Open Access Article
Open Access ArticleMain-chain flexibility and hydrophobicity of ionenes strongly impact their antimicrobial activity: an extended study on drug resistance strains and Mycobacterium†
Rafał Jerzy Kopiasz a,
Anna Zabost
a,
Anna Zabost b,
Magdalena Myszkaa,
Aleksandra Kuźmińskac,
Karolina Drężeka,
Jolanta Mierzejewska
b,
Magdalena Myszkaa,
Aleksandra Kuźmińskac,
Karolina Drężeka,
Jolanta Mierzejewska a,
Waldemar Tomaszewskia,
Agnieszka Iwańskab,
Ewa Augustynowicz-Kopećb,
Tomasz Ciachc and
Dominik Jańczewski
a,
Waldemar Tomaszewskia,
Agnieszka Iwańskab,
Ewa Augustynowicz-Kopećb,
Tomasz Ciachc and
Dominik Jańczewski a
a
aFaculty of Chemistry, Warsaw University of Technology, Noakowskiego 3, Warsaw 00-664, Poland. E-mail: dominik.janczewski@pw.edu.pl
bDepartment of Microbiology, National Tuberculosis and Lung Diseases Research Institute, Płocka 26, Warsaw 01-138, Poland
cFaculty of Chemical and Process Engineering, Warsaw University of Technology, Waryńskiego 1, Warsaw 00-645, Poland
First published on 15th September 2022
Abstract
The spread of antibiotic-resistant pathogens and the resurgence of tuberculosis disease are major motivations to search for novel antimicrobial agents. Some promising candidates in this respect are cationic polymers, also known as synthetic mimics of antimicrobial peptides (SMAMPs), which act through the membrane-lytic mechanism. Development of resistance toward SMAMPs is less likely than toward currently employed antibiotics; however, further studies are needed to better understand their structure–activity relationship. The main objective of this work is to understand the cross-influence of hydrophobicity, main-chain flexibility, and the topology of ionenes (polycations containing a cationic moiety within the main-chain) on activity. To fulfill this goal, a library of ionenes was developed and compared with previously investigated molecules. The obtained compounds display promising activity against the model microorganisms and drug-resistance clinical isolates, including Mycobacterium tuberculosis. The killing efficiency was also investigated, and results confirm a strong effect of hydrophobicity, revealing higher activity for molecules possessing the flexible linker within the polymer main-chain.
1. Introduction
The spread of antibiotic resistance among pathogens is a serious concern to the global healthcare and economy.1,2 One of the pathogens responsible for the highest mortality worldwide is Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis disease (TB). Humanity has been relatively successful in the limitation of TB in the last 100 years, however, the increasing number of rifampicin-resistant TB (RS-TB) and multidrug-resistant TB (MDR-TB) cases has led to the reemergence of this illness. In 2019, it was estimated that 10 million people fell ill with TB, about 1.2 million died due to TB, and almost half a million people developed RS-TB, of which 78% had MDR-TB.3,4 To avoid a global health crisis caused by highly infective drug-resistant bacteria, a strong effort to develop novel antibacterial agents is urgently needed.5 Unfortunately, there is an alarming scarcity of new antibiotics in the market.1 The high cost of research combined with a relatively low financial return, makes new antibiotic research not profitable enough for pharmaceutical companies.6–8 Therefore, academic studies in this area are critical.Membrane lytic compounds, such as antimicrobial peptides (AMPs)9,10 and their synthetic mimics (SMAMPs), are a promising class of antimicrobials. There are experimental evidences that SMAMPs display lower susceptibility to the resistance development than currently used antibiotics.11–13 Due to the more accessible and cheaper synthesis of SMAMPs and easy access to various chemical structures, these compounds seem to outperform AMPs to some extent. SMAMPs are characterized by the presence of positively charged and hydrophobic moieties and an overall amphiphilic character. These features facilitate the initial electrostatic interactions with a negatively charged cell membrane, and subsequent incorporation into a hydrophobic interior of the lipid bilayer, which leads to the membrane disruption and eventually cell death.14,15 According to the existing theory, a random coil conformation of a moderately hydrophobic amphiphilic polycation turns into globally amphiphilic upon contact with a cell membrane, which allows the molecule to incorporate into the membrane interior.14,16 Therefore, the elevated conformational freedom seems to facilitate such conformational changes and in turn influence antimicrobial potency.15 However, there is a scarcity of systematic studies reported on the impact of the polycationic main-chain flexibility on their activity. This problem has been broadly investigated in the case of AMPs, but in most of these studies, the flexibility modulation involved additional changes in the overall hydrophobicity.17–20 It may be expected that different conformers interact with a membrane with different strengths; therefore, polycationic mainchain topology, which determines the possible conformations, may also potentially affect the antimicrobial activity. It has been reported that the topology of ionenes (polycations with cationic groups inside the main-chain), in terms of an aryl group isomerism, strongly impacts their behavior in water solution,21–23 which supports the above hypothesis. Hydrophilic-lipophilic balance (HLB) is another important and well-studied structural parameter that strongly influences the conformational changes and penetration of the phospholipid bilayer. A large body of experimental data indicates that it is hard to establish a general rule describing the optimal level of HLB for high antimicrobial activity and selectivity. Each polycationic system requires an individual fine-tuning of this parameter.24–30 Among the studied polycationic architectures, we identified a class of relatively hydrophilic ionenes as especially interesting due to their excellent selectivity toward microbes over red blood cells (RBCs).31–38
Mycobacteria contain a complex and strongly hydrophobic cell wall consisting of the peptidoglycan-arabinogalactan-mycolic acid complex, with mycolic acids located at the outer layer.39,40 H. Chen et al., in their review, have pointed to the mycobacterial membrane as a novel target for anti-tuberculosis drugs, such as cationic amphiphiles.39 Low-molecular-weight cationic amphiphiles,41 cationic dendrimers,42 AMPs,43,44 cationic polymers,33,45–48 and gold nanoparticles coated with polycations,49 highly active against mycobacteria, have been reported to date. Most of those studies were performed on Mtb and Mycobacterium smegmatis (Mms), which is frequently employed as an avirulent and fast-growing model organism for Mtb, especially in mechanistic studies.50,51 Besides inhibitory growth activity, the abovementioned polycations display high mycobactericidal efficiency against Mtb and Msm at concentrations close to the minimum inhibitory concentration (MIC).41,45–47 Reported data have indicated a disruption of the mycobacterial membrane or morphological changes in a cell envelope under the influence of polycations.45–47 However, a possible intercellular mode of action was discussed.52 Intriguingly, the high selectivity toward mycobacteria over Staphylococcus aureus and Escherichia coli has been observed for some polycations.42,46–49 This reflects differences in the cell envelope structures of these bacteria, and highlights a possibility that cationic amphiphiles not active against Gram-negative and Gram-positive species may be highly active against Mtb.
In this work, we investigate the cross-influence of hydrophobicity, main-chain flexibility, and isomerism of ionenes on their antimicrobial activity against model microorganisms and drug-resistant clinical isolates, including mycobacteria. Besides the antimicrobial activity, the critical aggregation concentrations (CAC) and zeta-potential of the aggregates were also determined. To fulfill this task, a broad library of ionenes (Fig. 1a) characterized by an alternating amphiphilic pattern (Fig. 1b) was developed. The library consisted of already published structures and newly synthesized ones to summarize a broader investigation effort.
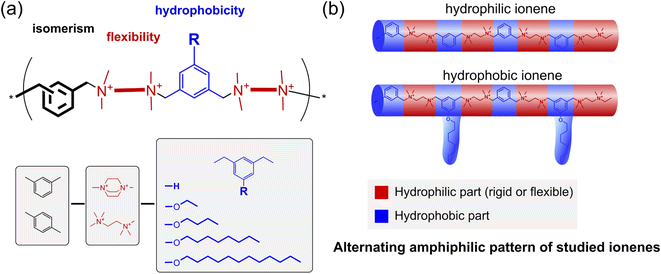 | ||
| Fig. 1 (a) The concept of the ionenes library, which allows for investigating the cross-influence between the isomerism, flexibility, and hydrophobicity on the antimicrobial activity; (b) a schematic illustration of the alternating amphiphilicity pattern of the studied ionenes based on recent work by Z. Chen et al.38 | ||
2. Experimental section
2.1 Materials
All chemical reagents were purchased from Sigma-Aldrich, TCI Chemicals, Fluorochem, or Chempur, and were used without further purification. Regenerated-cellulose dialysis tubing (Spectra/Por® 7, MWCO 1 kDa) was pre-treated according to the instruction provided by the manufacturer. Water for dialysis, antimicrobial assays, and SEC analysis was purified with a Milli-Q system (Millipore). Freeze-drying was performed using Labconco FreeZone ® 2.5 Liter Benchtop Freeze Dry Systems. Broths for antimicrobial assays were prepared using commercially available Mueller–Hinton Broth powder (Biocorp) and Sabouraud broth (SAB) powder containing 2% dextrose (Merck). As a solid medium for colony plate counting, commercially available Mueller–Hinton 2 containing agar (Biocorp) and Sabouraud containing agar (BioMaxima) powders were used. Buffered phosphate saline solution pH 7.4 (PBS) was prepared from 10× solution (Fisher BioReagents™) by dilution with Milli-Q water. American Type Culture Collection (ATCC) bacterial and yeast strains were used in this work: Escherichia coli (ATCC 8739), Staphylococcus aureus (ATCC 6538), Candida albicans (ATCC 10231), M. tuberculosis H37Rv strain (ATCC 25618), M. avium (ATCC 15769), and M. terrae (ATCC 15755). Clinical isolates: M. tuberculosis Spec. 210 (isoniazid resistant, INH-R), M. tuberculosis Spec. 192, K. pneumoniae (metallo-β-lactamase MBL), K. pneumoniae (extended-spectrum β-lactamase ESBL), E. faecium (vancomycin resistant, VRE), P. aeruginosa, S. aureus (methyciline resistant, MRSA), and A. baumanii were obtained from the collection of pathogenic strains of the Microbiology Department of the National Tuberculosis and Lung Diseases Research Institute (Warsaw, Poland). All of those strains were isolated from patients hospitalized at the National Tuberculosis and Lung Diseases Research Institute between 2019 and 2021, and kept frozen at −80 °C prior to use.2.2 Characterization of chemical compounds
The 1H and 13C NMR spectra were recorded using a Varian 400 MHz spectrometer and D2O or CDCl3 as solvents. 1H NMR chemical shifts were referenced to the residual signal of the protonated solvent (δ 7.26 for CDCl3 and 4.79 for D2O). 13C NMR chemical shifts were referenced to the solvent (δ 77.16 for CDCl3). Elemental analyses were performed using a Vario EL III CHNS Elemental instrument.SEC analysis was performed using an Agilent 1260 Infinity liquid chromatograph equipped with RID and UV DAD detector (detection at 268 nm), the PSS NOVEMA Max 5 μm analytical 300 × 8 mm column with precolumn (PSS GmbH), a mobile phase containing: 54/23/23 (v/v/v%) water/methanol/acetic acid and 0.5 M sodium acetate. All used chemicals were HPLC grade. To calibrate the SEC method, a series of poly(2-vinylpyridine) standards (PSS GmbH) in the range of molar masses of 620 Da–539 kDa were used. Samples, dissolved in the eluent at 5 mg mL−1 concentrations, were injected in 20 μL volumes. Analysis was performed at 50 °C at a flow rate of 0.4 mL min−1. The molar masses Mw, Mn, and dispersities ĐM were calculated using the Agilent GPC Addon Rev. B.01.02 software. The dynamic light scattering measurements were performed using a Zetasizer Nano from Malvern Instruments UK. Particle size (by scattering intensity) and zeta-potential were calculated using the software provided by Malvern. Dynamic light scattering was measured at a scattering angle of 173°. Zeta-potential was calculated from measured electrophoretic mobility by applying Smoluchowski's equation.
2.3 Material synthesis
Ionenes C0-T-p,53 C8-T-p,53 C0-T-m,31 1-alcoxy-3,5-bis(bromomethyl)benzenes (1-Cn)53 and α,α′-bis(1-azonia-4-azabicyclo[2.2.2]octyl)-para-xylene dibromide (2)31 were obtained as reported previously.α,α′-Bis(N,N,N′,N′-tetramethyl-ethylene-1-ammonium-2-amine)-meta-xylene dibromide (3). A solution of α,α′-dibromo-m-xylene (4.75 g, 18 mmol) in MeCN (120 mL) was added dropwise at room temperature to a stirred solution of TMEDA (16.3 mL, 12.5 g, 108 mmol) in MeCN (72 mL) over 30 min, and the reaction mixture was refluxed overnight. After cooling down to room temperature, the homogenous mixture was concentrated on a rotatory evaporator. The obtained oil was dissolved in MeCN (30 mL) and poured into diethyl ether (300 mL), facilitating precipitation of a sticky solid. The solid was washed using MeCN (40 mL) and cooled to −40 °C MeCN (100 mL). The obtained crystalline solid was then dried under vacuum overnight, yielding a strongly hygroscopic white solid (8.93 g, 77%). 1H NMR (400 MHz, D2O) δ: 2.25 (12H, s), 2.91 (4H, m), 3.06 (12H, s), 3.48 (4H, m), 4.59 (4H, s), 7.71 (4H, m); 13C NMR (400 MHz, D2O) δ: 137.29, 135.44, 130.37, 128.12, 67.93, 61.33, 50.83, 50.00, 44.40.
C2-D-p. A solution of 1-ethoxy-3,5-bis(bromomethyl)benzene (1-C2) (1.109 g, 3.60 mmol) in DMSO (4 mL) was added dropwise, at room temperature, to a stirred solution of α,α'-(1-azonia-4-azabicyclo[2.2.2]octyl)-p-xylene dibromide (2) (1.932 g, 3.96 mmol), DMSO (26 mL) and water (2 mL). The reaction mixture was stirred at room temperature for 24 h and poured into acetone (400 mL). The obtained suspension was centrifuged, and the supernatant was removed. Subsequently, the solid was preliminarily dried, dissolved in water (160 mL), transferred to dialysis tubing (MWC 1 kDa), and dialyzed against Milli-Q water for 3 days. The solution was freeze-dried, yielding 1.495 g (49%) of white powder. Found: C, 40.37; H, 6.36; N, 6.57%; (C30H44Br4N4O·5H2O)n requires: C, 40.65; H, 6.14; N, 6.32%. 1H NMR (400 MHz, D2O) (integrals for repeating units) δ: 1.33 (t, 3H), 3.12 (m, 6H, terminal), 3.44 (m, 6H, terminal), 4.03 (m, 26H), 4.53 (s, 4H, terminal group), 4.84 (m, 8H), 7.28 (s, 3H), 7.70 (m, 4H).
C4-D-p (20%). Found: C, 42.06; H, 6.78; N, 6.39%; (C32H48Br4N4O·5H2O)n requires: C, 42.03; H, 6.39; N, 6.13%. 1H NMR (400 MHz, D2O) (integrals for repeating unit) δ: 0.87 (t, 3H), 1.40 (m, 2H), 1.70 (m, 2H), 3.11 (m, 6H, terminal group), 3.44 (m, 6H, terminal group), 4.02 (m, 26H), 4.52 (s, 4H, terminal group), 4.84 (m, 8H), 7.28 (s, 3H), 7.70 (m, 4H).
C8-D-p (60%). Found: C, 44.03; H, 7.09; N, 5.86%; (C36H56Br4N4O·6H2O)n requires: C, 44.55; H, 6.85; N, 5.77%. 1H NMR (400 MHz, D2O) (integrals for repeating unit) δ: 0.89–1.74 (m, 15H), 3.13 (m, 6H, terminal), 3.43 (m, 6H, terminal), 4.03 (bs, 26H), 4.79 (s, 4H, terminal group), 4.79 (s, 8H), 7.29 (s, 3H), 7.76 (m, 4H).
C12-D-p (53%). Found: C, 48.85; H, 7.82; N, 5.82%; (C32H48Br4N4O·6H2O)n requires: C, 48.50; H, 7.12; N, 5.66%. 1H NMR (400 MHz, DMSO-d6) (integrals for repeating unit) δ: 0.83 (m, 3H), 1.22–1.37 (m, 18H), 1.70 (bs, 2H), 3.00 (m, 6H, terminal), 3.36 (m, 6H, terminal), 4.00 (m, 26H), 4.92 (s, 4H, terminal group), 4.87 (bs, 8H), 7.01–7.37 (m, 3H), 7.65–7.72 (m, 4H).
C0-D-p (30%). Found: C, 38.13; H, 6.17; N, 6.42%; (C36H64Br4N4O·7H2O)n requires: C, 38.29; H, 6.20; N, 6.38%. 1H NMR (400 MHz, D2O) (integrals for repeating unit) δ: 3.12 (m, 6H, terminal group), 3.44 (m, 6H, terminal group), 4.02 (m, 26H), 4.53 (s, 4H, terminal group), 4.84 (m, 8H), 7.28 (s, 3H), 7.64–7.73 (m, 8H).
C2-T-m. A solution of 1-ethyloxy-3,5-bis(bromomethyl)benzene (1-C2) (1.109 g, 3.60 mmol) in DMSO (4 mL) was added dropwise, at room temperature, to a stirred solution of 3 (1.964 g, 3.96 mmol), DMSO (5.6 mL) and water (2 mL). The reaction mixture was stirred at room temperature for 24 h and poured into acetone (400 mL). The obtained suspension was centrifuged, and the supernatant was removed. Subsequently, the solid was dried, dissolved in water (160 mL) and transferred to dialysis tubing (MWC 1 kDa) and dialyzed against Milli-Q water for 3 days. The solution was freeze-dried, yielding 1.495 g (49%) of white powder. Found: C, 41.54; H, 7.03; N, 6.55%; (C30H52Br4N4O·4H2O)n requires: C, 41.11; H, 6.90; N, 6.39%. 1H NMR (400 MHz, D2O) (integrals for repeating unit) δ: 1.34 (t, 3H), 2.33 (s, 6H, terminal group), 2.91 (m, 2H, terminal group), 3.07 (s, 6H, terminal group), 3.20 (bs, 24H), 3.45 (m, 2H, terminal group), 4.14–4.21 (m, 10H), 7.35–7.46 (m, 3H), 7.67–7.77 (m, 4H).
C4-T-m (34%). Found: C, 41.89; H, 7.18; N, 6.33%; (C32H56Br4N4O·4H2O)n requires: C, 42.49; H, 7.13; N, 6.19%. 1H NMR (400 MHz, D2O) (integrals for repeating unit) δ: 0.83 (t, 3H), 1.36 (m, 2H), 1.68 (m, 2H), 2.22 (s, 6H, terminal group), 2.90 (m, 2H, terminal group), 3.06 (s, 6H, terminal group), 3.20 (bs, 24H), 3.44 (m, 2H, terminal group), 4.08–4.20 (m, 10H), 7.35–7.45 (m, 3H), 7.68–7.76 (m, 4H).
C8-T-m (34%). Found: C, 43.79; H, 7.51; N, 5.84%; (C36H64Br4N4O·6H2O)n required: C, 43.38; H, 7.69; N, 5.62%. 1H NMR (400 MHz, D2O) (integrals for repeating unit) δ: 0.74 (t, 3H), 1.14–1.33 (m, 12H), 1.65 (m, 2H), 2.32 (s, 6H, terminal group), 3.06 (m, 2H, terminal group), 3.06 (s, 6H, terminal group), 3.20 (bs, 24H), 3.50 (m, 2H, terminal group), 4.09–4.20 (m, 10H), 7.37–7.44 (m, 3H), 7.68–7.76 (m, 4H).
C12-T-m (68%). Found: C, 46.15; H, 7.69; N, 5.09%; (C40H72Br4N4O·5H2O)n required: C, 46.43; H, 7.99; N, 5.41%. 1H NMR (400 MHz, CD3OD) (integrals for repeating unit) δ: 0.90 (t, 3H), 1.29–1.48 (m, 12H), 1.78 (m, 2H), 2.33 (s, 6H, terminal group), 2.88 (m, 2H, terminal group), 3.20 (s, 6H, terminal group), 3.41 (m, 24H), 3.58 (m, 2H, terminal group), 4.08 (bs, 2H), 4.42 (bs, 8H), 7.09–7.91 (m, 7H).
2.4 Minimum inhibitory concentration (MIC) determination for model strains
The broth microdilution method was applied following CLSI M07-A9 vol. 32 no. 2 (for bacteria) and CLSI M27-A2 vol. 22 no. 15 (for yeast) protocols. Single colonies of bacteria or yeast were used to inoculate 5 mL of Mueller–Hinton Broth (MHB) or Sabouraud broth, respectively, and the cultures were grown overnight at 37 °C with shaking (240 rpm; Lab Companion SI-600R, Ramsey, MN, USA). The polymer stock solutions of 5120 μg mL−1 prepared in Milli-Q water were diluted with a broth (MHB or SAB) to a concentration of 1024 μg mL−1, and used to prepare a series of different polymer concentrations (from 512 μg mL−1 to 2 μg mL−1; 100 μL each) in 96-well plates by two-fold dilution method. Subsequently, 100 μL of microbial suspension (2 × 105 CFU mL−1 for bacteria and 2 × 103 CFU mL−1 for yeast) was added to each well. Uninoculated broth, uninoculated broth with polymer solutions, and inoculated broth without any antimicrobial agent were used as controls. Four replicates were performed for each concentration of polymer and the control. The plates were incubated for 20 h at 37 °C. The optical density at 600 nm (OD600) was measured using a Synergy™ H4 Hybrid Microplate Reader, Biotech (Winooski, VT, USA). The recorded MIC value was the lowest concentration of the polymer at which no microbial growth was observed with the microplate reader.2.5 MIC against Mycobacterium
The assay was performed by the two-fold serial microdilution method (in 96-well microliter plates) using Middlebrook 7H9 Broth medium (Beckton Dickinson) containing 10% of OADC (Beckton Dickinson). The inoculum was prepared from fresh LJ culture in Middlebrook 7H9 Broth medium with OADC, adjusted to a no. 0.5 McFarland tube, and diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. The stock solution of a tested molecule was prepared in water and diluted in Middlebrook 7H9 Broth medium with OADC by four-fold the final highest concentration to be tested. Compounds were diluted serially in sterile 96-well microtiter plates using 100 μl Middlebrook 7H9 Broth medium with OADC. Concentrations of the tested agents ranged from 0.25 to 512 μg mL−1. A growth control containing no antibiotic and a sterile control without inoculation were also prepared on each plate. After incubation, (Mycobacterium avium for 7 days, and Mycobacterium terrae, Mycobacterium tuberculosis strains at 37 °C for 21 days), the MICs were visually assessed as the lowest concentration showing complete growth inhibition of the reference microbial strains. Isoniazid (INH) and Rifampicin (RMP) were used as the reference drugs.
20. The stock solution of a tested molecule was prepared in water and diluted in Middlebrook 7H9 Broth medium with OADC by four-fold the final highest concentration to be tested. Compounds were diluted serially in sterile 96-well microtiter plates using 100 μl Middlebrook 7H9 Broth medium with OADC. Concentrations of the tested agents ranged from 0.25 to 512 μg mL−1. A growth control containing no antibiotic and a sterile control without inoculation were also prepared on each plate. After incubation, (Mycobacterium avium for 7 days, and Mycobacterium terrae, Mycobacterium tuberculosis strains at 37 °C for 21 days), the MICs were visually assessed as the lowest concentration showing complete growth inhibition of the reference microbial strains. Isoniazid (INH) and Rifampicin (RMP) were used as the reference drugs.
2.6 MIC against clinical isolates
The assay was performed by the two-fold serial microdilution method (in 96-well microliter plates) using MHB medium (Sigma). The inoculum was prepared from fresh Columbia agar (BioMerieux) culture in MHB medium, adjusted to a no. 0.5 McFarland tube, and diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. The stock solution of a tested molecule was prepared in water and diluted in MHB medium by four-fold the final highest concentration to be tested. Compounds were diluted serially in sterile 96-well microtiter plates using 100 μl MHB medium with OADC. Concentrations of the tested agents ranged from 0.25 to 512 μg mL−1. A growth control containing no antibiotic and a sterile control without inoculation were also prepared on each plate. After incubation (bacterial strains at 35 °C for 24 h), the MICs were visually assessed as the lowest concentration showing complete growth inhibition of the reference microbial strains. Ciprofloxacin was used as the reference drug.
20. The stock solution of a tested molecule was prepared in water and diluted in MHB medium by four-fold the final highest concentration to be tested. Compounds were diluted serially in sterile 96-well microtiter plates using 100 μl MHB medium with OADC. Concentrations of the tested agents ranged from 0.25 to 512 μg mL−1. A growth control containing no antibiotic and a sterile control without inoculation were also prepared on each plate. After incubation (bacterial strains at 35 °C for 24 h), the MICs were visually assessed as the lowest concentration showing complete growth inhibition of the reference microbial strains. Ciprofloxacin was used as the reference drug.
2.7 Time-kill kinetics assay
A single colony of S. aureus was used to inoculate 5 mL of MHB, and the culture was grown overnight at 37 °C with shaking (240 rpm). Then, after a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution in fresh growth media, the cultures were further incubated at 37 °C with shaking until OD600 = 0.6 and diluted by 1
100 dilution in fresh growth media, the cultures were further incubated at 37 °C with shaking until OD600 = 0.6 and diluted by 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in fresh media again to ca. 105 CFU mL−1. The bacteria suspension (180 μL) was added to the water polymer solution (20 μL) at a concentration of 10, 20, and 40 × MIC, which resulted in the final polymer concentration of 1, 2, and 4 × MIC, respectively. The suspensions were incubated at 37 °C, 20 μL samples were collected after 5, 10, 20, 40, 60 min, and a series of tenfold dilutions using phosphate buffer solution were prepared. Growth medium-treated cultures served as a positive control of bacterial growth. The final dilutions (10 μL) were spread on agar plates with MHA, and left for incubation for 20 h at 37 °C. After incubation, the colony-forming units were counted.
100 in fresh media again to ca. 105 CFU mL−1. The bacteria suspension (180 μL) was added to the water polymer solution (20 μL) at a concentration of 10, 20, and 40 × MIC, which resulted in the final polymer concentration of 1, 2, and 4 × MIC, respectively. The suspensions were incubated at 37 °C, 20 μL samples were collected after 5, 10, 20, 40, 60 min, and a series of tenfold dilutions using phosphate buffer solution were prepared. Growth medium-treated cultures served as a positive control of bacterial growth. The final dilutions (10 μL) were spread on agar plates with MHA, and left for incubation for 20 h at 37 °C. After incubation, the colony-forming units were counted.
2.8 Hemolytic activity determination
Fresh blood was obtained from the Regional Center for Blood Donation and Blood Treatment in Warsaw. Samples were centrifuged (700g, 10 min), the supernatant plasma was rejected, and erythrocytes were washed with ice-cold phosphate-buffered saline (PBS) three times (by centrifuging, 700g, 10 min). After the final centrifugation, erythrocytes were diluted ten times with PBS. Polymer solutions were prepared in PBS buffer. Erythrocyte suspensions (500 μL) were added to polymer solutions (500 μL) at the investigated concentrations. Samples were incubated for 1 h at 37 °C, and then the samples were centrifuged (10 min, 700g). The hemoglobin release was subsequently measured in the supernatants using the spectrophotometric method (absorption at λ = 540 nm). PBS buffer and 0.2% Triton X-100 served as the negative and positive controls, respectively. Experiments were performed at the Faculty of Chemical and Process Engineering WUT with the local committee approval for research with human blood.3. Results and discussion
3.1 Polymer design, synthesis, and characterization
To investigate the cross-influence of the hydrophobicity, main-chain flexibility, and topology of ionenes on their antimicrobial and hemolytic activity, a library of ionenes composed of newly synthesized molecules and already reported structures31,53,54 was used (Fig. 1a). The molecules characterized by different topologies contain meta or para isomers of an aromatic linker, whereas the molecules characterized by different flexibilities contain 1,4-diazabicyclo[2.2.2]octane (DABCO) or tetramethylethylenediamine (TMEDA) subunit along the main-chain. The hydrophobicity was modulated by the length of the side alkyl group.Ionenes from the series Cn-D-m and Cn-T-p, and ionene C0-T-m, were obtained and characterized in previously published works.31,53,54 The synthesis and characterization of the flexible meta-ionenes (Cn-T-m) and rigid para-ionenes (Cn-D-p) are presented herein for the first time. To obtain the desired polycations monomers, 1-Cn, 2, and 3 were synthesized according to Schemes 1 and S1.† Dibromides 1-Cn were obtained by O-alkylation of 5-hydroxyisophthalate, and a subsequent ester reduction with lithium aluminum hydride (LAH) followed by bromination with PBr3 (Scheme S1†).53 Tertiary amines 2 and 3 were obtained by treating the para (1,4-) and meta (1,3-) isomers of bis(bromomethyl)benzene with an excess of DABCO31 and TMEDA, respectively. A polyaddition reaction between dibromides 1-Cn and diamines 2 and 3, using 10% molar excess, led to the final polycations. This approach enabled the obtaining of different ionenes characterized by a similar average molecular mass and well-defined terminal groups.31,55
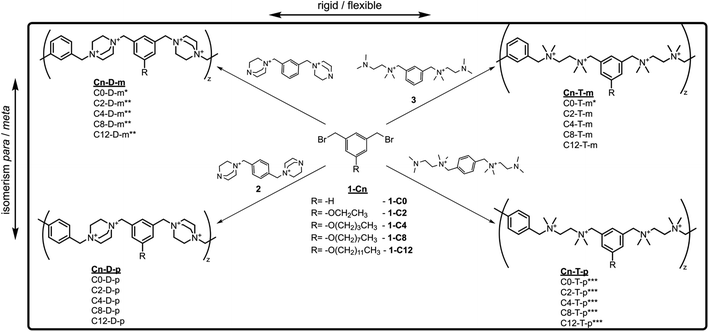 | ||
| Scheme 1 The synthetic pathway of the investigated ionenes; the polyaddition reactions were conducted in a DMSO/water mixture at room temperature using 10% molar excess of di-amines. * from ref. 31, ** from ref. 54, *** form ref. 53. | ||
The polymer structures were confirmed via 1H NMR spectroscopy. Owing to the known chemical shifts of the polymer terminal groups,31 the degree of polymerization (DP) and the number average molar mass (Mn, NMR) were calculated (Fig. 2 and Table 1). The DP falls within the range of 7.3–13.8. The ionenes were further characterized by size exclusion chromatography (SEC), which confirmed a similar molecular mass distribution (Fig. 2b). The number of the average molecular mass determined by SEC (Mn,SEC) remained in the range of 4.0–7.5 kDa for ionenes containing the pendant alkyl chain up to 8 carbon atoms, and the obtained values correlate well with Mn,NMR. For ionenes with the dodecyloxy pendant group, the Mn,SEC falls in the range of 2.3–3.3 kDa, and it is significantly lower than the Mn,NMR, which likely is a result of the hydrophobic interaction between the tested molecules and the stationary phase of the chromatographic column. Polymer dispersity (Đ) remains in the range between 1.38 to 1.85.
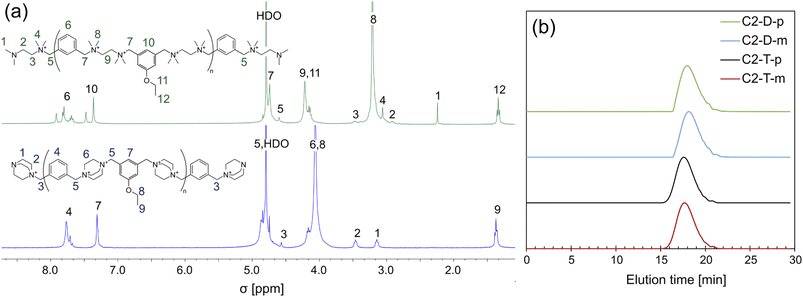 | ||
| Fig. 2 (a) Example of the 1H NMR spectra of the rigid and flexible ionenes, and (b) SEC traces of ionenes containing the ethylene pendant group. For full characterization of all ionenes, see ESI.† | ||
| Polymer | DPa | Mn,NMRb [kDa] | Mn,SECc [kDa] | Đc | CACd [μg mL−1] | Zeta potentiale [mV] | ||
|---|---|---|---|---|---|---|---|---|
| H2O | PBS | H2O | PBS | |||||
| a DP determined from 1H NMR spectra.b Mn,NMR calculated using DP and the molar mass of a repeating unit without bromides.c Mn,SEC and ĐM determined by size exclusion chromatography (SEC).d the critical aggregation concentration (CAC) was determined by pyrene method.e zeta potential of polymeric aggregates determined by DLS technique at a concentration of 2 mg mL−1.f Not determined. * from ref. 31. ** from ref. 54. *** from ref. 53. | ||||||||
| C0-D-m* | 10.6 | 4.6 | 4.8 | 1.73 | >2000 | >2000 | n.df | n.d.f |
| C2-D-m** | 11.8 | 5.6 | 4.6 | 1.77 | >2000 | >2000 | n.df | n.d.f |
| C4-D-m** | 10.4 | 5.2 | 5.3 | 1.63 | >2000 | >2000 | n.df | n.d.f |
| C8-D-m** | 9.0 | 5.0 | 4.5 | 1.73 | 360 | 38 | 65.0 ± 1.4 | 14.1 ± 1.6 |
| C12-D-m** | 11.9 | 7.3 | 2.3 | 1.43 | 20 | ≤4 | 67.0 ± 2.7 | 19.5 ± 2.1 |
| C0-D-p | 10.1 | 4.4 | 4.4 | 1.85 | >2000 | >2000 | n.df | n.df |
| C2-D-p | 11.6 | 5.5 | 5.2 | 1.79 | >2000 | >2000 | n.df | n.df |
| C4-D-p | 9.1 | 4.6 | 4.0 | 1.67 | >2000 | >2000 | n.df | n.df |
| C8-D-p | 11.0 | 6.2 | 4.1 | 1.80 | 1100 | 49 | 46.6 ± 1.3 | 17.3 ± 0.2 |
| C12-D-p | 9.2 | 5.7 | 2.3 | 1.38 | 53 | 15 | 60.4 ± 1.0 | 17.3 ± 1.1 |
| C0-T-m* | 7.3 | 3.2 | 4.0 | 1.48 | >2000 | >2000 | n.d.f | n.d.f |
| C2-T-m | 12.8 | 6.2 | 6.9 | 1.80 | >2000 | >2000 | n.d.f | n.d.f |
| C4-T-m | 13.0 | 6.7 | 6.7 | 1.82 | >2000 | >2000 | n.d.f | n.d.f |
| C8-T-m | 12.4 | 7.1 | 5.5 | 1.82 | 1600 | 600 | 82.1 ± 3.1 | 23.6 ± 1.6 |
| C12-T-m | 12.7 | 7.9 | 4.9 | 1.34 | 56 | 10 | 97.2 ± 1.7 | 31.3 ± 2.0 |
| C0-T-p*** | 12.2 | 5.4 | 7.3 | 1.78 | >2000 | >2000 | n.df | n.df |
| C2-T-p*** | 12.4 | 6.0 | 7.5 | 1.79 | >2000 | >2000 | n.d.f | n.d.f |
| C4-T-p*** | 10.9 | 5.6 | 6.7 | 1.84 | >2000 | >2000 | n.d.f | n.d.f |
| C8-T-p*** | 13.8 | 7.9 | 6.5 | 1.82 | >2000 | 920 | n.d.f | 18.2 ± 0.9 |
| C12-T-p*** | 10.6 | 6.6 | 3.3 | 1.46 | 80 | 16 | 64.1 ± 0.6 | 16.9 ± 1.5 |
The critical aggregation concentration (CAC) of the polymers in pure water and phosphate-buffered saline pH 7.4 (PBS) was determined using the pyrene method.56 Ionenes without an alkyl group (C0) and with C2 and C4 groups do not form aggregates at a concentration of up to 2000 μg mL−1 (Table 1), whereas C8 derivatives show CAC within a range of 38–1600 μg mL−1 and C12 derivatives within a range of less than 4 μg mL−1 to 110 μg mL−1. Besides an obvious relationship between the hydrophobicity and CAC, a systematic influence of the flexibility and isomerism can be perceived. More rigid DABCO-containing ionenes showed lower CAC values than their flexible counterparts. Comparing the ionenes containing meta- or para-substituted aryl linkers, lower CAC values may be observed for the former ones. This indicates that the increased stiffness of the polymeric main-chain and incorporation of meta isomers of the aryl moiety enhance the formation of aggregates. Polymeric aggregates were studied employing the dynamic light scattering technique (DLS) to determine their hydrodynamic size and zeta-potential. Size distributions for all studied polycations at a concentration above the CAC were multimodal. Therefore, they are not discussed herein. Zeta-potential measurements indicated a relatively high positive electrokinetic charge on the surface of the aggregates, which is higher for the more flexible TMEDA-containing ionenes and for the meta isomers in comparison to the para ones.
3.2 Antimicrobial and hemolytic activity
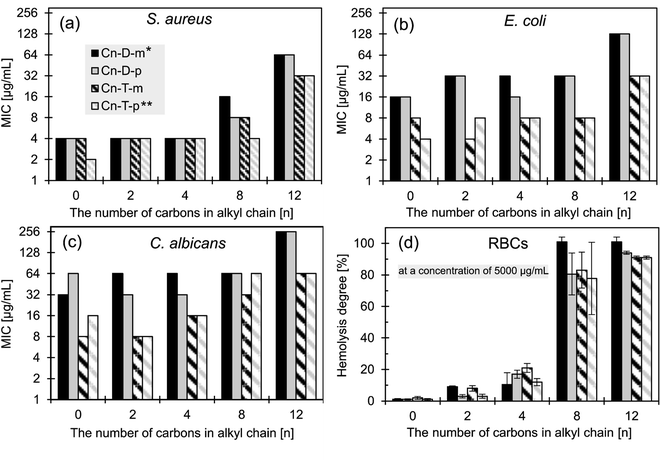 | ||
| Fig. 3 Antimicrobial activity of the studied ionenes toward model microorganisms: (a) S. aureus (ATTC 6538), (b) E. coli (ATTC 8739), and (c) C. albicans (ATCC 10231); (d) the degree of hemolysis (human RBCs) induced by the fixed polymers concentration (5000 μg mL−1 for n = 0, 2, 4, 8, and 2.5 mg mL−1 for n = 12). Data for polymers C0-D-m and C0-T-m from the ref. 31; * from the ref. 54, ** from the ref. 53. | ||
To facilitate further discussion, this section is divided into two short parts – in the first part, the influence of hydrophobicity and isomerism is discussed, whereas the second part covers the flexibility effect on the growth inhibitory activity.
It has been widely reported that the HLB of amphiphilic polycations must be fine-tuned to obtain the most potent antimicrobial macromolecules, and the optimal position is strongly dependent on the polycationic mainchain structure.15,63 For example, the Tew group reported the optimum HLB for polynorbornenes when propyl and butyl alkyl moieties were incorporated.64 Polycarbonates reported by the Hedrick and Yang groups were the most active with a hexyl moiety,26 whereas polyvinylpyridine derivatives reported by the Sen group displayed the lowest MIC with a butyl or hexyl group.24 In light of the published data, we can conclude that the optimal HLB value of the discussed ionenes herein is rather shifted toward hydrophilic structures.
The isomerism of aryl linkers displays a negligible influence on the antimicrobial activity, and no correlation is visible. The isomerism determines the possible conformations that the ionene molecules may attain. It hypothetically can modulate the interactions with a cell membrane and wall. The isomerism effect was observed for relatively hydrophilic ionenes without pendant groups in our previous work,31 and by Mayr et al.65 However, the results presented herein indicate that the effect is negligible for the ionenes containing higher hydrophobic side groups.
The elevated activity of the more flexible polymers could be explained by the hypothesis concerning the important role of the conformational freedom in the modulation of interactions between the polycations and the cell membrane.15 The flexible structure allows a molecule to attain a higher number of conformations separated with lower energy barriers. It may facilitate both a migration through a bacterial cell envelope and a formation of the most energetically privileged assembly between a polycation and a lipid bilayer upon binding to a water–lipid interface. However, when compared with the flexible ionenes (Table 1), the lower CAC values of the rigid ionenes, which point to their higher tendency to aggregation, and the lower zeta-potential of those aggregates may also be responsible for their lower activity.
| Polymer | MIC [μg mL−1] | |||||
|---|---|---|---|---|---|---|
| Gram-negative bacteria | Gram-positive bacteria | |||||
| K. pneumoniaea (MBL, NDM type) | K. pneumoniaea (ESBL) | P. aeruginosaa | A. baumanniia | S.aureusb (MRSA) | E. faeciumb (VRE) | |
| a Gram-negative bacteria.b Gram-positive bacteria.c Ciprofloxacin (CIP). | ||||||
| C0-D-p | 512 | 256 | >512 | >512 | 32 | >512 |
| C0-T-p | 64 | 128 | 64 | 64 | 32 | 16 |
| C8-D-p | >512 | >512 | >512 | >512 | >512 | >512 |
| C8-T-p | >512 | >512 | >512 | >512 | >512 | >512 |
| CIPc | >64 | 64 | >64 | >64 | >64 | >64 |
| Polymer | MIC [μg mL−1] | ||||
|---|---|---|---|---|---|
| M. tuberculosis H37RV | M. tuberculosis 210 (INH resistant) | M. tuberculosis 192 (INH susceptible) | M. terrae | M. avium | |
| a Isoniazid.b Rifampicin. | |||||
| C0-D-p | 32 | 64 | 64 | 16 | 512 |
| C0-T-p | 16 | 16 | 32 | 8 | 256 |
| C8-D-p | 16 | 16 | 32 | 32 | >512 |
| C8-T-p | 16 | 16 | 32 | 8 | >512 |
| INHa | ≤0.0625 | 2 | ≤0.0625 | 64 | 64 |
| RMPb | 0.5 | 16 | 0.5 | 4 | 32 |
The highly hydrophobic C8-D-p and C8-T-p showed no activity against non-Mycobacterium clinical strains within the tested concentration range, in contrast to the relatively hydrophilic C0-D-p and C0-T-p (Table 2). The rigid C0-D-p is moderately active against MRSA only, and displays limited activity against both K. pneumoniae strains. In contrast, the flexible C0-T-p is moderately active against all tested pathogens. In these studies, ciprofloxacin (CIP) was applied as a benchmark because both CIP and C0-ionenes show bactericidal rather than bacteriostatic activity.53 C0-T-p shows lower MICs than CIP for the majority of the tested isolates. Activity against MRSA (MIC 32 μg mL−1; Table 2) is significantly lower than against the model S. aureus from the ATCC collection (MIC 2–4 μg mL−1; Fig. 3a). The difference between MRSA and the model S. aureus strains lays in the structure of the penicillin-binding proteins (PBP).67 However, this feature is unlikely to induce the observed increase in the MICs. Some other structural features, e.g., a different cell membrane composition, may be responsible for the lower susceptibility of MRSA to ionenes. The structure–activity relationship observed for model microorganisms, namely, the lower antibacterial potency of the more hydrophobic and conformationally constrained ionenes, also applies to the pathogenic strains.
Both tested C8 ionenes and C0-T-p are comparably active toward Mtb, whereas C0-D-p is weaker (Table 3). When the activity toward M. terrae is considered, a higher activity of the flexible TMEDA-containing ionenes than that of the more rigid ones may be noticed. Two of the tested ionenes show very weak activity toward M. avium.
The hydrophobic modification of polymers, in the form of the C8 alkyl group, does not lower the ionene activity against mycobacteria (Table 3). The comparison between the MICs values of C0-D-p and C0-T-p against all studied mycobacteria indicates a higher activity of the flexible ionenes. A different response of mycobacteria to the presence of alkyl hydrophobic side chains, in comparison to all of the other tested microorganisms, can be associated with the cell envelope structure. Mycobacterium has a complex thick cell wall consisting of peptidoglycan, arabinogalactan, and hydrophobic mycolic acids covered by mycolates, phospholipids, and lipoglycans, whereas the cell walls of Gram-positive and Gram-negative bacteria predominantly consist of peptidoglycan.39 No HLB influence on antimycobacterial activity was observed for the discussed ionenes. However, other published studies provide rather inconclusive results in this respect. For example, in our previous paper, we observed an increase of such activity when the hydrophobicity of the polyethyleneimine derivatives was increased.48 On the other hand, P. Yavvari observed the opposite trend for poly-aspartamide derivatives.47 Ionenes C8-T-p and C8-D-p showed high selectivity toward mycobacteria over other tested clinical isolates. Such selectivity of polycations was also reported previously for the alkylated dimethyl-aminopropyl poly-aspartamides,47 poly(dimethylaminoethyl) methacrylate,46 and cationic phosphorus dendrimers.42 Activity against Mtb, for the discussed herein polymers, remains at similar levels to the previously reported ionenes (MIC values 2–40 μg mL−1).33,42,45,47,48 For example, an ionene with a structure similar to C0-T-p, studied by J. Tan et al., displayed a MIC value of 16 μg mL−1 against different Mtb strains.33
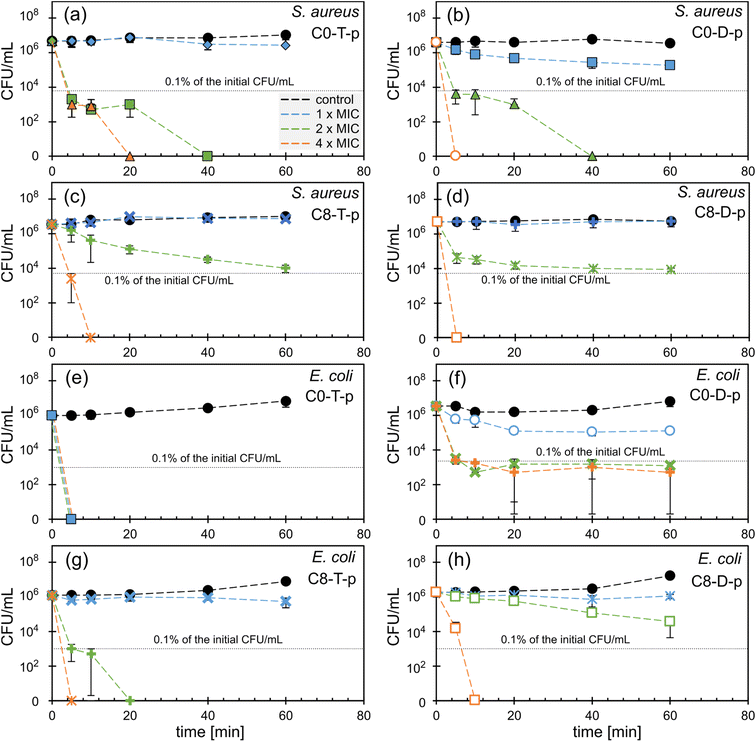
Ionenes C0-T-p, C8-T-p, and C8-D-p exert bacteriostatic activity against S. aureus since they do not reduce colony-forming units (CFU) during 1 h incubation at the concentration of 1 × MIC (Fig. 4a, c and d). Only C0-D-p shows bactericidal activity at 1 × MIC, reducing the initial CFU by 95% after 60 min of incubation (Fig. 4b). At the concentration of 2× MIC, ionenes C0-T-p and C0-D-p kill more than 99.9% of bacteria in the sample after 5 min incubation (Fig. 4a and b). In contrast, their more hydrophobic analogs, containing the C8 group, work significantly slower. After 60 min of incubation, CFU was reduced to almost 0.1% of its initial value (Fig. 4c and d). These hydrophobic ionenes require the concentration of 4 × MIC to eradicate all S. aureus from the sample.
Polymer C0-T-p shows the highest effectiveness against E. coli, killing all bacteria at the concentration of 1 × MIC (4 μg mL−1) after just 5 min of incubation (Fig. 4e). The less flexible analog, C0-D-p, also reduces the initial CFU by 97% after 60 min at the concentration of 1 × MIC (16 μg mL−1). However, at 2 and 4 × MIC, the reduction reaches 99.9% after 5 min (Fig. 4f). In contrast to the C0 polymers, C8 derivatives show bacteriostatic activity only at the concentration of 1 × MIC (Fig. 4g and h). The more flexible C8-T-p eliminates all bacteria at 2 × MIC, whereas the stiff C8-D-p requires as much as 4 × MIC to get a similar effect within the studied time range.
All tested C0 ionenes displayed higher killing rates than their hydrophobic C8 analogs. C0-T-p is particularly active with clear bactericidal activity at a concentration of 4 μg mL−1 and MBC close to MIC values. Considering the influence of the main-chain flexibility, one may notice that this parameter does not significantly impact activity toward S. aureus. However, the more flexible TMEDA-containing ionenes exert stronger and faster bactericidal potency toward E. coli than their rigid analogs. Flexibility of the main chain affects the killing efficiency and bacteria growth inhibition for both S. aureus and E. coli in a similar way (Fig. 3). This effect is much more pronounced for E. coli than S. aureus. The fast killing kinetics of the presented ionenes prove their contact mechanism of action, which was previously investigated for this class of polycations in other works.36,53,54
4. Conclusions
The comparison of the MIC values against the model microorganisms determined for the ionenes obtained in this work (Cn-T-m and Cn-D-p series), and for previously published compounds (Cn-T-p53 and Cn-D-m54 series), allowed us to discuss the cross-influence of hydrophobicity, mainchain flexibility, and topology on the antimicrobial activity. The topology, resulting from different isomers of the aryl linker and located within the polymeric main-chain, seems not to affect the growth inhibitory activity, whereas the increased hydrophilicity and main-chain flexibility increases the antimicrobial activity. The elevated hydrophobicity also increases toxicity against red blood cells. The impact of these parameters is clearly visible in the case of C. albicans and E. coli, and less against S. aureus. Killing kinetics studies showed that strongly hydrophobic C8 ionenes are less bactericidal against both tested bacteria than their more hydrophilic C0 analogs, whereas the main-chain stiffness affects the killing rate against E. coli only. Further studies on selected ionenes revealed good activity toward drug resistance clinical isolates, and confirmed that the polymeric chain flexibility of ionenes should be considered besides their hydrophobicity to obtain more potent antimicrobial agents.The studied ionenes show good activity toward mycobacteria species, including Mtb, which is an important result in light of the returning tuberculosis disease threat. Interestingly, in contrast to other tested microorganisms, the excessive hydrophobicity of ionenes does not affect the potency of the polycations toward mycobacteria. It indicates that the polycations developed against Mycobacteria require a unique HLB, which should be separately optimized. Data gathered on the clinical strains and bactericidal kinetics experiments confirmed that C0-T-p is the most active and promising structure in the investigated library.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was financially supported by the National Science Center, Poland (PoCoDi project, grant 2015/18/E/ST5/00222), and by the Faculty of Chemistry, Warsaw University of Technology.References
- Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline, World Health Organization, Geneva, 2019, Licence: CC BY-NC-SA 3.0 IGO Search PubMed.
- J. J. De Waele, M. Akova, M. Antonelli, R. Canton, J. Carlet, D. De Backer, G. Dimopoulos, J. Garnacho-Montero, J. Kesecioglu, J. Lipman, M. Mer, J. A. Paiva, M. Poljak, J. A. Roberts, J. Rodriguez Bano, J. F. Timsit, J. R. Zahar and M. Bassetti, Intensive Care Med., 2018, 44, 189–196 CrossRef PubMed.
- J. Chakaya, M. Khan, F. Ntoumi, E. Aklillu, R. Fatima, P. Mwaba, N. Kapata, S. Mfinanga, S. E. Hasnain, P. D. M. C. Katoto, A. N. H. Bulabula, N. A. Sam-Agudu, J. B. Nachega, S. Tiberi, T. D. McHugh, I. Abubakar and A. Zumla, Int. J. Infect. Dis., 2021, 113, S7–S12 CrossRef CAS PubMed.
- Global tuberculosis report 2020, World Health Organization, 2020, Geneva, Licence: CC BY-NC-SA 3.0 Search PubMed.
- M. Lakemeyer, W. Zhao, F. A. Mandl, P. Hammann and S. A. Sieber, Angew. Chem., Int. Ed., 2018, 57, 14440–14475 CrossRef CAS PubMed.
- B. Spellberg, R. Guidos, D. Gilbert, J. Bradley, H. W. Boucher, W. M. Scheld, J. G. Bartlett and J. Edwards, Clin. Infect. Dis., 2008, 46, 155–164 CrossRef.
- C. Nathan and O. Cars, N. Engl. J. Med., 2014, 371, 1761–1763 CrossRef PubMed.
- L. J. V. Piddock, Lancet Infect. Dis., 2012, 12, 249–253 CrossRef PubMed.
- H. Wang, M. Niu, T. Xue, L. Ma, X. Gu, G. Wei, F. Li and C. Wang, J. Mater. Chem. B, 2022, 10, 1858–1874 RSC.
- O. Stachurski, D. Neubauer, I. Małuch, D. Wyrzykowski, M. Bauer, S. Bartoszewska, W. Kamysz and E. Sikorska, Bioorg. Med. Chem., 2019, 27, 115129 CrossRef CAS PubMed.
- Y. Yuan, F. Zhou, H. Su and Y. Zhang, Sci. Rep., 2019, 9, 11885 CrossRef PubMed.
- Y. Yuan, S. Liang, J. Li, S. Zhang and Y. Zhang, J. Mater. Chem. B, 2019, 7, 5620–5625 RSC.
- N. Wiradharma, M. Khan, L.-K. Yong, C. A. E. Hauser, S. V. Seow, S. Zhang and Y.-Y. Yang, Biomaterials, 2011, 32, 9100–9108 CrossRef CAS PubMed.
- M. S. Ganewatta and C. Tang, Polymer, 2015, 63, A1–A29 CrossRef CAS.
- C. Ergene, K. Yasuhara and E. F. Palermo, Polym. Chem., 2018, 9, 2407–2427 RSC.
- Y. Yang, Z. Cai, Z. Huang, X. Tang and X. Zhang, Polym. J., 2018, 50, 33–44 CrossRef CAS.
- S.-B. T. A. Amos, L. S. Vermeer, P. M. Ferguson, J. Kozlowska, M. Davy, T. T. Bui, A. F. Drake, C. D. Lorenz and A. J. Mason, Sci. Rep., 2016, 6, 37639 CrossRef CAS.
- L. Liu, Y. Fang and J. Wu, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 2479–2486 CrossRef CAS PubMed.
- A. Oddo, T. T. Thomsen, H. M. Britt, A. Løbner-Olesen, P. W. Thulstrup, J. M. Sanderson and P. R. Hansen, ACS Med. Chem. Lett., 2016, 7, 741–745 CrossRef CAS PubMed.
- T. Rončević, D. Vukičević, N. Ilić, L. Krce, G. Gajski, M. Tonkić, I. Goić-Barišić, L. Zoranić, Y. Sonavane, M. Benincasa, D. Juretić, A. Maravić and A. Tossi, J. Med. Chem., 2018, 61, 2924–2936 CrossRef.
- J. Bachl, O. Bertran, J. Mayr, C. Alemán and D. Díaz Díaz, Soft Matter, 2017, 13, 3031–3041 RSC.
- M. Häring, S. Grijalvo, D. Haldar, C. Saldías and D. D. Díaz, Eur. Polym. J., 2019, 115, 221–224 CrossRef.
- M. G. Saborío, O. Bertran, S. Lanzalaco, M. Häring, L. Franco, J. Puiggalí, D. D. Díaz, F. Estrany and C. Alemán, Soft Matter, 2018, 14, 6374–6385 RSC.
- V. Sambhy, B. R. Peterson and A. Sen, Angew. Chem., Int. Ed., 2008, 47, 1250–1254 CrossRef CAS PubMed.
- T. Eren, A. Som, J. R. Rennie, C. F. Nelson, Y. Urgina, K. Nüsslein, E. B. Coughlin and G. N. Tew, Macromol. Chem. Phys., 2008, 209, 516–524 CrossRef CAS.
- W. Chin, C. Yang, V. W. L. Ng, Y. Huang, J. Cheng, Y. W. Tong, D. J. Coady, W. Fan, J. L. Hedrick and Y. Y. Yang, Macromolecules, 2013, 46, 8797–8807 CrossRef CAS.
- D. S. S. M. Uppu, S. Samaddar, J. Hoque, M. M. Konai, P. Krishnamoorthy, B. R. Shome and J. Haldar, Biomacromolecules, 2016, 17, 3094–3102 CrossRef CAS PubMed.
- E. F. Palermo and K. Kuroda, Biomacromolecules, 2009, 10, 1416–1428 CrossRef CAS PubMed.
- C. Ergene and E. F. Palermo, J. Mater. Chem. B, 2018, 6, 7217–7229 RSC.
- K. You, B. Gao, M. Wang, X. Wang, K. C. Okoro, A. Rakhimbekzoda and Y. Feng, J. Mater. Chem. B, 2022, 10, 1005–1018 RSC.
- R. J. Kopiasz, W. Tomaszewski, A. Kuźmińska, K. Chreptowicz, J. Mierzejewska, T. Ciach and D. Jańczewski, Macromol. Biosci., 2020, 20, 2000063 CrossRef CAS.
- C. Krumm, S. Trump, L. Benski, J. Wilken, F. Oberhaus, M. Köller and J. C. Tiller, ACS Appl. Mater. Interfaces, 2020, 12, 21201–21209 CrossRef CAS.
- J. P. K. Tan, J. Tan, N. Park, K. Xu, E. D. Chan, C. Yang, V. A. Piunova, Z. Ji, A. Lim, J. Shao, A. Bai, X. Bai, D. Mantione, H. Sardon, Y. Y. Yang and J. L. Hedrick, Macromolecules, 2019, 52, 7878–7885 CrossRef CAS.
- S. Venkataraman, J. P. K. Tan, S. T. Chong, C. Y. H. Chu, E. A. Wilianto, C. X. Cheng and Y. Y. Yang, Biomacromolecules, 2019, 20, 2737–2742 CrossRef CAS PubMed.
- W. Lou, S. Venkataraman, G. Zhong, B. Ding, J. P. K. Tan, L. Xu, W. Fan and Y. Y. Yang, Acta Biomater., 2018, 78, 78–88 CrossRef CAS PubMed.
- S. Liu, R. J. Ono, H. Wu, J. Y. Teo, Z. C. Liang, K. Xu, M. Zhang, G. Zhong, J. P. K. Tan, M. Ng, C. Yang, J. Chan, Z. Ji, C. Bao, K. Kumar, S. Gao, A. Lee, M. Fevre, H. Dong, J. Y. Ying, L. Li, W. Fan, J. L. Hedrick and Y. Y. Yang, Biomaterials, 2017, 127, 36–48 CrossRef CAS PubMed.
- S. Bai, J. Wang, K. Yang, C. Zhou, Y. Xu, J. Song, Y. Gu, Z. Chen, M. Wang, C. Shoen, B. Andrade, M. Cynamon, K. Zhou, H. Wang, Q. Cai, E. Oldfield, S. C. Zimmerman, Y. Bai and X. Feng, Sci. Adv., 2021, 7, 1–17 Search PubMed.
- Z. Chen, C. Zhou, Y. Xu, K. Wen, J. Song, S. Bai, C. Wu, W. Huang, Q. Cai, K. Zhou, H. Wang, Y. Wang, X. Feng and Y. Bai, Biomaterials, 2021, 275, 120858 CrossRef CAS PubMed.
- H. Chen, S. A. Nyantakyi, M. Li, P. Gopal, D. B. Aziz, T. Yang, W. Moreira, M. Gengenbacher, T. Dick and M. L. Go, Front. Microbiol., 2018, 9, 1627 CrossRef PubMed.
- P. J. Brennan, Tuberculosis, 2003, 83, 91–97 CrossRef CAS.
- S. Bansal, M. Singh, S. Kidwai, P. Bhargava, A. Singh, V. Sreekanth, R. Singh and A. Bajaj, MedChemComm, 2014, 5, 1761–1768 RSC.
- S. Mignani, V. D. Tripathi, D. Soam, R. P. Tripathi, S. Das, S. Singh, R. Gandikota, R. Laurent, A. Karpus, A.-M. Caminade, A. Steinmetz, A. Dasgupta, K. K. Srivastava and J.-P. Majoral, Biomacromolecules, 2021, 22, 2659–2675 CrossRef CAS PubMed.
- A. W. Simonson, A. S. Mongia, M. R. Aronson, J. N. Alumasa, D. C. Chan, A. Lawanprasert, M. D. Howe, A. Bolotsky, T. K. Mal, C. George, A. Ebrahimi, A. D. Baughn, E. A. Proctor, K. C. Keiler and S. H. Medina, Nat. Biomed. Eng., 2021, 5, 467–480 CrossRef CAS PubMed.
- S. Ramón-García, R. Mikut, C. Ng, S. Ruden, R. Volkmer, M. Reischl, K. Hilpert and C. J. Thompson, Antimicrob. Agents Chemother., 2013, 57, 2295–2303 CrossRef PubMed.
- L. M. Timofeeva, N. A. Kleshcheva, M. O. Shleeva, M. P. Filatova, Y. A. Simonova, Y. A. Ermakov and A. S. Kaprelyants, Appl. Microbiol. Biotechnol., 2015, 99, 2557–2571 CrossRef CAS PubMed.
- D. J. Phillips, J. Harrison, S.-J. Richards, D. E. Mitchell, E. Tichauer, A. T. M. Hubbard, C. Guy, I. Hands-Portman, E. Fullam and M. I. Gibson, Biomacromolecules, 2017, 18, 1592–1599 CrossRef CAS.
- P. S. Yavvari, S. Gupta, D. Arora, V. K. Nandicoori, A. Srivastava and A. Bajaj, Biomacromolecules, 2017, 18, 2024–2033 CrossRef CAS.
- D. Kozon, J. Mierzejewska, T. Kobiela, A. Grochowska, K. Dudnyk, A. Głogowska, A. Sobiepanek, A. Kuźmińska, T. Ciach, E. Augustynowicz-Kopeć and D. Jańczewski, Macromol. Biosci., 2019, 19, 1900254 CrossRef CAS.
- S.-J. Richards, K. Isufi, L. E. Wilkins, J. Lipecki, E. Fullam and M. I. Gibson, Biomacromolecules, 2018, 19, 256–264 CrossRef CAS PubMed.
- M. U. Shiloh and P. A. DiGiuseppe Champion, Curr. Opin. Microbiol., 2010, 13, 86–92 CrossRef CAS PubMed.
- N. Lelovic, K. Mitachi, J. Yang, M. R. Lemieux, Y. Ji and M. Kurosu, J. Antibiot., 2020, 73, 780–789 CrossRef CAS PubMed.
- A. Sharma, A. A. Pohane, S. Bansal, A. Bajaj, V. Jain and A. Srivastava, Chem.–Eur. J., 2015, 21, 3540–3545 CrossRef CAS PubMed.
- R. J. Kopiasz, A. Rukasz, K. Chreptowicz, R. Podgórski, A. Kuźmińska, J. Mierzejewska, W. Tomaszewski, T. Ciach and D. Jańczewski, Colloids Surf., B, 2021, 207, 112016 CrossRef CAS PubMed.
- R. J. Kopiasz, N. Kulbacka, K. Drężek, R. Podgórski, I. Łojszczyk, J. Mierzejewska, T. Ciach, E. Augustynowicz-Kopeć, A. Głogowska, A. Iwańska, W. Tomaszewski and D. Jańczewski, Macromol. Biosci., 2022, 2200094 CrossRef CAS.
- J. M. Layman, E. M. Borgerding, S. R. Williams, W. H. Heath and T. E. Long, Macromolecules, 2008, 41, 4635–4641 CrossRef CAS.
- J. Aguiar, P. Carpena, J. A. Molina-Bolívar and C. Carnero Ruiz, J. Colloid Interface Sci., 2003, 258, 116–122 CrossRef CAS.
- A. Strassburg, F. Kracke, J. Wenners, A. Jemeljanova, J. Kuepper, H. Petersen and J. C. Tiller, Macromol. Biosci., 2015, 15, 1710–1723 CrossRef CAS.
- M. S. Ganewatta, M. A. Rahman, L. Mercado, T. Shokfai, A. W. Decho, T. M. Reineke and C. Tang, Bioact. Mater., 2018, 3, 186–193 CrossRef PubMed.
- K. E. S. Locock, T. D. Michl, J. D. P. Valentin, K. Vasilev, J. D. Hayball, Y. Qu, A. Traven, H. J. Griesser, L. Meagher and M. Haeussler, Biomacromolecules, 2013, 14, 4021–4031 CrossRef CAS PubMed.
- R. Liu, X. Chen, S. Chakraborty, J. J. Lemke, Z. Hayouka, C. Chow, R. A. Welch, B. Weisblum, K. S. Masters and S. H. Gellman, J. Am. Chem. Soc., 2014, 136, 4410–4418 CrossRef CAS PubMed.
- N. A. R. Gow and B. Hube, Curr. Opin. Microbiol., 2012, 15, 406–412 CrossRef CAS.
- N. Malanovic and K. Lohner, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 936–946 CrossRef CAS.
- E. F. Palermo, K. Lienkamp, E. R. Gillies and P. J. Ragogna, Angew. Chem., Int. Ed., 2019, 58, 3690–3693 CrossRef CAS PubMed.
- G. J. Gabriel, J. A. Maegerlein, C. F. Nelson, J. M. Dabkowski, T. Eren, K. Nüsslein and G. N. Tew, Chem.–Eur. J., 2009, 15, 433–439 CrossRef CAS PubMed.
- J. Mayr, J. Bachl, J. Schlossmann and D. D. Díaz, Int. J. Mol. Sci., 2017, 18, 303 CrossRef PubMed.
- S. N. Riduan, Y. Yuan, F. Zhou, J. Leong, H. Su and Y. Zhang, Small, 2016, 12, 1928–1934 CrossRef CAS PubMed.
- P. D. Stapleton and P. W. Taylor, Sci. Prog., 2002, 85, 57–72 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: NMR spectra, synthetic scheme. See https://doi.org/10.1039/d2ra04121a |
| This journal is © The Royal Society of Chemistry 2022 |