Open Access Article
Open Access ArticlePyrogallol, Corilagin and Chebulagic acid target the “fuzzy coat” of alpha-synuclein to inhibit the fibrillization of the protein†
Mandar Bopardikar a,
Sri Rama Koti Ainavarapu
a,
Sri Rama Koti Ainavarapu *a and
Ramakrishna V. Hosur*b
*a and
Ramakrishna V. Hosur*b
aDepartment of Chemical Sciences, Tata Institute of Fundamental Research, Homi Bhabha Road, Colaba, Mumbai 400005, India. E-mail: koti@tifr.res.in
bUM-DAE Centre for Excellence in Basic Sciences, University of Mumbai, Kalina Campus, Santacruz, Mumbai 400098, India. E-mail: rvhosur53@gmail.com
First published on 14th December 2022
Abstract
The accumulation of the intrinsically disordered protein alpha-synuclein (αSyn) in the form of insoluble fibrillar aggregates in the central nervous system is linked to a variety of neurodegenerative disorders such as Parkinson's disease, Lewy body dementia, and multiple system atrophy. Here we show that Pyrogallol, Corilagin and Chebulagic acid, compounds containing a different number of catechol rings, are independently capable of delaying and reducing the extent of αSyn fibrillization. The efficiency of inhibition was found to correlate with the number of catechol rings. Further, our NMR studies reveal that these compounds interact with the N-terminal region of αSyn which is unstructured even in the fibrillar form of the protein and is known as the “fuzzy coat” of fibrils. Thus, Corilagin and Chebulagic acid target the fuzzy coat of αSyn and not the amyloid core which is a common target for the inhibition of protein fibrillization. Our results indicate that the N-terminus also plays a key role in the fibrillization of αSyn.
Introduction
Neurodegenerative disorders account for a significant and increasing proportion of deaths and disabilities worldwide. Neurodegenerative disorders such as Parkinson's disease (PD), lewy body dementia (LBD) and multiple system atrophy (MSA) are characterized pathologically by the accumulation of alpha-synuclein (αSyn) protein in the form of insoluble aggregates, known as Lewy Bodies (LB), in the cytoplasm of dopaminergic neurons in the central nervous system.1–3 Familial early onset PD is linked with the duplication or triplication of the gene encoding αSyn (SNCA).4 These observations suggest a strong correlation between αSyn fibrillization and the occurrence of neurodegenerative disorders like PD, LBD and MSA which are collectively known as synucleinopathies.αSyn‡ is an intrinsically disordered protein (IDP) that is made up of 140 amino acid residues.5 αSyn consists of an amphipathic N-terminal domain (residues 1–60), a hydrophobic central part known as NAC (non-amyloid β component) region (residues 61–95) and an acidic C-terminal region (residues 96–140).6 Interestingly, the Lewy Body variant of Alzheimer's Disease is characterized by the accumulation of a 35 residues long peptide, with the exact same sequence as that of the NAC region of αSyn, along with amyloid β peptide in the form of cytotoxic insoluble aggregates.7,8
There is no preventive therapy available for synucleinopathies as yet. However, inhibition of αSyn fibrillization with the help of chemical reagents represents a likely therapeutic strategy. In the past few years, several studies have reported the inhibition of αSyn fibrillization by small organic molecules,9–35 nanomaterials,36–38 herbal formulations39–43 and chaperone proteins.44–46 However, there are relatively fewer reports which provide an in-depth residue-level understanding of how these reagents inhibit αSyn fibrillization.14,15,17,20,45–48 A vast majority of the small organic molecules that are capable of inhibiting αSyn fibrillization are catechols.9 Here, we have investigated the effect of Pyrogallol (PGL), Corilagin (CLN) and Chebulagic acid (CA) (Fig. 1) on αSyn fibrillization. The choice of these molecules was driven by the presence of different number of catechol rings. PGL, CLN and CA consist of one, three and four catechol rings respectively. The point here is to understand qualitatively, the dependence of amyloid inhibition potential of catechol compounds on the number of catechol rings. Also, in a previous study we had shown that triphala, a herbal preparation, is effective in inhibiting αSyn fibrillization and that PGL, CLN and CA are ingredients of triphala.39 Therefore, in the current study we wanted to investigate the effect of each of these molecules on αSyn fibrillization.
PGL is known to possess antiproliferative activity towards human tumor cell lines.49 Also, it has been reported that PGL induces G2-M arrest in human lung cancer cells and inhibits tumor growth in animal model system.50 Another study has shown that PGL regulates expression of pro-inflammatory genes in bronchial epithelial cells.51 On the other hand, CLN is capable of alleviating hepatic fibrosis caused by egg granuloma in Schistosoma japonicum infection.52 A study has shown that CLN exhibits anti-hyperalgesic activity in thermal and chemical stimulation cased nociception mice models.53 It has been demonstrated that CLN produces anti-inflammatory effect in herpes simplex virus (HSV)-1 induced encephalitis.54 In the case of CA, it is known that this compound exerts inhibitory effect on neuraminidase-mediated influenza A viral release55 and also possesses broad-spectrum antiviral activity against viruses that use glycosaminoglycans for entry.56 CA inhibits tumor necrosis factor-α induced pro-angiogenic and pro-inflammatory activities in retinal capillary endothelial cells by inhibiting the phosphorylation of p38 group of mitogen-activated protein (MAP) kinases, extracellular signal-regulated kinase (ERK) and nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB).57 CA is a highly potent inhibitor of topoisomerase I.58
As mentioned above, there are several reports showing the therapeutic potential of PGL, CLN and CA against different disorders and infections. However, the effect of these compounds on αSyn fibrillization has not been studied so far. In the current study, we find that they are independently capable of delaying and reducing the extent of fibrillization of αSyn. Further, our NMR studies provide residue-level insights into the mode of interaction of these compounds with αSyn.
Materials and methods
Protein overexpression and purification
αSyn gene inserted into a pT7-7 vector construct, with the capability to withstand ampicillin, was expressed in Escherichia coli (E. coli) BL21 (DE3) cells. A glycerol stock of these E. coli cells, containing αSyn plasmid, was inoculated into 10 ml of LB containing 100 μg ml−1 of freshly prepared ampicillin. The culture was allowed to grow overnight at 37 °C with shaking (180 rpm). The overnight culture was then transferred to 1 litre of LB or M9 culture containing 100 μg ml−1 ampicillin. The resulting culture was grown until an OD600 of 0.8–1.0 was reached. At this point, the culture was instigated for protein expression with the help of 1 mM IPTG for 6–10 h. Eventually, the cells were pelleted down from the culture and then stored at −80 °C. αSyn protein was purified by a method reported earlier.59,60 The method goes as follows: initially, frozen cell pellet obtained from 1 litre culture was resuspended in Tris buffer (50 mM Tris, 10 mM EDTA and 150 mM NaCl) pH 8. Subsequently, sonication was performed for 10 min. Then, the suspension was placed in a boiling water-bath for 20 min. This was followed by centrifugation for 10 min. After centrifugation, the precipitate was discarded and the supernatant was collected into a fresh tube. Streptomycin sulphate (136 μl of 10% solution per ml of supernatant) and glacial acetic acid (228 μl per ml of supernatant) were added to the supernatant. The resulting mixture was centrifuged for 10 min. Again, the supernatant was collected, whereas, the precipitate was discarded. Now, saturated ammonium sulphate was added to the supernatant in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) proportion which causes protein precipitation. Precipitated protein was washed with ammonium sulphate solution (50% saturated). The pellet was resuspended in 3 ml of 100 mM ammonium acetate and re-precipitated with the help of an equal volume of ethanol. The previous step was performed a total of three times. The precipitate was then solubilized in 100 mM ammonium acetate. Ultimately, the solution was frozen in liquid nitrogen and lyophilized. Prior to usage in experiments, the lyophilized protein was further purified by solubilizing it in PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl and 0.01% (w/v) sodium azide) pH 7.4, followed by centrifugation and then performing size exclusion chromatography for the supernatant using a HiLoad 16/600 Superdex75 column (GE Healthcare, UK) with the help of BioLogic DuoFlow FPLC system (Bio-Rad, USA). The protein purity of the fractions was analyzed by SDS-PAGE. All of those fractions which contained only pure αSyn were pooled together and concentrated with the help of 3 kDa Amicon ultra centrifugal filter units (Merck KGaA, Germany). Protein concentration was determined spectrophotometrically by UV absorption measurements using ε280 = 5960 M−1 cm−1.
1 (v/v) proportion which causes protein precipitation. Precipitated protein was washed with ammonium sulphate solution (50% saturated). The pellet was resuspended in 3 ml of 100 mM ammonium acetate and re-precipitated with the help of an equal volume of ethanol. The previous step was performed a total of three times. The precipitate was then solubilized in 100 mM ammonium acetate. Ultimately, the solution was frozen in liquid nitrogen and lyophilized. Prior to usage in experiments, the lyophilized protein was further purified by solubilizing it in PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl and 0.01% (w/v) sodium azide) pH 7.4, followed by centrifugation and then performing size exclusion chromatography for the supernatant using a HiLoad 16/600 Superdex75 column (GE Healthcare, UK) with the help of BioLogic DuoFlow FPLC system (Bio-Rad, USA). The protein purity of the fractions was analyzed by SDS-PAGE. All of those fractions which contained only pure αSyn were pooled together and concentrated with the help of 3 kDa Amicon ultra centrifugal filter units (Merck KGaA, Germany). Protein concentration was determined spectrophotometrically by UV absorption measurements using ε280 = 5960 M−1 cm−1.
Thioflavin T (ThT) fluorescence assay
The process of fibrillization was initiated with 150 μM αSyn in 2 ml Protein LoBind Eppendorf tube in PBS pH 7.4. The protein samples either in the presence or absence of equimolar concentration of PGL, CLN or CA were exposed to mechanical agitation at 65 rpm and 37 °C. At different time points, 5 μl aliquots were taken out from each of the samples. Each aliquot was added to 245 μl of 15 μM ThT solution in PBS pH 7.4. Fluorescence was immediately recorded for the resulting solution with an Agilent spectrofluorometer. The excitation wavelength and emission wavelength range were 444 nm and 455–500 nm respectively. For all the measurements, both the excitation and emission bandwidths were 5 nm. In the end, the normalized fluorescence intensity at 485 nm was plotted against time.ThT fluorescence data analysis
The ThT fluorescence data sets reporting the fibrillization of αSyn in the absence and presence of PGL or CLN were fitted with the following equation:
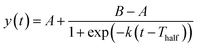 | (1) |
| Tlag = Thalf − 2/k | (2) |
All the data analysis was performed with the help of in-house written python programs.
Transmission electron microscopy (TEM)
At the end of the fibrillization process, each sample was diluted to 30 μM with 0.22 micron filtered distilled water. These samples were then spotted onto a carbon-coated Formvar grid and incubated for 5 min. Further, staining was performed with a freshly prepared 1% (w/v) aqueous solution of uranyl acetate, filtered through 0.22 μm syringe filter, by incubation for 1 min. Eventually, the samples were washed with 0.22 micron filtered distilled water. The samples were then allowed to dry overnight. Finally, images were acquired using a 200 kV FEI Tecnai-20 transmission electron microscope (TEM).Nuclear magnetic resonance (NMR) spectroscopy
NMR experiments were performed on 800 MHz Bruker Avance NMR spectrometer equipped with cryogenically cooled triple-resonance probe. All the NMR measurements were performed in PBS, pH 7.4 at a temperature of 280 K if not otherwise indicated. 1H–15N BEST-HSQC spectra were recorded for 108 μM 15N isotopically enriched αSyn in the presence and absence of different concentrations of PGL, CLN or CA with inter-scan delays of 200 ms and 2048 × 512 points for the 1H and 15N dimension respectively. Spectra were recorded in phase-sensitive mode and quadrature detection was performed with the help of States-TPPI method.61 The 1H and 15N dimensions were zero-filled to 4096 and 1024 points, respectively. Sine squared bell apodization with SSB = 2 was used as a window function for both dimensions. Data was processed with the help of Topspin 4.0 (Bruker, USA) and analysed with CCPN62 (Collaborative Computing Project for NMR, University of Leicester, UK). Chemical shift assignments were obtained by transfer from BMRB (accession number 16300) and other previous reports.46 Chemical shift perturbations (CSP) were calculated as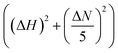 where ΔN and ΔH represent the difference in the nitrogen and proton chemical shifts of αSyn in the presence of PGL, CLN or CA with respect to that of pure αSyn, respectively.
where ΔN and ΔH represent the difference in the nitrogen and proton chemical shifts of αSyn in the presence of PGL, CLN or CA with respect to that of pure αSyn, respectively.
Results and discussion
Pyrogallol, Corilagin and Chebulagic acid inhibit αSyn fibrillization
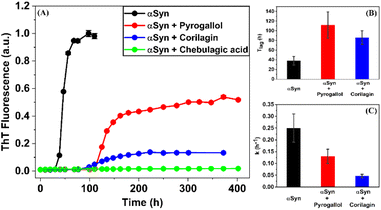
The process of fibrillization of αSyn was monitored with the help of thioflavin T (ThT) binding assay63–65 in a time-dependent manner. αSyn fibrillization is a nucleation-controlled polymerization66 process which shows a sigmoidal curve with three different phases – (i) nucleation or lag, (ii) elongation and (iii) saturation. It was observed that an equimolar concentration of PGL or CLN increased the duration of the nucleation phase or lag phase (Fig. 2(A)). In other words, both PGL and CLN delayed the onset of αSyn fibrillization. Also, there was reduction in ThT fluorescence in the saturation phase to 52% and 13% in the presence of PGL and CLN, respectively (Fig. 2(A)), indicating that a smaller amount of the protein underwent the transition from natively unstructured conformation into the amyloid fibrillar form as compared to the protein in the absence of these molecules. In order to check for any interfering effects of PGL, CLN or CA on the ThT fluorescence assay due to (a) absorbance in the wavelength region where ThT fluoresces or (b) absorbance in the wavelength region where ThT absorbs, UV-visible spectrophotometric measurements were performed (see ESI Methods). It was observed that PGL, CLN or CA do not absorb in this spectral region (Fig. S1†). Therefore, the reduced ThT fluorescence (Fig. 2(A)) may be attributed to the inhibition of αSyn fibrillization by PGL, CLN or CA in the respective cases.Further quantitative insights into the effect of PGL and CLN on αSyn fibrillization were obtained by fitting the ThT data sets with sigmoidal function (eqn (1) in Materials and Methods) to obtain the values of some of the important parameters (eqn (2) in Materials and Methods) characterizing the process of amyloid formation – duration of lag phase (Tlag), inflection point (Thalf) which is also the time required to reach 50% of maximal fluorescence and apparent first-order rate constant (k). For αSyn alone, Tlag was 38 (h) (Fig. 2(B) and Table 1). In the presence of PGL and CLN, the value of Tlag increased to 112 (h) and 86 (h) respectively (Fig. 2(B) and Table 1). Thus, the presence of PGL and CLN clearly caused a delay in the initiation of αSyn fibrillization. On the other hand, the value of k was 0.25 (h−1) for αSyn, which decreased to 0.13 (h−1) and 0.047 (h−1) in the presence of PGL and CLN respectively (Fig. 2(C) and Table 1). This shows that PGL reduced the rate of αSyn fibrillization to nearly half its value, whereas, CLN caused more than five times reduction. In the case of CA, the efficiency of inhibition was so high that there was no observable increase in ThT fluorescence even up to 17 days of incubation. Our ThT fluorescence experiments suggest that the efficiency of these compounds to inhibit αSyn fibrillization is in the following order:
| Experiment | Tlag (h) | Thalf (h) | k (h−1) |
|---|---|---|---|
| a Values of parameters in Table 1 were obtained by fitting the data in Fig. 2(A) with eqn (1) and Tlag was calculated with the help of eqn (2). The errors represent twice the standard deviation (2σ) from the mean. | |||
| αSyn | 38 ± 9 | 45.8 ± 0.9 | 0.25 ± 0.06 |
| αSyn + Pyrogallol | 112 ± 27 | 127 ± 2 | 0.13 ± 0.03 |
| αSyn + Corilagin | 86 ± 14 | 128 ± 3 | 0.047 ± 0.007 |
| αSyn + Chebulagic acid | — | — | — |
Chebulagic acid > Corilagin > Pyrogallol.
In order to confirm these observations, transmission electron microscopy (TEM) studies were performed to assess the effect of these compounds on αSyn fibril morphology and density. It was observed that PGL, CLN and CA cause a decrease in the fibril density (Fig. 3). However, the most pronounced effect was observed in the case of CA, where the reduction in fibril density was highest (Fig. 3). Thus, our TEM studies qualitatively support the trend shown by ThT fluorescence data.
Corilgain and Chebulagic acid mainly interact with the N-terminal region of αSyn
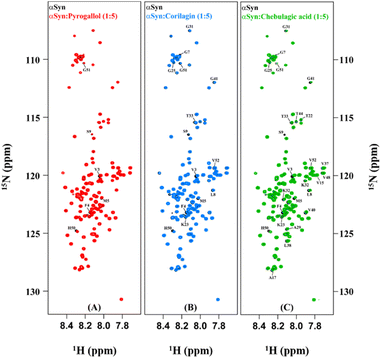
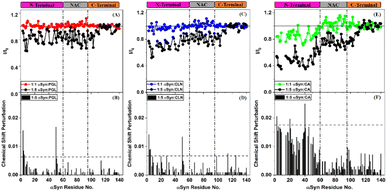
In order to understand the mechanism of inhibition of αSyn fibrillization by PGL, CLN and CA, 1H–15N HSQC experiments were performed on uniformly 15N isotopically enriched protein in the absence and presence of each of these compounds. However, before we understand the nature of interaction of these amyloid inhibitors with αSyn by NMR, it is important to note that the addition of each of these molecules to PBS solution introduced pH changes. It was observed the pH difference between the ‘protein + ligand’ solution and the protein solution was ∼0.3, ∼0.5 and ∼0.7 in the case of PGL, CLN and CA respectively (Fig. S2†). Now it is possible that such a pH change in the solution conditions might affect the NMR intensities and thereby interfere with the experiment. Upon investigation, we indeed observed a drastic deviation in the intensities of αSyn peaks when the pH of the ‘protein + ligand’ solution and the protein solution was not adjusted to the same value as compared to when there was no pH difference between these two solutions (Fig. S2†). Interestingly, even a pH change, as small as 0.3 (in the case of PGL) was found to have a profound influence on the intensities of 1H–15N resonances of αSyn (Fig. S2(A) and (B)†). This shows that protein backbone resonances could be extremely sensitive to the solvent conditions, especially pH which is possibly due to the labile nature of the backbone amide protons. Therefore, all our NMR experiments were performed with utmost care taking into consideration these drastic effects of pH conditions.An overlay of the two-dimensional NMR spectra of αSyn in the absence and presence of each amyloid inhibitor provided crucial insight into the location of residues that were affected the most by that particular compound (Fig. 4). It was observed that most of these residues belonged to the N-terminus in all the three cases (Fig. 4(A–C)). In order to obtain further insight, the intensity ratio and CSP for each individual well-resolved resonance were calculated for αSyn in the presence of each compound and plotted against the corresponding residue position in the protein primary sequence (Fig. 5). When equimolar amounts of PGL were used, the peak intensities were unaffected (Fig. 5(A)). However, when five molar equivalent concentration of PGL was used, peak broadening was observed for residues in the N-terminal domain and NAC region (Fig. 5(A)). The peak intensities dropped to nearly 83% in these domains of αSyn (Fig. 5(A) and Table S1†). This observation suggests that it is the N-terminal domain and NAC region of the protein which interact with PGL.
In the case of CLN, similar peak broadening was observed in the N-terminus and NAC domain when five molar equivalent concentration of CLN was used (Fig. 5(C)). However, the reduction in intensities for N-terminus was to a higher extent up to 73% as compared to NAC region 82% (Fig. 5(C) and Table S1†). The extent of signal attenuation for N-terminal region was found to be statistically higher than NAC domain (Table S1†). This suggests that the N-terminus is a preferred interaction domain for CLN over the NAC region of αSyn. The small magnitude of CSPs (<0.01 ppm) in the presence of PGL and CLN (Fig. 5(B) and (D)) is suggestive of the weak and transient nature of the interactions between each of these molecules and the protein.
On the other hand, the addition of equimolar amount of CA to αSyn resulted in a slight reduction in intensity for N-terminal residues (Fig. 5(E)). At a higher concentration of CA, that is αSyn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CA 1
CA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 molar stoichiometry, a drastic reduction in the peak intensities for residues in the N-terminus was observed (Fig. 5(E)). In fact, intensity ratios dropped to nearly 50% for residues V3-V26 and A29-G51 (Fig. 5(E)). The average signal attenuation of 51% for N-terminus was found to be statistically much lower (Table S1†) as compared to 76% for NAC region (Fig. 5(E) and Table S1†) indicating that CA prefers interaction with N-terminal region over NAC domain. Clearly, the effect of CA was much more pronounced as compared to that of PGL and CLN. Also, the CSPs were higher in the case of CA (Fig. 5(F)) in comparison to those observed due to PGL and CLN (Fig. 5(B) and (D) respectively).
5 molar stoichiometry, a drastic reduction in the peak intensities for residues in the N-terminus was observed (Fig. 5(E)). In fact, intensity ratios dropped to nearly 50% for residues V3-V26 and A29-G51 (Fig. 5(E)). The average signal attenuation of 51% for N-terminus was found to be statistically much lower (Table S1†) as compared to 76% for NAC region (Fig. 5(E) and Table S1†) indicating that CA prefers interaction with N-terminal region over NAC domain. Clearly, the effect of CA was much more pronounced as compared to that of PGL and CLN. Also, the CSPs were higher in the case of CA (Fig. 5(F)) in comparison to those observed due to PGL and CLN (Fig. 5(B) and (D) respectively).
Significance of the disordered N-terminus of αSyn in the process of fibrillization
Protein fibrillization is a complex and heterogeneous process. During this process, monomeric protein self-assembles along a multitude of pathways to form a wide variety of species such as off-pathway oligomers, on-pathway oligomers that transform into fibrils in the due course of time, protofibrils and fibrils. In the case of αSyn, the fibrillar core consists of a rigid Greek-key motif which is flanked by the unstructured C- and N-termini.67,68 Current ways of inhibiting protein fibrillization mainly target the amyloid core and include sequestration of monomers to prevent self-assembly,69,70 treatment with reagents that cause fibril clustering which reduces fragmentation of fibrils and consequently conceals binding site(s) for the addition of monomeric protein71,72 and using chaperones or engineered proteins to choke fibril surfaces.73–75 However, recent years have witnessed a growing interest in understanding the role of unstructured regions in fibril elongation. Truncating the disordered N- and C-termini modulates the aggregation kinetics of αSyn suggesting that these unstructured regions are not mere bystanders but actively participate in the process of fibrillization.76–79 In fact, Yang et. al. showed that the first 11 residues in the unstructured N-terminal region of αSyn form a motif for the recruitment of monomeric protein to fibril.80Here we have shown that Pyrogallol, Corilagin and Chebulagic acid inhibit αSyn fibrillization by interacting with the extreme N-terminal region of the protein. Our results, especially in the case of Chebulagic acid, clearly show that the N-terminus of αSyn plays a crucial role in the process of fibrillization of the protein. Similar to our findings, Burmann et. al. had observed that six different chaperone proteins inhibit αSyn fibrillization by binding to a common motif consisting of 12 residues in the N-terminus.46 Also, a recent study showed that an off-pathway oligomer of αSyn acts as an auto-inhibitor of fibrillization by recruiting monomeric protein through an N-terminal binding site comprising of the first 11 residues.80 All of these evidences highlight the significance of the N-terminal domain of αSyn in the process of fibrillization of the protein.
Conclusions
Our results demonstrate that Pyrogallol, Corilagin and Chebulagic acid inhibit αSyn fibrillization. Interestingly, each of these amyloid inhibitors interact with αSyn in the N-terminal and NAC regions of the protein. The C-terminal domain, which is highly acidic in nature, remains unaffected. In the case of Corilagin and Chebulagic acid, the interaction is more pronounced in the N-terminus than the NAC region. Corilagin and Chebulagic acid target the unstructured part of αSyn which belongs to the extreme N-terminal region of the protein, comprising of residues 3–26 and interact relatively weakly with the amyloid core, consisting of residues 37–43, 52–59, 62–66, 68–77 and 90–95, which is a common target for inhibiting fibrillization.81 Further studies need to be undertaken in order to understand the role of N-terminus in αSyn fibrillization in greater detail which could lead to the development of better therapeutic approaches for amyloid inhibition in the future.The compounds utilized in this study showed an efficacy to inhibit αSyn fibrillization in the following order: Chebulagic acid > Corilagin > Pyrogallol. Interestingly, the number of catechol rings in these molecules follows the same order, that is, Chebulagic acid, Corilagin and Pyrogallol consist of four, three and one catechol rings respectively. This observation possibly indicates that the capability of catechols to inhibit αSyn fibrillization could be dependent on the number of catechol rings. Further studies investigating the effect of a systematically designed library of catechols need to be pursued to confirm this observation. On the other hand, the fact that the choice of Pyrogallol, Corilagin and Chebulagic acid was also driven by these compounds being ingredients of triphala, herbal preparation which was previously shown to be effective in inhibiting αSyn fibrillization,39 strongly suggests that natural product-based drug discovery is an exercise worth perceiving towards the identification of new and effective amyloid inhibitors.
Author contributions
M. B., S. R. K. A. and R. V. H. conceived the project. M. B. performed all the experiments and analyzed the results. M. B. wrote the manuscript. All the authors were involved in the interpretation of results. S. R. K. A. and R. V. H. corrected the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
αSyn plasmid was a kind gift from Prof. Vinod Subramaniam, Vrije Universiteit Amsterdam. The authors are thankful to Shilpa Shirodkar, Jayesh Parmar and Lalit Borde for their help in TEM imaging. The authors are grateful to the National Facility for High-Field NMR at TIFR, Mumbai. The authors would like to thank Sudipta Maiti, Janeka Gartia and Anusri Bhattacharya for discussions during the course of the study. The help received from Ashutosh Kumar and Rajlaxmi Panigrahi in NMR data analysis is highly appreciated. S. R. K. A. acknowledges the financial support of the Department of Atomic Energy (DAE), India under Project No. 12-R&DTFR-5.10-0100.Notes and references
- J. Q. Trojanowski, M. Goedert, T. Iwatsubo and V. M. Y. Lee, Cell Death Differ., 1998, 5, 832–837 CrossRef CAS PubMed.
- M. Hashimoto and E. Masliah, Brain Pathol., 1999, 9, 707–720 CrossRef CAS PubMed.
- D. M. Skovronsky, V. M. Y. Lee and J. Q. Trojanowski, Annu. Rev. Pathol.: Mech. Dis., 2006, 1, 151–170 CrossRef CAS PubMed.
- A. B. Singleton, M. Farrer, J. Johnson, A. Singleton, S. Hague, J. Kachergus, M. Hulihan, T. Peuralinna, A. Dutra, R. Nussbaum, S. Lincoln, A. Crawley, M. Hanson, D. Maraganore, C. Adler, M. R. Cookson, M. Muenter, M. Baptista, D. Miller, J. Blancato, J. Hardy and K. Gwinn-Hardy, Science, 2003, 302, 841 CrossRef CAS PubMed.
- P. H. Weinreb, W. Zhen, A. W. Poon, K. A. Conway and P. T. Lansbury Jr, Biochemistry, 1996, 35, 13709–13715 CrossRef CAS PubMed.
- D. J. Moore, A. B. West, V. L. Dawson and T. M. Dawson, Annu. Rev. Neurosci., 2005, 28, 57–87 CrossRef CAS PubMed.
- K. Uéda, H. Fukushima, E. Masliah, Y. Xia, A. Iwai, M. Yoshimoto, D. A. Otero, J. Kondo, Y. Ihara and T. Saitoh, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 11282–11286 CrossRef PubMed.
- O. M. El-Agnaf, R. Jakes, M. D. Curran, D. Middleton, R. Ingenito, E. Bianchi, A. Pessi, D. Neill and A. Wallace, FEBS Lett., 1998, 440, 71–75 CrossRef CAS PubMed.
- K. A. Conway, J. C. Rochet, R. M. Bieganski and P. T. Lansbury Jr, Science, 2001, 294, 1346–1349 CrossRef CAS PubMed.
- J. Li, M. Zhu, S. Rajamani, V. N. Uversky and A. L. Fink, Chem. Biol., 2004, 11, 1513–1521 CrossRef CAS PubMed.
- M. Caruana, T. Högen, J. Levin, A. Hillmer, A. Giese and N. Vassallo, FEBS Lett., 2011, 585, 1113–1120 CrossRef CAS PubMed.
- B. Ahmad and L. J. Lapidus, J. Biol. Chem., 2012, 287, 9193–9199 CrossRef CAS PubMed.
- M. Zhu, S. Han and A. L. Fink, Biochim. Biophys. Acta, 2013, 1830, 2872–2881 CrossRef CAS PubMed.
- D. E. Ehrnhoefer, J. Bieschke, A. Boeddrich, M. Herbst, L. Masino, R. Lurz, S. Engemann, A. Pastore and E. E. Wanker, Nat. Struct. Mol. Biol., 2008, 15, 558–566 CrossRef CAS PubMed.
- X. Meng, L. A. Munishkina, A. L. Fink and V. N. Uversky, Biochemistry, 2009, 48, 8206–8224 CrossRef CAS PubMed.
- X. Meng, L. A. Munishkina, A. L. Fink and V. N. Uversky, Parkinson's Dis., 2010, 2010, 1–16 CrossRef PubMed.
- G. R. Lamberto, A. Binolfi, M. L. Orcellet, C. W. Bertoncini, M. Zweckstetter, C. Griesinger and C. O. Fernandez, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 21057–21062 CrossRef CAS PubMed.
- L. Breydo, J. W. Wu and V. N. Uversky, Biochim. Biophys. Acta, 2012, 1822, 261–285 CrossRef CAS PubMed.
- P. K. Singh, V. Kotia, D. Ghosh, G. M. Mohite, A. Kumar and S. K. Maji, ACS Chem, Neurosci., 2013, 4, 393–407 CAS.
- M. Perni, C. Galvagnion, A. Maltsev, G. Meisl, M. B. Müller, P. K. Challa, J. B. Kirkegaard, P. Flagmeier, S. I. Cohen, R. Cascella, S. W. Chen, R. Limbocker, P. Sormanni, G. T. Heller, F. A. Aprile, N. Cremades, C. Cecchi, F. Chiti, E. A. Nollen, T. P. Knowles, M. Vendruscolo, A. Bax, M. Zasloff and C. M. Dobson, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E1009–E1017 CrossRef CAS PubMed.
- N. N. Jha, R. Kumar, R. Panigrahi, A. Navalkar, D. Ghosh, S. Sahay, M. Mondal, A. Kumar and S. K. Maji, ACS Chem, Neurosci., 2017, 8, 2722–2733 CAS.
- T. Ibrahim and J. McLaurin, Biochem. Biophys. Res. Commun., 2016, 469, 529–534 CrossRef CAS PubMed.
- A. A. Deeg, A. M. Reiner, F. Schmidt, F. Schueder, S. Ryazanov, V. C. Ruf, K. Giller, S. Becker, A. Leonov, C. Griesinger, A. Giese and W. Zinth, Biochim. Biophys. Acta, 2015, 1850, 1884–1890 CrossRef CAS PubMed.
- N. Ahsan, S. Mishra, M. K. Jain, A. Surolia and S. Gupta, Sci. Rep., 2015, 5, 9862 CrossRef CAS PubMed.
- K. Ji, Y. Zhao, T. Yu, Z. Wang, H. Gong, X. Yang, Y. Liu and K. Huang, Food Funct., 2016, 7, 409–416 RSC.
- N. N. Jha, D. Ghosh, S. Das, A. Anoop, R. S. Jacob, P. K. Singh, N. Ayyagari, I. N. Namboothiri and S. K. Maji, Sci. Rep., 2016, 6, 28511 CrossRef CAS PubMed.
- N. Ahsan, I. A. Siddique, S. Gupta and A. Surolia, Eur. J. Med. Chem., 2018, 143, 1174–1184 CrossRef CAS PubMed.
- R. Chakraborty, S. Sahoo, N. Halder, H. Rath and K. Chattopadhyay, ACS Chem, Neurosci., 2019, 10, 573–587 CAS.
- D. Yedlapudi, G. S. Joshi, D. Luo, S. V. Todi and A. K. Dutta, Sci. Rep., 2016, 6, 38510 CrossRef CAS PubMed.
- J. Pujols, S. Peña-Díaz, D. F. Lázaro, F. Peccati, F. Pinheiro, D. González, A. Carija, S. Navarro, M. Conde-Giménez, J. García, S. Guardiola, E. Giralt, X. Salvatella, J. Sancho, M. Sodupe, T. F. Outeiro, E. Dalfó and S. Ventura, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 10481–10486 CrossRef CAS PubMed.
- S. Ghosh, A. Kundu and K. Chattopadhyay, Sci. Rep., 2018, 8, 5481 CrossRef PubMed.
- D. Ghosh, S. Mehra, S. Sahay, P. K. Singh and S. K. Maji, Int. J. Biol. Macromol., 2017, 100, 37–54 CrossRef CAS PubMed.
- M. J. Daniels, J. B. Nourse Jr, H. Kim, V. Sainati, M. Schiavina, M. G. Murrali, B. Pan, J. J. Ferrie, C. M. Haney, R. Moons, N. S. Gould, A. Natalello, R. Grandori, F. Sobott, E. J. Petersson, E. Rhoades, R. Pierattelli, I. Felli, V. N. Uversky, K. A. Caldwell, G. A. Caldwell, E. S. Krol and H. Ischiropoulos, Sci. Rep., 2019, 9, 2937 CrossRef PubMed.
- S. S. Save, K. Rachineni, R. V. Hosur and S. Choudhary, Int. J. Biol. Macromol., 2019, 141, 585–595 CrossRef CAS PubMed.
- A. Mahapatra, S. Sarkar, S. C. Biswas and K. Chattopadhyay, Chem. Commun., 2019, 55, 11052–11055 RSC.
- N. Joshi, S. Basak, S. Kundu, G. De, A. Mukhopadhyay and K. Chattopadhyay, Langmuir, 2015, 31, 1469–1478 CrossRef CAS PubMed.
- N. Taebnia, D. Morshedi, S. Yaghmaei, F. Aliakbari, F. Rahimi and A. Arpanaei, Langmuir, 2016, 32, 13394–13402 CrossRef CAS PubMed.
- A. Mahapatra, S. Sarkar, S. C. Biswas and K. Chattopadhyay, ACS Chem, Neurosci., 2020, 11, 3442–3454 CAS.
- M. Bopardikar, A. Bhattacharya, V. M. R. Kakita, K. Rachineni, L. C. Borde, S. Choudhary, S. R. K. Ainavarapu and R. V. Hosur, RSC Adv., 2019, 9, 28470–28477 RSC.
- S. Honarmand, B. Dabirmanesh, M. Amanlou and K. Khajeh, PLoS One, 2019, 14, e0217801 CrossRef CAS PubMed.
- M. T. Ardah, S. S. Ghanem, S. A. Abdulla, G. Lv, M. M. Emara, K. E. Paleologou, N. N. Vaikath, J.-H. Lu, M. Li, K. Vekrellis, D. Eliezer and O. M. A. El-Agnaf, BMC Complementary Med. Ther., 2020, 20, 73 CrossRef PubMed.
- D. Morshedi, F. Aliakbari, A. Tayaranian-Marvian, A. Fassihi, F. Pan-Montojo and H. Pérez-Sánchez, J. Food Sci., 2015, 80, H2336–H2345 CrossRef CAS PubMed.
- H. Javed, M. F. Nagoor Meeran, S. Azimullah, A. Adem, B. Sadek and S. K. Ojha, Front. Pharmacol., 2019, 9, 1555 CrossRef PubMed.
- A. Kakinen, I. Javed, A. Faridi, T. P. Davis and P. C. Ke, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 1803–1809 CrossRef CAS PubMed.
- G. Bellomo, S. Bologna, L. Cerofolini, S. Paciotti, L. Gatticchi, E. Ravera, L. Parnetti, M. Fragai and C. Luchinat, J. Phys. Chem. B, 2019, 123, 4380–4386 CrossRef CAS PubMed.
- B. M. Burmann, J. A. Gerez, I. Matečko-Burmann, S. Campioni, P. Kumari, D. Ghosh, A. Mazur, E. E. Aspholm, D. Šulskis, M. Wawrzyniuk, T. Bock, A. Schmidt, S. G. D. Rüdiger, R. Riek and S. Hiller, Nature, 2020, 577, 127–132 CrossRef CAS PubMed.
- J. N. Rao, V. Dua and T. S. Ulmer, Biochemistry, 2008, 47, 4651–4656 CrossRef CAS PubMed.
- R. Ahmed, J. Huang, D. K. Weber, T. Gopinath, G. Veglia, M. Akimoto, A. Khondker, M. C. Rheinstadter, V. Huynh, R. G. Wylie, J. C. Bozelli Jr, R. M. Epand and G. Melacini, J. Am. Chem. Soc., 2020, 142, 9686–9699 CrossRef CAS PubMed.
- M. T. Khan, I. Lampronti, D. Martello, N. Bianchi, S. Jabbar, M. S. Choudhuri, B. K. Datta and R. Gambari, Int. J. Oncol., 2002, 21, 187–192 CAS.
- C. J. Yang, C. S. Wang, J. Y. Hung, H. W. Huang, Y. C. Chia, P. H. Wang, C. F. Weng and M. S. Huang, Lung Cancer, 2009, 66, 162–168 CrossRef PubMed.
- E. Nicolis, I. Lampronti, M. C. Dechecchi, M. Borgatti, A. Tamanini, N. Bianchi, V. Bezzerri, I. Mancini, M. G. Giri, P. Rizzotti, R. Gambari and G. Cabrini, Int. Immunopharmacol., 2008, 8, 1672–1680 CrossRef CAS PubMed.
- Y. F. Huang, S. L. Zhang, F. Jin, D. Cheng, Y. P. Zhou, H. R. Li, Z. M. Tang, J. Xue, W. Cai, J. H. Dong and L. Zhao, Int. J. Immunopathol. Pharmacol., 2013, 26, 85–92 CrossRef CAS PubMed.
- J. Moreira, L. C. Klein-Júnior, V. Cechinel Filho and F. de Campos Buzzi, J. Ethnopharmacol., 2013, 146, 318–323 CrossRef CAS PubMed.
- Y. J. Guo, L. Zhao, X. F. Li, Y. W. Mei, S. L. Zhang, J. Y. Tao, Y. Zhou and J. H. Dong, Eur. J. Pharmacol., 2010, 635, 79–86 CrossRef CAS PubMed.
- P. Li, R. Du, Y. Wang, X. Hou, L. Wang, X. Zhao, P. Zhan, X. Liu, L. Rong and Q. Cui, Front. Microbiol., 2020, 11, 182 CrossRef PubMed.
- L. T. Lin, T. Y. Chen, S. C. Lin, C. Y. Chung, T. C. Lin, G. H. Wang, R. Anderson, C. C. Lin and C. D. Richardson, BMC Microbiol., 2013, 13, 1–15 CrossRef PubMed.
- S. Shanmuganathan and N. Angayarkanni, Vasc. Pharmacol., 2018, 108, 23–35 CrossRef CAS PubMed.
- S. M. Hecht, D. E. Berry, L. J. MacKenzie, R. W. Busby and C. A. Nasuti, J. Nat. Prod., 1992, 55, 401–413 CrossRef CAS PubMed.
- R. Jakes, M. G. Spillantini and M. Goedert, FEBS Lett., 1994, 345, 27–32 CrossRef CAS PubMed.
- M. J. Volles and P. T. Lansbury Jr, J. Mol. Biol., 2007, 366, 1510–1522 CrossRef CAS PubMed.
- D. Marion, M. Ikura, R. Tschudin and A. Bax, J. Magn. Reson., 1989, 85, 393–399 CAS.
- S. P. Skinner, R. H. Fogh, W. Boucher, T. J. Ragan, L. G. Mureddu and G. W. Vuister, J. Biomol. NMR, 2016, 66, 111–124 CrossRef CAS PubMed.
- M. Biancalana and S. Koide, Biochim. Biophys. Acta, 2010, 1804, 1405–1412 CrossRef CAS PubMed.
- M. R. Krebs, E. H. Bromley and A. M. Donald, J. Struct. Biol., 2005, 149, 30–37 CrossRef CAS PubMed.
- C. Xue, T. Y. Lin, D. Chang and Z. Guo, R. Soc. Open Sci., 2017, 4, 160696 CrossRef PubMed.
- M. F. Bishop and F. A. Ferrone, Biophys. J., 1984, 46, 631–644 CrossRef CAS PubMed.
- M. D. Tuttle, G. Comellas, A. J. Nieuwkoop, D. J. Covell, D. A. Berthold, K. D. Kloepper, J. M. Courtney, J. K. Kim, A. M. Barclay, A. Kendall, W. Wan, G. Stubbs, C. D. Schwieters, V. Y. M. Lee, J. M. George and C. M. Rienstra, Nat. Struct. Mol. Biol., 2016, 23, 409–415 CrossRef CAS PubMed.
- B. Li, P. Ge, K. A. Murray, P. Sheth, M. Zhang, G. Nair, M. R. Sawaya, W. S. Shin, D. R. Boyer, S. Ye, D. S. Eisenberg, Z. H. Zhou and L. Jiang, Nat. Commun., 2018, 9, 1–10 CrossRef PubMed.
- E. A. Mirecka, H. Shaykhalishahi, A. Gauhar, Ş. Akgül, J. Lecher, D. Willbold, M. Stoldt and W. Hoyer, Angew. Chem., Int. Ed., 2014, 53, 4227–4230 CrossRef CAS PubMed.
- E. D. Agerschou, P. Flagmeier, T. Saridaki, C. Galvagnion, D. Komnig, L. Heid, V. Prasad, H. Shaykhalishahi, D. Willbold, C. M. Dobson, A. Voigt, B. Falkenburger, W. Hoyer and A. K. Buell, Elife, 2019, 8, e46112 CrossRef PubMed.
- C. M. Cremers, D. Knoefler, S. Gates, N. Martin, J. U. Dahl, J. Lempart, L. Xie, M. R. Chapman, V. Galvan, D. R. Southworth and U. Jakob, Mol. Cell, 2016, 63, 768–780 CrossRef CAS PubMed.
- H. T. Lam, M. C. Graber, K. A. Gentry and J. Bieschke, Biochemistry, 2016, 55, 675–685 CrossRef CAS PubMed.
- T. Scheidt, U. Łapińska, J. R. Kumita, D. R. Whiten, D. Klenerman, M. R. Wilson, S. I. A. Cohen, S. Linse, M. Vendruscolo, C. M. Dobson, T. P. J. Knowles and P. Arosio, Sci. Adv., 2019, 5, eaau3112 CrossRef CAS PubMed.
- H. Shaykhalishahi, A. Gauhar, M. M. Wördehoff, C. S. Grüning, A. N. Klein, O. Bannach, M. Stoldt, D. Willbold, T. Hard and W. Hoyer, Angew. Chem., Int. Ed., 2015, 54, 8837–8840 CrossRef CAS PubMed.
- E. D. Agerschou, V. Borgmann, M. M. Wördehoff and W. Hoyer, Chem. Sci., 2020, 11, 11331–11337 RSC.
- Z. A. Sorrentino, N. Vijayaraghavan, K. M. Gorion, C. J. Riffe, K. H. Strang, J. Caldwell and B. I. Giasson, J. Biol. Chem., 2018, 293, 18914–18932 CrossRef CAS PubMed.
- M. Terada, G. Suzuki, T. Nonaka, F. Kametani, A. Tamaoka and M. Hasegawa, J. Biol. Chem., 2018, 293, 13910–13920 CrossRef CAS PubMed.
- Z. A. Sorrentino, E. Hass, N. Vijayaraghavan, K. M. Gorion, C. J. Riffe, J. K. S. Dhillon and B. I. Giasson, Neurosci. Lett., 2020, 732, 135017 CrossRef CAS PubMed.
- K. P. Wu and J. Baum, J. Am. Chem. Soc., 2010, 132, 5546–5547 CrossRef CAS PubMed.
- X. Yang, B. Wang, C. L. Hoop, J. K. Williams and J. Baum, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2017452118 CrossRef CAS PubMed.
- M. Vilar, H. T. Chou, T. Lührs, S. K. Maji, D. Riek-Loher, R. Verel, G. Manning, H. Stahlberg and R. Riek, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 8637–8642 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Comparison between the electronic absorption profile of ThT and the catechols, Pyrogallol (PGL), Corilagin (CLN) and Chebulagic acid (CA) (Fig. S1), 1H–15N HSQC signal attenuation (I/I0) of αSyn in various conditions (Fig. S2) and statistical analyses of 1H–15N HSQC intensity ratios of αSyn (Table S1). See DOI: https://doi.org/10.1039/d2ra04358k |
| ‡ Accession code: alpha-synuclein (Homo sapiens) UniProtKB-P37840 (SYUA_HUMAN). |
| This journal is © The Royal Society of Chemistry 2022 |