Open Access Article
Open Access ArticlePromotion of cytoplasmic localization of oligonucleotides by connecting cross-linked duplexes†
Yu Hirano‡
 and
Yasuo Komatsu‡
and
Yasuo Komatsu‡ *
*
Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), 2-17-2-1 Tsukisamu-Higashi, Toyohira-ku, Sapporo 062-8517, Japan. E-mail: komatsu-yasuo@aist.go.jp
First published on 30th August 2022
Abstract
We previously reported that antisense oligonucleotides (ASOs) flanked by duplexes can suppress microRNA (miRNA) function with high efficiency for a long duration. In this study, we examined the effect of the double-stranded structure on the subcellular localization of ASOs. Double strands were cross-linked to prevent dissociation into single strands, and this cross-linked duplex (CD) was connected at the 5′ or 3′ termini of an antisense-targeting miRNA-21 (AS). The subcellular distribution of fluorescently labelled ASOs was analyzed following transfection into cells. While single-stranded AS molecules promptly moved to the nucleus, AS with the CD at the 5′-end (5′CD-AS) interestingly showed significantly higher cytoplasmic localization. The 3′-CD-modified AS (3′CD-AS) was degraded from the 5′-end of the AS, but the degradation was prevented by 5′-end chemical modifications, thereby allowing the imaging of the cytoplasmic localization. The CD modification significantly promoted the cytoplasmic localization of ASOs and enabled the effective knockdown of miRNA existing in cytoplasm. These results reveal that the duplex structure has promising potential to control the subcellular distribution of ASOs.
Introduction
Single stranded antisense oligonucleotides (ASOs) are widely used for controlling gene expression in a sequence-specific manner.1–3 ASOs are divided into two categories based on their functional mechanisms: Ribonuclease H1 (RNase H1)-mediated RNA cleavage and steric blocking. Endogenous RNase H1 binds to an RNA–DNA heteroduplex and degrades the RNA4; however, fully 2′-modified ASOs cannot induce RNase H-mediated RNA cleavage.5 Gapmer ASOs containing deoxynucleotides flanked by several modified nucleotides on both ends can induce RNase H1-mediated RNA cleavage at the gap region.6 The 2′-modifications can improve ASO/RNA binding affinity and nuclease resistance.7 Among these gapmer ASOs, β-L-locked nucleic acid (LNA) has attracted considerable interst.8,9 The phosphodiester (PO) backbone linkage in an ASO is also frequently replaced by a phosphorothioate (PS) linkage to enhance nuclease resistance and binding with various proteins, such as plasma proteins.10–14These chemical modifications affect cellular uptake and intracellular trafficking of ASOs.15 PS-modified ASOs (PS-ASOs) are taken up by scavenger receptors, which mediate ASO internalization into the cells.16 PS-ASOs can be selectively delivered into liver hepatocytes by conjugation with N-acetyl galactosamine (GalNac), because asialoglycoprotein receptor mediates the uptake of GalNac-conjugated PS-ASOs into hepatocytes.3,17 However, the effects of intracellular distribution on ASO activity is not fully understood.15,16 The intracellular trafficking and localization of ASOs can be monitored using fluorescence microscopy.18–20 Single-stranded PO-ASOs and PS-ASOs accumulate in the nucleus following lipofection or microinjection.18–20 In contrast, after cells internalize gapmer PS-ASOs with LNA modifications in the absence of transfection reagents, the PS-ASOs become localized in the perinuclear region of the cytoplasm.19 Moreover, when gapmer PS-ASOs are introduced into cells without transfection reagents, ASO subcellular localization is affected by the 2′-modifications at both ends.15,21 Although the subcellular localization of PS-ASOs depends on different binding proteins,22,23 single-stranded PO-ASOs rarely interact with proteins in cells and are primarily distributed to the nucleus.18,24
MicroRNAs (miRNAs) are small endogenous noncoding RNAs (20–23 nucleotides) that regulate gene expression. In the nucleus, miRNAs are transcribed as pri-miRNA, and processed into a hairpin-shaped pre-miRNA. Next, the pre-miRNA is exported to the cytoplasm and one of the strands of the duplex associates with the Argonaute protein to form a miRNA-RISC complex that engages the target mRNA to mediate gene silencing.25,26 The discovery that miRNAs have important roles in various physiological processes and the pathogenesis of diseases26,27 has led to ASOs targeted toward miRNA, which are also known as antagomiRs, antimir, or anti-microRNA oligonucleotides, to become an important tool for investigating miRNA function. ASOs bind to miRNA through complementary sequences, thereby inhibiting the function of the miRNA in cells.28–31 Additionally, the double-stranded duplexes flanking the antimir improve the inhibitory effects.32–34 Recently, we constructed an ASO flanked by an interstrand cross-linked duplex (CD) of 2′-O-methylated RNA and found that the CD attenuated the anti-miRNA activity of the ASO.35 Compared with other structured ASOs, ASO flanked by CDs at the 5′- and 3′-termini (CD-AS) exhibit higher inhibitory activity in cells. For example, CD-AS effectively suppressed the proliferation of breast cancer cells.36,37 In addition, the activity of CD-AS constructs is target-specific, and the expression level of PTEN mRNA, which is a target of miR-21, was shown to be up-regulated by CD-AS in breast cancer cells.36 Moreover, anti-miRNA activity, nuclease resistance, and the miRNA modification profile in a CD-AS differ significantly based on the CD-connected positions in the ASO. However, the relationship between the inhibitory activity and subcellular distribution of ASOs has not been eluciated.
In this study, we investigated the effect of duplex structures on the subcellular localization of ASOs. The cytosol localization of AS having the CD at the 5′ end was much larger than that of AS with the CD at the 3′ end. Moreover, the cytoplasmic localization of ASOs targeted toward miRNA significantly correlated with their miRNA inhibitory activities. These results suggest the importance of the duplex structure in controlling the subcellular localization of oligonucleotides.
Results and discussion
Subcellular localization of oligonucleotides with a CD at the 5′- or 3′-end
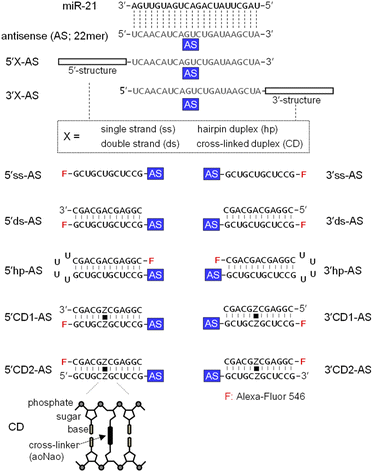
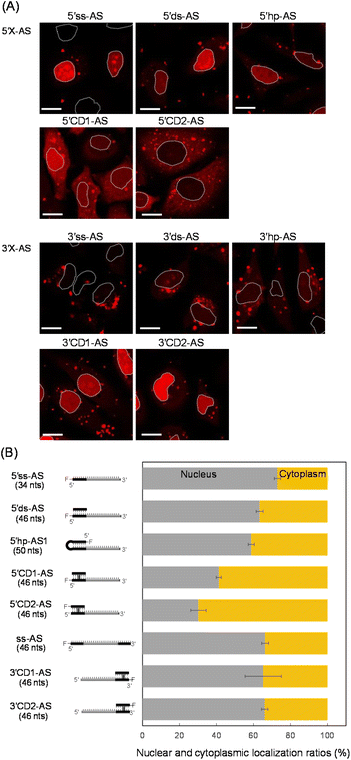
MicroRNA-21 (miR-21), a cancer-promoting miRNA, is overexpressed in solid tumors.38 The effect of miR-21 can be suppressed by an ASO that has a complementary sequence.36,39,40 We previously examined the anti-miRNA activities of 2′-O-methylated ASOs in which a 22-mer antisense (AS) was flanked by perfectly matched double-strand or a CD at the 5′- or 3′-end of the AS. The CD was constructed from the bis-aminooxy linker mediated conjugation of two abasic sites in the center of the complementary 12-mer duplex.35,41 The 12-mer 2′-O-methylated CD adopted a typical A-form structure that was identical to the standard duplex of Watson–Crick base pairs. The rigid conformation of the CD confers the potential to be highly resistant to nucleases and to thereby stabilize neighboring hybridization. A 5′CD-AS having the CD at 5′-end of AS has been shown to knockdown miR-21 more efficiently than a 3′CD-AS having the CD at 3′-end,35 although the reason for the difference in the anti-miRNA activity was unclear. In this study, we focused on the subcellular localization of ASOs with or without double-stranded structures. ASs having single strand (ss) (5′ss-AS), double strand (ds) (5′ds-AS), hairpin loop (hp) (5′hp-AS) or CD (5′CD1-AS) at the 5′-end were prepared (Fig. 1, 5′X-AS). All of these oligonucleotides were synthesized from 2′-O-methyl ribonucleotides with phosphodiester internucleotide (PO) linkages. The 5′-end of the 34- or 50-mer strands containing an AS was labelled with a fluorescein (Alexa-Fluor 546). We also prepared 5′CD2-AS that contained the 3′-end fluorescein labelled 12-mer in the CD (Fig. 1) to examine the effect of the labelling position.These oligonucleotides were transfected into HeLa cells. After 6 h, ASO-transfected cells were stained with Hoechst 33342 and calcein-AM to image the nucleus and cytoplasm, respectively. All fluorescent images were analyzed using confocal microscopy (Fig. 2A and S1†) after 6 h transfection, when subcellular localizations of ASOs could be detected and almost all cells were viable (Fig. S1†). We detected several red bright foci (Fig. 2A) that showed endosomes or lysosomes as seen in the lipofection-mediated transfections.20,42 Based on the calcein-AM and Hoechst 33342 fluorescence staining images (Fig. S1†), the total fluorescence intensities of Alexa-Fluor 546 in the nucleus and cytoplasm were quantified for approximately 100 cells, and the nuclear and cytoplasmic localization ratios of each oligonucleotide were determined, respectively (Fig. 2B). An AS with single strand and double strand at the 5′-end (5′ss-AS and 5′ds-AS) displayed approximately two-fold more total fluorescence intensity in the nucleus than in the cytoplasm (Fig. 2B). The 5′hp-AS with a hairpin loop at 5′-end had a similar nuclear localization ratio (59%) as that of 5′ds-AS (Fig. 2B).
The 5′CD1-AS, with the CD at the 5′-end, significantly decreased the nuclear localization ratio from approximately 73% for 5′ss-AS to 41% (Fig. 2B). Additionally, the 5′CD2-AS had a similar nuclear localization ratio (30%) as that of 5′CD1-AS (Fig. 2B). Since the 5′ss-AS consisted of 34 nucleotides (nts), this had a lower molecular weight than that of 5′CD1-AS, a single stranded 46 nts (ss-AS, Fig. S2†) having a 12-mer ss at both 5′- and 3′-ends of the AS (Fig. S1†). Although ss-AS had the same molecular weight as 5′CD1-AS (46 nts), ss-AS had a high nucleus localization (66%) similar to that of 5′ss-AS (Fig. 2B). Notably, 5′CD1-AS displayed a distinctly different behavior in cells from that of 5′ds-AS and 5′hp-AS that contained the standard duplex, despite the similar molecular weights of 46 and 50 nts, respectively (Fig. 2B). This difference in subcellular localization was derived from the duplex structures adjacent to the AS. The CD of 5′CD1-AS was stable in cells because of the interstrand cross-link, whereas the duplexes of 5′ds-AS and 5′hp-AS were dissociated or degraded to provide short single strands such as 5′ss-AS (Fig. 2). The 5′CD-AS maintained cytoplasmic localization 48 h after transfection (Fig. S3†); however, 5′ss-AS and 5′ds-AS significantly reduced the fluorescence intensities in cells after 6 h, due to the degradation.
We then prepared 3′ss-AS, 3′ds-AS, 3′hp-AS, 3′CD1-AS, and 3′CD2-AS, based on the 22 nt AS with ss, ds, hp, and CD at the 3′-end, respectively (Fig. 1, 3′X-AS), and analysed their intracellular localizations (Fig. 2A and S1†). Similar to 5′-modified ASs, all 3′-modified ASs produced several bright red foci in the cytoplasm (Fig. 2A); however, the fluorescence intensities of 3′ss-AS, 3′ds-AS, and 3′hp-AS were very weak, suggesting degradation in cells (Fig. 2A). In contrast, both 3′CD1-AS and 3′CD2-AS were clearly detected in the nucleus with a localization ratio of approximately 65% (Fig. 2B). The 3′CD-AS maintained nuclear localization 48 h after transfection (Fig. S3†). Interestingly, this result differed from that observed with 5′CD-AS despite their identical molecular weight.
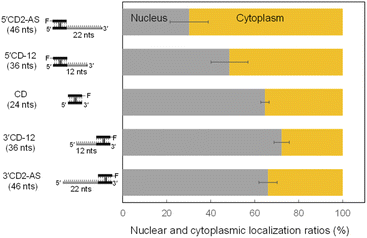
The AS sequence of 5′CD2-AS and 3′CD2-AS was then changed to the mismatched AS (mAS), the AS of microRNA-16 (R16), and a scrambled sequence (NC), respectively (Fig. S2†). The 5′CD- or 3′CD-linked oligonucleotides maintained the same localization profile as the parent molecule (5′CD2-AS or 3′CD2-AS, Fig. S4†), confirming that the AS sequence does not affect the subcellular localization. Moreover, the AS sequence was shortened from 22 nts to 0 or 12 nts (CD, 5′CD-12, 3′CD-12; Fig. S2†). The percentages of nucleus localization of 5′CD-12 (36 nts) slightly increased to 49% (Fig. 3) from 30% of 5′CD2-AS (46 nts). The CD (24 nts) without the dangling sequence showed a nucleus localization (65%, Fig. 3) as high as the single-stranded ss-AS (Fig. 2B). In contrast, 3′CD-12 (36 nts) interestingly showed no change in the nucleus localization ratio from 3′CD2-AS or CD (Fig. 3), with a different localization profile from 5′CD-linked sequences that demonstrated a dependence on molecular weight.
5′-End modifications of single-stranded oligonucleotides
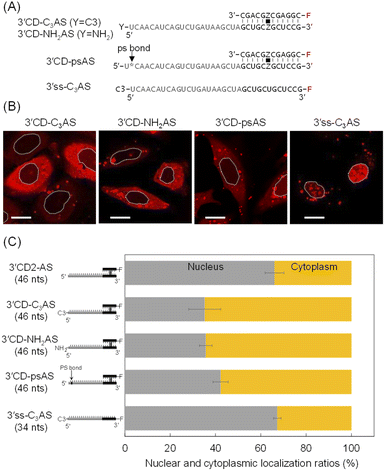
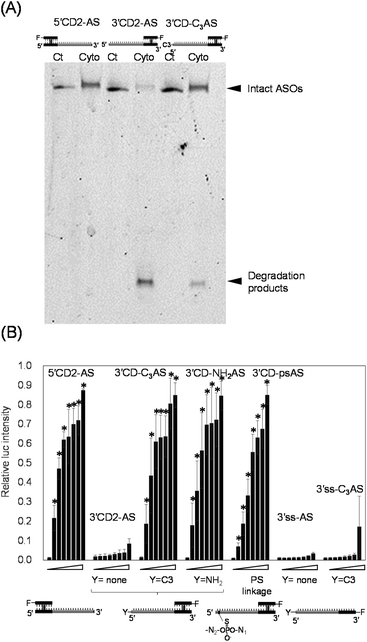
The intracellular localization of 3′CD-linked single strands was independent of the length or the sequence of the dangling regions. Therefore, we hypothesized that the dangling AS region of 3′CD-AS might be degraded from the 5′-terminal without any protecting groups. Nuclease resistance was conferred to AS by attaching a non-nucleotidyl propyl (C3)- or an amino (NH2)-linker to the 5′-end to create 3′CD-C3AS and 3′CD-NH2AS, respectively (Fig. 4A). The subcellular localization of these oligonucleotides was then examined. Surprisingly, both 3′CD-C3AS and 3′CD-NH2AS were dominantly localized in the cytoplasm (Fig. 4B), reducing the nuclear localization ratios to approximately 35% (Fig. 4C), which is the same as those of 5′CD-AS. Additionally, the phosphodiester bond between the first and second nucleotides (5′UC3′) of the AS was changed to a PS bond, which is highly resistant to nuclease digestion, to create 3′CD-psAS (Fig. 4A). After transfection into cells, 3′CD-psAS was mainly detected in the cytoplasm, similar to 3′CD-C3AS, 3′CD-NH2AS or 5′CD-AS with a protected 5′-end on the AS (Fig. 4B and C). The dangling AS of 3′CD-AS may be degraded in the cytoplasm by a 5′-3′exonuclease such as XRN,43 resulting in the production of the CD domain, i.e., which was nuclease resistant due to the rigid conformation. Since the CD domain is localized in the nucleus, as shown in Fig. 3, 3′CD-AS exhibited localization in the nucleus. These results indicate that the protection from the 5′–3′ cleavage is an important factor for ensuring prolonged antisense effects in cells. To evaluate the effect of 5′ -end modification on localization of a single-stranded oligonucleotide, the 5′ -end of 3′ss-AS was modified with a propyl-linker to create 3′ss-C3AS (Fig. 4A). Although 3′ss-AS without the 5′ -terminal protection could not provide images because of prompt degradation (Fig. 2A), 3′ss-C3AS could be detected in the nucleus (Fig. 4B and C). As shown in Fig. 2, 5′ss-AS which had an Alexa-Fluor residue at the 5′ -end, also exhibited nuclear localization. Taken together, these results indicated that single-stranded oligonucleotides became stable in cells because of 5′ -end protection but they are delivered to the nucleus unlike 3′CD-C3AS.We then directly analysed the stability of oligonucleotides with or without 5′ -end protection in the cytoplasm. After 5′CD2-AS, 3′CD2-AS, and 3′CD-C3AS were transfected into cells, their respective cytoplasmic fractions were recovered, followed by polyacrylamide gel analysis. 5′CD2-AS was intact (lane cyto in Fig. 5A and S5†), 3′CD2-AS was degraded, whereas most 3′CD-C3AS molecules remained stable in the cytoplasm with a small number of degraded products (Fig. 5A). These results were consistent with those derived from the imaging analyses. The stability of single-stranded 3′ss-AS, 3′ss-C3AS, and ss-AS was also examined. Most 3′ss-AS molecules without 5′-protection were degraded in the cytoplasm (Fig. S6†). In contrast, 3′ss-C3AS and ss-AS with the propyl and Alexa-Fluor residues at the 5′-end (Fig. S6†), respectively, were resistant to degradation. These results confirmed that a 5′-end modification such as a CD or the non-nucleotidyl linker could protect the single-stranded AS region from degradation in the cytoplasm.
Inhibition of miRNAs is associated with cytoplasmic localization of ASOs
There was increased cytoplasm localization of 3′CD-yASs (y = C3, NH2, ps) due to the y-modifications at the 5′-end of the AS (Fig. 4). We then evaluated the anti-miRNA activities of the oligonucleotides 48 h after transfection using a dual-luciferase assay. A target sequence complementary to the mature miR-21 was inserted into the 3′-UTR of the Renilla luciferase (hRluc) gene of the psiCHECK-2 vector, which contained both hRluc and firefly luciferase (Fluc) genes in a single plasmid. When the vector and ASOs are introduced into cells, the effects of endogenous miR-21 function can be monitored by measuring the relative expression of hRluc and Fluc. The ratio of hRluc/Fluc were normalized using the ratio of psiCHECK-2-treated cells without ASO. The 3′CD2-AS without the 5′-end modification displayed little suppression of miR-21, although 3′CD-yASs could suppress miR-21 to the same degree as 5′CD2-AS (Fig. 5B). The same results were also obtained for unlabelled 3′CD-yASs (Fig. S7†). Single-stranded 3′ss-C3AS with a high nuclear localization ratio did not suppress miR-21. These results confirmed that the anti-miRNA effects of ASOs are closely associated with the cytoplasmic localization ratio of the ASOs. The miRNAs are loaded into the Argonaute protein to form a miRNA-induced silencing complex (miRISC), which then binds the 3′-UTR of mRNA in the cytoplasm.25,44,45 Therefore, the increase in ASO concentration in the cytoplasm is advantageous for the association between ASOs and their target miRNAs.Fig. 6 summarizes these results. The dangling AS part of 3′CD-AS was degraded in the cytoplasm, producing the CD domain, which is resistant to nuclease degradation because of the interstrand cross-linking. When the CD was transfected into cells, the CD was translocated to the nucleus (Fig. 3). Thus, 3′CD-AS was apparently localized in the nucleus (I in Fig. 6). Single-stranded ss-AS, 5′ss-AS, and 3′ss-C3AS with 5′-end protections were stable in the cytoplasm (II in Fig. 6) but most of them moved to the nucleus. Both 5′CD-AS and 3′CD-yAS (y = C3, NH2 and ps) were stable in cytoplasm because of the CD and the y moiety at the 5′-end of the AS and were suppressed from migrating to the nucleus (III in Fig. 6). These localization profiles increased the concentrations of 5′CD-AS and 3′CD-yAS in the cytoplasm, resulting in the effective knockdown of miRNA acting in cytoplasm.
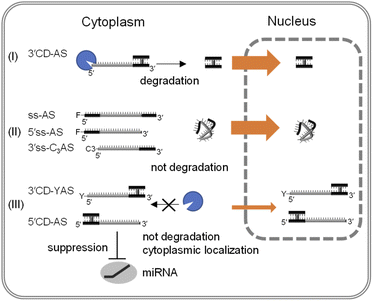 | ||
| Fig. 6 Schematic representation of the intracellular distribution of single-stranded ASOs and ASOs with CD. | ||
Notably, 5′CD-AS, 3′CD-yAS, and ss-AS showed different localization profiles despite their identical molecular weight of 46 nts; therefore, the differences in localization were likely due to the differences in structure. A single-stranded oligonucleotide adopts a flexible and compactly folded random-coil structure. As the result, a single-stranded oligonucleotide might pass through nuclear pores from the cytoplasm. In contrast, the single dangling strand may be preorganized to have an extended form by the connection with a CD due to the rigid rod-like structure of the CD.46 Therefore, the CD-connected AS having with a bulky conformation may be suppressed from been translocated to the nucleus. These results suggest that the CD modification can promote the accumulation of single-stranded ASOs in the cytoplasm.
Conclusions
Phosphorothioate (PS) linkages have been widely used to confer nuclease resistance to oligonucleotides.7 Since a PS linkage has a high affinity for protein, protein-binding greatly affects the intracellular function or distribution of PS-modified ASOs (PS-ASOs).10–12 Additionally, unfavorable protein-binding with PS-ASOs sometimes causes toxicological effects.22,47 In contrast, oligonucleotides with wild-type phosphodiester (PO) linkages have minimal interaction with proteins, but single-stranded PO-ASOs are susceptible to nuclease digestion. Previously, we demonstrated that connecting the antisense sequence with a CD could overcome PO-ASOs instability.35In this study, we examined the intracellular distributions of CD-connected ASs with PO-linkages. After transfection into cells, all single-stranded ASs promptly moved into the nucleus, whereas the CD-connected ASs were suppressed from such translocation, increasing the amount of CD-connected AS in the cytoplasm. We believe that CD-connected ASs would be advantageous in vivo by targeting RNAs existing in the cytoplasm. Importantly, dangling ASs with a free hydroxyl 5′-terminal were susceptible to degradation in the cytoplasm. The AS instability was compensated for by introducing a CD or 5′-end chemical modifications. We propose that the AS will be organized to form an extended structure by connecting with the CD due to the rigid structure of this and the high stacking effect on the neighboring strand.46 The resultant extended conformation may contribute to suppression of the transportation of the oligonucleotide to the nucleus. In contrast, a single-stranded AS has a flexible and random-coil structure that enables its rapid translocation to the nucleus.
Various types of RNAs localize differently in cells. If ASOs can be delivered to intracellular sites specific for target RNAs, dose and side-effects such as the off-targeting of ASOs, can be significantly reduced. This study reveals that a stable duplex has the potential to regulate the intracellular distribution of oligonucleotides. Therefore, the results obtained here can provide important insights into controlling the localization of oligonucleotides in cells.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr Yasuhiro Mie for helpful discussion. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI [Grant-in-Aid for Scientific Research (No. 16H05106 and No. 20H03372)]. Funding for open access charge: JSPS.References
- A. Khvorova and J. K. Watts, Nat. Biotechnol., 2017, 35, 238–248 CrossRef CAS PubMed.
- D. R. Scoles, E. V. Minikel and S. M. Pulst, Neurol.: Genet., 2019, 5, e323 CAS.
- T. C. Roberts, R. Langer and M. J. A. Wood, Nat. Rev. Drug Discovery, 2020, 19, 673–694 CrossRef CAS PubMed.
- M. Nowotny, S. A. Gaidamakov, R. J. Crouch and W. Yang, Cell, 2005, 121, 1005–1016 CrossRef CAS PubMed.
- B. P. Monia, E. A. Lesnik, C. Gonzalez, W. F. Lima, D. McGee, C. J. Guinosso, A. M. Kawasaki, P. D. Cook and S. M. Freier, J. Biol. Chem., 1993, 268, 14514–14522 CrossRef CAS PubMed.
- S. T. Crooke, X. H. Liang, B. F. Baker and R. M. Crooke, J. Biol. Chem., 2021, 296, 100416 CrossRef CAS PubMed.
- J. Kurreck, Eur. J. Biochem., 2003, 270, 1628–1644 CrossRef CAS PubMed.
- M. A. Campbell and J. Wengel, Chem. Soc. Rev., 2011, 40, 5680–5689 RSC.
- T. Yamaguchi, M. Horiba and S. Obika, Chem. Commun., 2015, 51, 9737–9740 RSC.
- S. T. Crooke, T. A. Vickers and X. H. Liang, Nucleic Acids Res., 2021, 48, 5235–5253 CrossRef PubMed.
- D. A. Brown, S. H. Kang, S. M. Gryaznov, L. DeDionisio, O. Heidenreich, S. Sullivan, X. Xu and M. I. Nerenberg, J. Biol. Chem., 1994, 269, 26801–26805 CrossRef CAS.
- X. H. Liang, H. Sun, W. Shen and S. T. Crooke, Nucleic Acids Res., 2015, 43, 2927–2945 CrossRef CAS PubMed.
- E. Koller, T. M. Vincent, A. Chappell, S. De, M. Manoharan and C. F. Bennett, Nucleic Acids Res., 2011, 39, 4795–4807 CrossRef CAS PubMed.
- N. Oka and T. Wada, Chem. Soc. Rev., 2011, 40, 5829–5843 RSC.
- S. T. Crooke, S. Y. Wang, T. A. Vickers, W. Shen and X. H. Liang, Nat. Biotechnol., 2017, 35, 230–237 CrossRef CAS PubMed.
- R. L. Juliano, Nucleic Acids Res., 2016, 44, 6518–6548 CrossRef PubMed.
- A. J. Debacker, J. Voutila, M. Catley, D. Blakey and N. Habib, Mol. Ther., 2020, 28, 1759–1771 CrossRef CAS PubMed.
- T. L. Fisher, T. Terhorst, X. Cao and R. W. Wagner, Nucleic Acids Res., 1993, 21, 3857–3865 CrossRef CAS PubMed.
- D. Castanotto, M. Lin, C. Kowolik, L. A. Wang, X. Q. Ren, H. S. Soifer, T. Koch, B. R. Hansen, H. Oerum, B. Armstrong, Z. G. Wang, P. Bauer, J. Rossi and C. A. Stein, Nucleic Acids Res., 2015, 43, 9350–9361 CrossRef CAS PubMed.
- Z. U. Rehman, D. Hoekstra and I. S. Zuhorn, ACS Nano, 2013, 7, 3767–3777 CrossRef PubMed.
- X. H. Liang, W. Shen, H. Sun, G. A. Kinberger, T. P. Prakash, J. G. Nichols and S. T. Crooke, Nucleic Acids Res., 2016, 44, 3892–3907 CrossRef CAS PubMed.
- W. Shen, C. L. De Hoyos, M. T. Migawa, T. A. Vickers, H. Sun, A. Low, T. A. Bell, M. Rahdar, S. Mukhopadhyay, C. E. Hart, M. Bell, S. Riney, S. F. Murray, S. Greenlee, R. M. Crooke, X. hai Liang, P. P. Seth and S. T. Crooke, Nat. Biotechnol., 2019, 37, 640–650 CrossRef CAS PubMed.
- J. K. Bailey, W. Shen, X. H. Liang and S. T. Crooke, Nucleic Acids Res., 2017, 45, 10649–10671 CrossRef CAS PubMed.
- P. Lorenz, T. Misteli, B. F. Baker, C. F. Bennett and D. L. Spector, Nucleic Acids Res., 2000, 28, 582–592 CrossRef CAS PubMed.
- H. Kobayashi and Y. Tomari, Biochim. Biophys. Acta, Gene Regul. Mech., 2016, 1859, 71–81 CrossRef CAS PubMed.
- A. Stroynowska-Czerwinska, A. Fiszer and W. J. Krzyzosiak, Cell. Mol. Life Sci., 2014, 71, 2253–2270 CrossRef CAS PubMed.
- D. P. Bartel, Cell, 2009, 136, 215–233 CrossRef CAS PubMed.
- J. Weiler, J. Hunziker and J. Hall, Gene Ther., 2006, 13, 496–502 CrossRef CAS PubMed.
- K. A. Lennox and M. A. Behlke, Gene Ther., 2011, 18, 1111–1120 CrossRef CAS PubMed.
- K. Yoshioka, T. Kunieda, Y. Asami, H. Guo, H. Miyata, K. Yoshida-Tanaka, Y. Sujino, W. Y. Piao, H. Kuwahara, K. Nishina, R. I. Hara, T. Nagata, T. Wada, S. Obika and T. Yokota, Nucleic Acids Res., 2019, 47, 7321–7332 CrossRef CAS PubMed.
- M. Takahashi, N. Yamada, H. Hatakeyama, M. Murata, Y. Sato, N. Minakawa, H. Harashima and A. Matsuda, Nucleic Acids Res., 2013, 41, 10659–10667 CrossRef CAS PubMed.
- B. Robertson, A. B. Dalby, J. Karpilow, A. Khvorova, D. Leake and A. Vermeulen, Silence, 2010, 1, 10 CrossRef PubMed.
- T. Haraguchi, H. Nakano, T. Tagawa, T. Ohki, Y. Ueno, T. Yoshida and H. Iba, Nucleic Acids Res., 2012, 40, e58 CrossRef CAS PubMed.
- A. M. Abdelhady, Y. Hirano, K. Onizuka, H. Okamura, Y. Komatsu and F. Nagatsugi, Bioorg. Med. Chem. Lett., 2021, 48, 128257 CrossRef CAS PubMed.
- Y. Mie, Y. Hirano, K. Kowata, A. Nakamura, M. Yasunaga, Y. Nakajima and Y. Komatsu, Mol. Ther. Nucleic Acids, 2018, 10, 64–74 CrossRef CAS PubMed.
- S. Okumura, Y. Hirano and Y. Komatsu, Nucleosides, Nucleotides Nucleic Acids, 2020, 39, 225–235 CrossRef CAS PubMed.
- S. Okumura, Y. Hirano and Y. Komatsu, Sci. Rep., 2021, 11, 11467 CrossRef CAS PubMed.
- A. M. Krichevsky and G. Gabriely, J. Cell. Mol. Med., 2009, 13, 39–53 CrossRef CAS PubMed.
- M. L. Si, S. Zhu, H. Wu, Z. Lu, F. Wu and Y. Y. Mo, Oncogene, 2007, 26, 2799–2803 CrossRef CAS PubMed.
- X. Pan, Z. X. Wang and R. Wang, Cancer Biol. Ther., 2010, 10, 1224–1232 CrossRef CAS PubMed.
- K. Ichikawa, N. Kojima, Y. Hirano, T. Takebayashi, K. Kowata and Y. Komatsu, Chem. Commun., 2012, 48, 2143–2145 RSC.
- M. Hirsch and M. Helm, Nucleic Acids Res., 2015, 43, 4650–4660 CrossRef CAS PubMed.
- V. K. Nagarajan, C. I. Jones, S. F. Newbury and P. J. Green, Biochim. Biophys. Acta, Gene Regul. Mech., 2013, 1829, 590–603 CrossRef CAS PubMed.
- H. Siomi and M. C. Siomi, Nat. Cell Biol., 2009, 11, 1049–1051 CrossRef CAS PubMed.
- L. Stalder, W. Heusermann, L. Sokol, D. Trojer, J. Wirz, J. Hean, A. Fritzsche, F. Aeschimann, V. Pfanzagl, P. Basselet, J. Weiler, M. Hintersteiner, D. V Morrissey and N. C. Meisner-Kober, EMBO J., 2013, 32, 1115–1127 CrossRef CAS PubMed.
- Y. Mie, K. Kowata, N. Kojima and Y. Komatsu, Langmuir, 2012, 28, 17211–17215 CrossRef CAS PubMed.
- M. T. Migawa, W. Shen, W. Brad Wan, G. Vasquez, M. E. Oestergaard, A. Low, C. L. De Hoyos, R. Gupta, S. Murray, M. Tanowitz, M. Bell, J. G. Nichols, H. Gaus, L. Xue-Hai, E. E. Swayze, S. T. Crooke and P. P. Seth, Nucleic Acids Res., 2019, 47, 5465–5479 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra04375k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |