in absorption indicated the occurrence of static quenching due to formation of complex at ground state between ds-DNA and N-GQDs. As the dynamic quenching only affects the excited state of fluorophore and absorption spectra does not change.30 The effect of temperature on static quenching is shown in Fig. 3D. By increasing temperature, the efficiency of static quenching is decreased because increasing temperature tends to disfavor the binding interaction that causes static quenching. Efficiency of static quenching is increased if there is great affinity between N-GQDs and UV damaged ds-DNA for each other.
3.3 N-GQDs interaction with ds-DNA
The interaction of N-GQDs with ds-DNA was investigated by UV-Vis spectroscopy. The absorption spectra of N-GQDs and N-GQDs–ds-DNA complex before and after irradiation of UV were shown in Fig. 3C. In UV-Vis spectroscopy, If a hypochromic effect is observed in the spectrum, it indicate intercalation type of interaction of object with the DNA while in case of a hyperchromic effect, the intercalative and electrostatic type of interactions of object with the DNA are observed.31,32 In the UV-Vis spectrum, two intense bands were observed (at 234 and 337 nm) for N-GQDs. When they interact with ds-DNA and UV damaged ds-DNA, hyperchromism and hypochromism are observed at 234 and 337 nm, respectively. These changes in the absorption peaks of N-GQDs confirm the electrostatic and intercalative type of interaction between UV damaged ds-DNA and N-GQDs complex.333.4 Effect of pH, glucose and NaCl on stability of biosensor
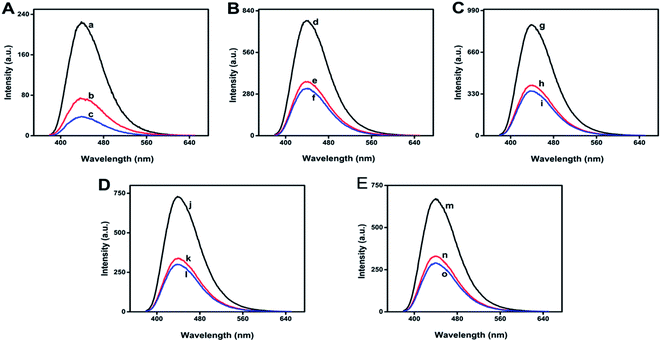
To explore the practical application of this biosensor, the effect of pH on fluorescence intensity of N-GQDs and on the UV damage ds-DNA detection process was investigated over the pH range of 4–12. The result is shown in Fig. 4A–E. The pH was adjusted using NaOH (1 mol L−1) and HCl (1 mol L−1). It was observed that the fluorescence intensity of N-GQDs and its quenching by ds-DNA is pH dependent. First, the florescent intensity of N-GQDs increased from pH 4 to 8 followed by a prominent decrease from pH 8 to 12. The maximum fluorescence intensity was observed at pH 8 where N-GQDs could detect UV damaged ds-DNA more efficiently. In general, the fluorescence intensity of N-GQDs is stable in a wide range of pH24 and hence could be useful to detect UV damaged ds-DNA in wide range pH.In a control experiment, the fluorescence intensity of N-GQD was quenched by glucose, which conduced the presence of glucose in physiological environment could be crucial for damaged DNA detection. Hence, the effect of glucose on the fluorescence intensity of N-GQDs and N-GQD–ds-DNA complex was studied as shown in Fig. 5A.
In the presence of glucose, UV irradiation caused less damage in ds-DNA which corroborated minor quenching in the fluorescence intensity of N-GQDs. In the absence of glucose, the fluorescence quenching was more, as can be evidenced in Fig. 5A. Moreover, the effect of NaCl on the fluorescence intensity of N-GQDs and N-GQD–ds-DNA complex were studied as shown in Fig. 5B. The fluorescence intensity of N-GQDs was slightly quenched by NaCl solution indicating that N-GQDs were stable and did not aggregate even under high ionic conditions.28 In the presence of ionic solution, ds-DNA did not damage under UV irradiation because sodium ions provided stability to it by neutralizing the negative change on its backbones. After neutralization of negative charge on DNA, the repulsive coulombic interactions between the phosphates was reduced and DNA molecules remained in their native structure.34
4 Conclusion
In conclusion, we have developed a novel and convenient fluorescence sensor by using N-GQDs to achieve the rapid detection of UV induced ds-DNA damage. The prepared N-GQDs exhibited excellent optical properties and wavelength independent FL behavior. The fluorescence intensity of N-GQDs was quenched by static quenching of UV damaged DNA through formation of N-GQDs with individual base proteins of DNA and their photolesions via intercalative and electrostatic interactions. Based on this principle, UV damaged DNA can be detected under different physiological conditions. The present study provides a simple, fast, and cost-effective fluorescence based DNA detection method that could further be researched for development of other bio-sensing platforms.Conflicts of interest
There are no conflicts to declare.Acknowledgements
MHN acknowledges the financial support provided by HEC (20-4993/R&D/HEC/14/614, 20-16284/NRPU/R&D/HEC/2021).References
- U. S. Srinivas, B. W. Q. Tan, B. A. Vellayappan and A. D. Jeyasekharan, Redox Biol., 2019, 25, 101084 CrossRef CAS
.
- S. Chakarov, R. Petkova, G. C. Russev and N. Zhelev, BioDiscovery, 2014, 11, e8957 Search PubMed
.
- N. Chatterjee and G. C. Walker, Environ. Mol. Mutagen., 2017, 58, 235–263 CrossRef CAS
.
- W. J. Schreier, P. Gilch and W. Zinth, Annu. Rev. Phys. Chem., 2015, 66, 497–519 CrossRef CAS PubMed
.
- A. P. Schuch and C. F. M. Menck, J. Photochem. Photobiol., B, 2010, 99, 111–116 CrossRef CAS
.
- M. Yousefzadeh, C. Henpita, R. Vyas, C. Soto-Palma, P. Robbins and L. Niedernhofer, Elife, 2021, 10, e62852 CrossRef CAS PubMed
.
- J. W. Harper and S. J. Elledge, Mol. Cell, 2007, 28, 739–745 CrossRef CAS
.
- A. R. Lehmann, Biochimie, 2003, 85, 1101–1111 CrossRef CAS PubMed
.
- L. H. Mullenders, Photochem. Photobiol. Sci., 2018, 17, 1842–1852 CrossRef CAS PubMed
.
- J. Peccia and M. Hernandez, Appl. Environ. Microbiol., 2002, 68, 2542–2549 CrossRef CAS
.
- R. Rouget, Y. Auclair, M. Loignon, E. B. Affar and E. A. Drobetsky, J. Biol. Chem., 2008, 283, 5533–5541 CrossRef CAS
.
- S. Mouret, P. Bogdanowicz, M. J. Haure, N. Castex-Rizzi, J. Cadet, A. Favier and T. Douki, Photochem. Photobiol., 2011, 87, 109–116 CrossRef CAS PubMed
.
- N. Goto, G. Bazar, Z. Kovacs, M. Kunisada, H. Morita, S. Kizaki, H. Sugiyama, R. Tsenkova and C. Nishigori, Sci. Rep., 2015, 5, 1–13 Search PubMed
.
- Y. Jiang, C. Ke, P. A. Mieczkowski and P. E. Marszalek, Biophys. J., 2007, 93, 1758–1767 CrossRef CAS PubMed
.
- C. Yan, X. Hu, P. Guan, T. Hou, P. Chen, D. Wan, X. Zhang, J. Wang and C. Wang, J. Mater. Sci., 2020, 55, 1198–1215 CrossRef CAS
.
- M. Kortel, B. D. Mansuriya, N. Vargas Santana and Z. Altintas, Micromachines, 2020, 11, 866 CrossRef PubMed
.
- Z. Qian, J. Ma, X. Shan, H. Feng, L. Shao and J. Chen, Chem.–Eur. J., 2014, 20, 2254–2263 CrossRef CAS PubMed
.
- H. M. Kashani, T. Madrakian and A. Afkhami, New J. Chem., 2017, 41, 6875–6882 RSC
.
- F. Lu, Y.-h. Zhou, L.-h. Wu, J. Qian, S. Cao, Y.-f. Deng and Y. Chen, Int. J. Opt., 2019, 2019, 8724320 Search PubMed
.
- Y. Li, Y. Zhao, H. Cheng, Y. Hu, G. Shi, L. Dai and L. Qu, J. Am. Chem. Soc., 2012, 134, 15–18 CrossRef CAS PubMed
.
- L. Fan, M. Zhu, X. Lee, R. Zhang, K. Wang, J. Wei, M. Zhong, D. Wu and H. Zhu, Part. Part. Syst. Char., 2013, 30, 764–769 CrossRef CAS
.
- J. Ju, R. Zhang, S. He and W. Chen, RSC Adv., 2014, 4, 52583–52589 RSC
.
- X. Zhu, X. Zuo, R. Hu, X. Xiao, Y. Liang and J. Nan, Mater. Chem. Phys., 2014, 147, 963–967 CrossRef CAS
.
- F. Cai, X. Liu, S. Liu, H. Liu and Y. Huang, RSC Adv., 2014, 4, 52016–52022 RSC
.
- Z. Cai, F. Li, P. Wu, L. Ji, H. Zhang, C. Cai and D. F. Gervasio, Anal. Chem., 2015, 87, 11803–11811 CrossRef CAS
.
- X. Hou, Y. Li and C. Zhao, Aust. J. Chem., 2015, 69, 357–360 CrossRef
.
- X. Sun, H.-J. Li, N. Ou, B. Lyu, B. Gui, S. Tian, D. Qian, X. Wang and J. Yang, Molecules, 2019, 24, 344 CrossRef PubMed
.
- L. Lin, M. Rong, S. Lu, X. Song, Y. Zhong, J. Yan, Y. Wang and X. Chen, Nanoscale, 2015, 7, 1872–1878 RSC
.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, 2006 Search PubMed
.
- Q. Lu, C. Chen, S. Zhao, F. Ge and D. Liu, Int. J. Food Prop., 2016, 19, 2481–2494 CrossRef CAS
.
- M. Yıldız, Ö. Karpuz, C. T. Zeyrek, B. Boyacıoğlu, H. Dal, N. Demir, N. Yıldırım and H. Ünver, J. Mol. Struct., 2015, 1094, 148–160 CrossRef
.
- H. Ünver, B. Boyacıoğlu, C. T. Zeyrek, M. Yıldız, N. Demir, N. Yıldırım, O. Karaosmanoğlu, H. Sivas and A. Elmalı, J. Mol. Struct., 2016, 1125, 162–176 CrossRef
.
- B. Şenel, N. Demir, G. Büyükköroğlu and M. Yıldız, Saudi Pharmaceut. J., 2019, 27, 846–858 CrossRef
.
- Z.-J. Tan and S.-J. Chen, Biophys. J., 2006, 90, 1175–1190 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2022 |