Open Access Article
Open Access ArticleNiII and CoII binary and ternary complexes of 3-formylchromone: spectroscopic characterization, antimicrobial activities, docking and modeling studies
Noha Nabil,
Omima M. I. Adly ,
Magdy Shebl
,
Magdy Shebl ,
Ali Taha
,
Ali Taha and
Fatma Samy
and
Fatma Samy *
*
Department of Chemistry, Faculty of Education, Ain Shams University, Roxy, Cairo 11566, Egypt. E-mail: noha.nabil1986@yahoo.com; omima_adly@edu.asu.edu.eg; magdyshebl@edu.asu.edu.eg; alitaha@edu.asu.edu.eg; fatmasame@edu.asu.edu.eg; Fax: +20 0222581243; Tel: +20 1096418414
First published on 19th October 2022
Abstract
Reactions of 3-formylchromone (L) with Ni(II) and Co(II) ions having different anions (acetate, perchlorate, nitrate, and chloride) yielded a series of binary and ternary octahedral complexes with the general formula [MLnL′mXy(S)a]Zy·bS, where M = Ni or Co, n = 1–3, L′ = auxiliary ligand = 8-hydroxyquinoline or 1,10-phenanthroline, m = 1 or 2, X = acetate or chloride, y = 0 or 2, S = H2O or MeOH, a = 0–2, Z = nitrate or perchlorate and b = 0–1.5. Elemental and thermal analyses and infra-red, electronic, mass, magnetic susceptibility and molar conductivity measurements were successfully utilized to characterize the structures of the chromone complexes. The chromone ligand acts as a neutral bidentate ligand through its formyl and γ-pyrone oxygen atoms. The obtained complexes were formed with molar ratios 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 M
3 M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L for the binary and 1
L for the binary and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 M
1 M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L
L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L′ for the ternary complexes. The kinetic parameters of the thermal degradation steps were estimated and explained using the Coats–Redfern equations. The synthesized complexes showed antimicrobial activity with higher activity toward Candida albicans and Bacillus subtilis. Docking studies showed good agreement with the antimicrobial activity. Molecular modeling of the synthesized complexes was performed using Hyperchem at the PM3 level and the calculated structures correlate with the experimental data.
L′ for the ternary complexes. The kinetic parameters of the thermal degradation steps were estimated and explained using the Coats–Redfern equations. The synthesized complexes showed antimicrobial activity with higher activity toward Candida albicans and Bacillus subtilis. Docking studies showed good agreement with the antimicrobial activity. Molecular modeling of the synthesized complexes was performed using Hyperchem at the PM3 level and the calculated structures correlate with the experimental data.
1. Introduction
Chromones are important natural compounds with a γ-pyrone heterocyclic ring structure. They are known for inhibiting enzymes, including mushroom tyrosinase and protein kinase-C.1 Plant leaves and flowers are sources and contain chromone as a pigment. They are essential for the production of various xanthones, transition metal chelates, and heterocyclic compounds with oxygen.2 The flexible reactive synthon chromone-3-carbaldehyde (3-formychromone) has attracted much attention of late.3 This molecule can react as an aldehyde, a Michael acceptor, a heterodiene, and a dienophile since it has three highly electrophilic carbons (C-2 and C-4 of the pyrone moiety and the carbon atom of the formyl group).3 Due to their three electrophilic sites (C1, C3, and C4), 3-formylchromones are versatile, practical, and commercial building blocks that are frequently used in the creation of functionalized fused heterocycles.4 The two electrophilic sites (C1 and C4) may react with bis-nucleophilic reagents, such as enamine ketones/esters, heterocyclic ketene aminals (HKAs), or 1,1-enediamine (EDAMs)4 among others,4 to produce the site-selective synthesis of fused heterocycles. Because it comprises a wide range of pharmacologically active chemicals, such as antimicrobial, anti-inflammatory, antiallergic, anti-HIV, antitumor, antiplaque, antioxidant and neuroprotective,5–9 as well as acting as a fluorescent marker, the bicyclic chromone moiety has been categorized as a favorable structure in drug discovery.2 Protein kinases P13Ks, CDK, and TNF signaling, among other cancer targets, are known to be inhibited by chromone derivatives. The chromone derivative LY294002 was discovered to be a kinase inhibitor through inhibiting the phosphatidylinositol-3-kinase (PI3K) signaling pathway.10 The cyclin-dependent kinase (CDK) inhibitor flavopirid, a different chromen-4-one molecule, was discovered to be capable of inhibiting the cell cycle and inducing apoptosis.10 Another set of substituted chromones that provided site-specificity for the ABCG2 protein were discovered to be powerful inhibitors of breast cancer strains. The 5-(4-bromobenzyloxy) 2-(2-(5-methoxyindolyl) ethyl-1-carbonyl)-4H-chromen-4-one compound in this series showed strong inhibition and minimal toxicity, showing a noticeably high therapeutic profile.10 There are a few intriguing literature reports on biological investigations, such as the DNA binding and cytotoxicity profile of copper(II)-appended chromone therapeutic candidates that have demonstrated varied responses with diverse cytotoxicity potencies against various phenotypes of malignancies.10Mixed-ligand complexes have lately gained considerable attention due to their important roles in biological processes.11 The presence of heterocyclic nitrogen donor ligands, such as 1,10-phenanthroline or oxine (8-hydroxyquinoline), has been shown to significantly enhance the various pharmacological impacts of metal complexes.12
Nickel and cobalt elements are generally required for life as nickel is involved in a variety of enzymes13 and cobalt in vitamin B-12 and a number of therapeutics.14 Their complexes have different biological applications;15–19 some are used as cisplatin alternatives in cancer treatment20 and some as catalysts in the oxygenation processes of organic molecules.21
In an extension of our continuing interest in the synthesis, spectroscopic characterization and biological activity of chromone-based ligands and their complexes,22 a series of binary and ternary Ni(II)- and Co(II)-3-formylchromone complexes were synthesized and successfully characterized. Different Ni(II)- and Co(II)-salts (acetate, perchlorate, nitrate, and chloride) were employed to synthesize binary complexes, while auxiliary ligands (Scheme 1), such as N,O-donor oxine or N,N-donor 1,10-phenanthroline, were utilised to synthesize mixed-ligand complexes. Various analytical and spectroscopic techniques were used to characterize the prepared complexes. Theoretical studies were performed on Hyperchem 7.52 software at the PM3 level and the theoretical findings were compared and correlated with the experimental data. The ligand and its complexes were tested for antibacterial activity.
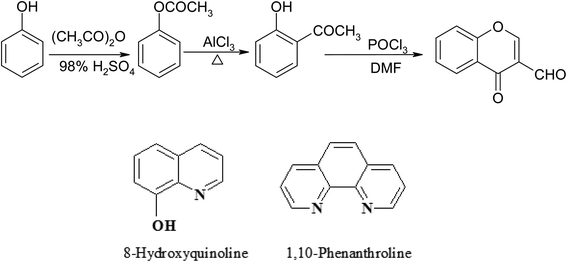 | ||
| Scheme 1 Synthesis of 3-formylchromone and auxiliary ligands 8-hydroxyquinoline and 1,10-phenanthroline. | ||
2. Experimental
2.1. Measurements
IR spectra of the chromone ligand and its complexes were recorded as KBr discs on a FT IR Nicolet IS10 spectrometer. The percentages of C, H and N were estimated on a Vario El-Elementar at the Ministry of Defense, Chemical War Department. A Stuart SMP3 melting point apparatus was utilized to record the decomposition temperatures of the complexes. After decomposing an exact weight of the solid complex with concentrated nitic acid, neutralising with ammonia and titrating with Na2EDTA, the Ni(II) and Co(II) ions were analysed as described elsewhere.23 On a GC-2010 Shimadzu gas chromatography mass spectrometer, mass spectra of the metal–chromone complexes were recorded. Electronic spectral measurements were performed as solutions in dimethylformamide and/or Nujol mulls on a Jasco model V-550 UV/Vis spectrophotometer. The Corning conductivity meter NY 14831 model 441 was employed to test the molar conductivity of 10−3 M solid complex solutions in dimethylformamide. Magnetic susceptibility measurements for the chromone complexes were done at room temperature with an MKI Johnson Matthey magnetic susceptibility balance (Alpha). The obtained effective magnetic moment values were corrected using Pascal's constants for the diamagnetism of the atoms included in the prepared chelates.24 On a Shimadzu-50 thermal analyzer, TGA measurements were taken from ambient temperature to 800 °C under N2 at a heating rate of 10 °C min−1. X-ray diffraction powder patterns of the ligand and its complexes (2, 5, 6, 7, 10 and 11) were recorded at room temperature on a Philips X'Pert powder diffractometer using the Bragg–Brentano configuration and CuKα radiation λ = 1.5406 Å at the Central Metallurgical Research Institute (CMRDI). The disc diffusion method was used to investigate the biological activities of the chromone ligand and its metal complexes.2.2. Materials
3-Formylchromone (Scheme 1) was synthesized using a method described in the literature.25 Ni(OAc)2·4H2O, Ni(NO3)2·6H2O, Ni(ClO4)2·6H2O, NiCl2·6H2O, Co(OAc)2·4H2O, Co(NO3)2·6H2O, CoCl2·6H2O, 8-hydroxyquinoline, oxine (8-HQ), 1,10-phenanthroline, Na2EDTA, nitric acid, mureoxide, and ammonium hydroxide were purchased from either BDH or Merck. The organic solvents were directly employed without distillation and were reagent grade compounds.2.3. Synthesis of the metal complexes
The following synthetic procedures are detailed as representative instances and the other complexes were obtained similarly.2.4. Antimicrobial activity
Using the conventional disc-agar diffusion technique,26,27 the biological activities of the chromone ligand and its complexes were examined. The biological activity was tested against Gram positive bacteria Staphylococcus aureus (ATCC 25923) and Bacillus subtilis (ATCC 6635), Gram negative bacteria Salmonella typhimurium (ATCC 14028) and Escherichia coli (ATCC 25922) and fungal strains Candida albicans (ATCC 10231) and Aspergillus fumigatus. In the case of the Gram-positive and Gram-negative bacteria, the antibiotics chloramphenicol and cephalothin were employed as references, respectively, while in the case of the fungi, cycloheximide was used.2.5. Molecular docking and modeling
Molecular docking studies for the compounds were tested against S. aureus nucleoside diphosphate kinase, PDB ID: 3Q89, using the MOE 2014.0901 software.Geometrical optimization and conformation analysis were performed using the PM3 forcefield as implemented in Hyperchem 7.52 (ref. 28) in order to acquire a better understanding of the molecular structure of the chromone ligand and its complexes.
3. Results and discussion
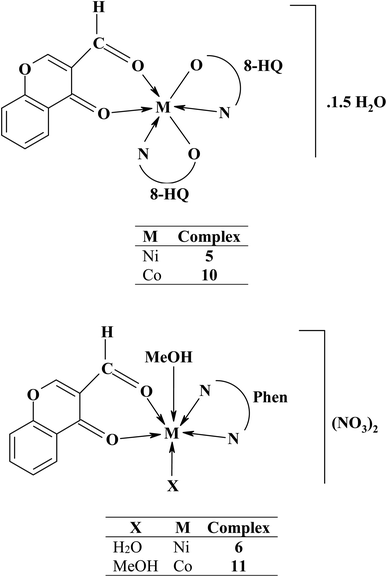
To investigate the influence of counter ions on the products, the chromone ligand was reacted with numerous salts of nickel(II) and cobalt(II) ions (acetate, perchlorate, nitrate, and chloride). Reactions of nickel(II) and cobalt(II) nitrates with the chromone ligand in the presence of auxiliary ligands (8-hydroxyquinoline or 1,10-phenanthroline) gave mixed-ligand complexes. The obtained complexes have good storage qualities, i.e. they are non-hygroscopic and sparingly soluble in water and most common organic solvents except dimethylformamide. Table 1 summarizes the main analytical and physical data of the chromone ligand along with its nickel(II) and cobalt(II) complexes.| No. | Reaction | Complex | Color | Yield (%) | M. P. (°C) | Elemental analysis, % found/(calc.) | |||
|---|---|---|---|---|---|---|---|---|---|
| M. F. [F. Wt] | C | H | N/S | M | |||||
| a Not determined. | |||||||||
| 1 | L + Ni(OAc)2·4H2O | [Ni(L)2(OAc)2]·H2O | Light green | 50 | >300 | 52.86 | 4.52 | — | 10.70 (10.81) |
| C24H20O11Ni [543.13] | (53.08) | (3.71) | |||||||
| 2 | L + Ni(NO3)2·6H2O | [Ni(L)3](NO3)2 · 0.5MeOH | Light green | 48 | 253–257 | 52.82 | 3.30 | 4.10 | 7.92 |
| C30.5H20N2O15.5Ni [721.21] | (50.80) | (2.80) | (3.88) | (8.14) | |||||
| 3 | L + Ni(ClO4)2·6H2O | [Ni(L)2(H2O)2](ClO4)2·H2O | Green | 45 | a | 36.41 | 2.30 | — | a |
| C20H18O17Cl2Ni [ 659.97] | (36.40) | (2.75) | (8.90) | ||||||
| 4 | L + NiCl2.6H2O | [Ni(L)2Cl2(H2O)2] | Pale lemon | 83 | >300 | 50.01 | 2.90 | — | 12.10 |
| C20H12O6Cl2Ni [ 477.93] | (50.26) | (2.53) | (12.28) | ||||||
| 5 | L + Ni(NO3)2·6H2O + 8-HQ | [Ni(L)(8-HQ)2]·1.5H2O | Dark green | 57 | >300 | 60.80 | 3.73 | 5.60 | 10.40 |
| C28H21N2O6.5Ni [548.20] | (61.35) | (3.86) | (5.11) | (10.71) | |||||
| 6 | L + Ni(NO3)2·6H2O + Phen | [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 | Pale green | 45 | 156 | 47.30 | 3.87 | 7.60 | 9.61 |
| C23H19N3O12Ni [588.13] | (46.97) | (3.26) | (7.14) | (9.98) | |||||
| 7 | L + Co(OAc)2·4H2O | [Co(L)2(OAc)2]·1.5H2O | Light orange | 47 | >300 | 52.44 | 4.14 | — | 10.30 |
| C24H21O11.5Co [552.36] | (52.19) | (3.83) | (10.67) | ||||||
| 8 | L + Co(NO3)2·6H2O | [Co(L)3](NO3)2·MeOH | Orange | 61 | >300 | 50.90 | 3.40 | 3.40 | 7.70 |
| C31H22N2O16Co [737.46] | (50.49) | (3.01) | (3.80) | (7.99) | |||||
| 9 | L + CoCl2·6H2O | [Co(L)2Cl2] | Orange | 40 | >300 | 50.70 | 3.00 | — | 12.10 |
| C20H12O6Cl2Co [478.15] | (50.24) | (2.53) | (12.33) | ||||||
| 10 | L + Co(NO3)2·6H2O + 8-HQ | [Co(L)(8-HQ)2]·1.5H2O | Dark brown | 87 | >300 | 60.90 | 3.75 | 5.70 | 10.50 |
| C28H21N2O6.5Co [548.42] | (61.32) | (3.86) | (5.11) | (10.75) | |||||
| 11 | L + Co(NO3)2·6H2O + Phen | [Co(L)(Phen)(MeOH)2](NO3)2 | Rose | 50 | 130 | 48.10 | 3.77 | 7.31 | 9.65 |
| C24H21N3O12Co [602.38] | (47.85) | (3.51) | (6.98) | (9.78) | |||||
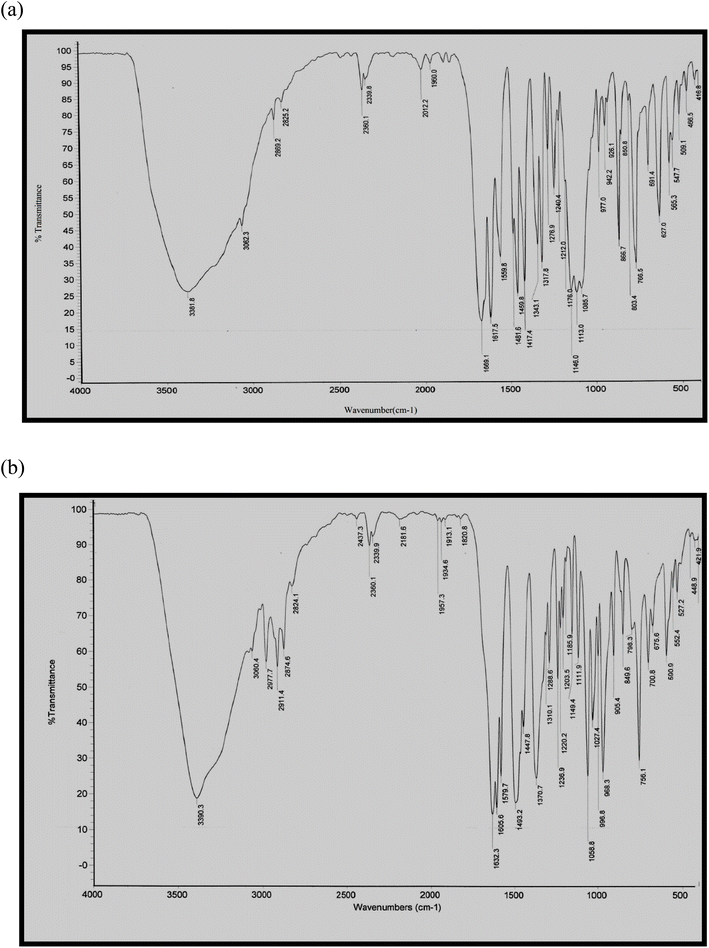
3.1. IR spectra
The IR spectra of some representative metal complexes are depicted in Fig. 1a and b and the characteristic IR spectral bands are tabulated in Table 2. The IR spectra of the complexes are compared to that of the ligand to identify the coordinating sites and their extent. This comparison revealed the following: (i) the spectra of the complexes exhibited strong and broad bands in the range 3296-3535 cm−1 which can be ascribed to the ν(OH) of the coordinated or non-coordinated water and/or methanol molecules in the complexes. (ii) The strong bands located at 1694 and 1643 cm−1 assigned to ν(HC = O) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), respectively, of the chromone ligand exhibited red shifts in the complexes, revealing the involvement of these groups in complexation.29 The extent of carbonyl coordination could be seen by the negative slope of the linear correlation of ΔEgab, which is analogous with the stability of the complex (vide infra), versus the stretching frequency of carbonyl. For complexes 1, 3, 5, 8, and 11, ΔEgab (eV) = 65.2–0.0367 ν(C
O), respectively, of the chromone ligand exhibited red shifts in the complexes, revealing the involvement of these groups in complexation.29 The extent of carbonyl coordination could be seen by the negative slope of the linear correlation of ΔEgab, which is analogous with the stability of the complex (vide infra), versus the stretching frequency of carbonyl. For complexes 1, 3, 5, 8, and 11, ΔEgab (eV) = 65.2–0.0367 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)/cm−1, r = 0.96; i.e., as the extent of carbonyl coordination with the metal ion increases, the carbonyl bond is elongated and the frequency shifts lower, as shown by the negative slope. (iii) Participation of the anions and their modes of bonding was investigated using the infra-red spectral data. The acetato-complexes (1 and 7) displayed new bands at 1493 and 1490 (1) and 1318 and 1319 cm−1 (7), which may be respectively attributed to the νas(COO−) and νs(COO−) of the acetate group.30 The separation between the two bands, Δν = (νas − νs), demonstrates the monodentate nature of the acetate group.31 In complexes 2, 6, 8 and 11, the new bands observed in the range 1425–1447 cm−1 may be those of the ionic nitrate group.32 In complex 3, the new bands observed at 1086 and 627 cm−1 may be related to the ν3 and ν4 vibrations, respectively, of the ionic perchlorate group.33 The direct coordination of the acetate anion as well as the ionic nature of the nitrate and perchlorate anions is supported by molar conductivity data (see Section 3.2). (iv) The spectra of ternary complexes (5, 6, 10 and 11) displayed new bands in the range 1501–1518 cm−1 which may be ascribed to the coordination of the C
O)/cm−1, r = 0.96; i.e., as the extent of carbonyl coordination with the metal ion increases, the carbonyl bond is elongated and the frequency shifts lower, as shown by the negative slope. (iii) Participation of the anions and their modes of bonding was investigated using the infra-red spectral data. The acetato-complexes (1 and 7) displayed new bands at 1493 and 1490 (1) and 1318 and 1319 cm−1 (7), which may be respectively attributed to the νas(COO−) and νs(COO−) of the acetate group.30 The separation between the two bands, Δν = (νas − νs), demonstrates the monodentate nature of the acetate group.31 In complexes 2, 6, 8 and 11, the new bands observed in the range 1425–1447 cm−1 may be those of the ionic nitrate group.32 In complex 3, the new bands observed at 1086 and 627 cm−1 may be related to the ν3 and ν4 vibrations, respectively, of the ionic perchlorate group.33 The direct coordination of the acetate anion as well as the ionic nature of the nitrate and perchlorate anions is supported by molar conductivity data (see Section 3.2). (iv) The spectra of ternary complexes (5, 6, 10 and 11) displayed new bands in the range 1501–1518 cm−1 which may be ascribed to the coordination of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N group of the 8-HQ or 1,10-Phen ligands to the nickel or cobalt.34 (v) The new bands observed in the ranges 501–567 and 424–450 cm−1 may be attributed to ν(Ni- or Co–O) and ν(Ni- or Co–N), respectively.35,36
N group of the 8-HQ or 1,10-Phen ligands to the nickel or cobalt.34 (v) The new bands observed in the ranges 501–567 and 424–450 cm−1 may be attributed to ν(Ni- or Co–O) and ν(Ni- or Co–N), respectively.35,36
| No. | Complex | IR spectra (cm−1) | |||||
|---|---|---|---|---|---|---|---|
| ν(OH) | ν(HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
ν(M–O) | ν(M–N) | Other bands | ||
| L | — | 1694 | 1643 | — | — | — | |
| 1 | [Ni(L)2(OAc)2]·H2O | 3535 | 1629 | 1606 | 528 | — | 1493 νas(COO−), 1318 νs(COO−); (monodentate OAc−) |
| 2 | [Ni(L)3](NO3)2·0.5MeOH | 3296 | 1628 | 1607 | 528 | — | 1447; ν(NO3−) (ionic) |
| 3 | [Ni(L)2(H2O)2](ClO4)2·H2O | 3382 | 1669 | 1618 | 509 | — | 1086, 627; ν(ClO4−) (ionic) |
| 4 | [Ni(L)2Cl2(H2O)2] | 3390 | 1632 | 1606 | 527 | — | |
| 5 | [Ni(L)(8-HQ)2]·1.5H2O | 3385 | 1629 | 1581 | 501 | 450 | 1504; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 8-HQ N) 8-HQ |
| 6 | [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 | 3357 | 1673 | 1619 | 567 | 428 | 1427; ν(NO3−) (ionic), 1518; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) Phen N) Phen |
| 7 | [Co(L)2(OAc)2]·1.5H2O | 3523 | 1625 | 1606 | 528 | — | 1490 νas(COO−), 1319 νs(COO−); (monodentate OAc−) |
| 8 | [Co(L)3](NO3)2·MeOH | 3523 | 1626 | 1605 | 528 | — | 1446; ν(NO3−) (ionic) |
| 9 | [Co(L)2Cl2] | 3521 | 1628 | 1605 | 528 | — | |
| 10 | [Co(L)(8-HQ)2]·1.5H2O | 3385 | 1633 | 1604 | 539 | 424 | 1501; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 8-HQ N) 8-HQ |
| 11 | [Co(L)(Phen)(MeOH)2](NO3)2 | 3309 | 1646 | 1613 | 505 | 427 | 1425; ν(NO3−) (ionic), 1517; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) Phen N) Phen |
3.2. Conductivity measurements
Based on the molar conductance values of the complexes in dimethylformamide (Table 3), the complexes can be divided into two classes: (i) non-electrolytes (complexes 1, 4, 5, 7, 9 and 10) with molar conductance values in the range 6.40–44.8 Ω−1 cm2 mol−1 and (ii) 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 electrolytes (complexes 2, 3, 6, 8 and 11) with molar conductance values in the range 103.2–125 Ω−1 cm2 mol−1.37 This finding agrees with the weak coordinating capability of the nitrate and perchlorate anions as compared to the strong coordinating capability of the acetate anion. Furthermore, this finding is in agreement with the infrared spectral data (see Section 3.1), illustrating the coordinated nature of the acetate group and the ionic nature of the nitrate and perchlorate groups.
1 electrolytes (complexes 2, 3, 6, 8 and 11) with molar conductance values in the range 103.2–125 Ω−1 cm2 mol−1.37 This finding agrees with the weak coordinating capability of the nitrate and perchlorate anions as compared to the strong coordinating capability of the acetate anion. Furthermore, this finding is in agreement with the infrared spectral data (see Section 3.1), illustrating the coordinated nature of the acetate group and the ionic nature of the nitrate and perchlorate groups.
| No. | Complex | Electronic spectral bandsa (nm), λmaxa (nm)/(εmax, L cm−1 mol−1) | μeff (B.M.) | Conductancea (Ω−1 cm2 mol−1) |
|---|---|---|---|---|
| a Solutions in DMF (10−3 M).b Nujol mull. | ||||
| L | 271 (4340), 353 (4700), 387 (3415) | — | — | |
| 1 | [Ni(L)2(OAc)2]·H2O | (500 sh)b | 2.80 | 10.6 |
| 2 | [Ni(L)3](NO3)2 · 0.5MeOH | (500 sh, 613 sh)b | 3.20 | 106.3 |
| 3 | [Ni(L)2(H2O)2](ClO4)2·H2O | (470 sh, 504, 605sh)b | 2.83 | 125.0 |
| 4 | [Ni(L)2Cl2(H2O)2] | (400, 502, 630)b | 2.43 | 18.5 |
| 5 | [Ni(L)(8-HQ)2]·1.5H2O | (493, 693)b | 2.00 | 44.2 |
| 6 | [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 | (504, 605)b | 2.30 | 122.8 |
| 7 | [Co(L)2(OAc)2]·1.5H2O | (505 sh)b | 4.74 | 9.5 |
| 8 | [Co(L)3](NO3)2·MeOH | (510)b | 5.21 | 103.2 |
| 9 | [Co(L)2Cl2] | (532 sh)b | 4.34 | 6.40 |
| 10 | [Co(L)(8-HQ)2]·1.5H2O | (488)b | 2.00 | 44.8 |
| 11 | [Co(L)(Phen)(MeOH)2](NO3)2 | (500 sh)b | 3.86 | 108.1 |
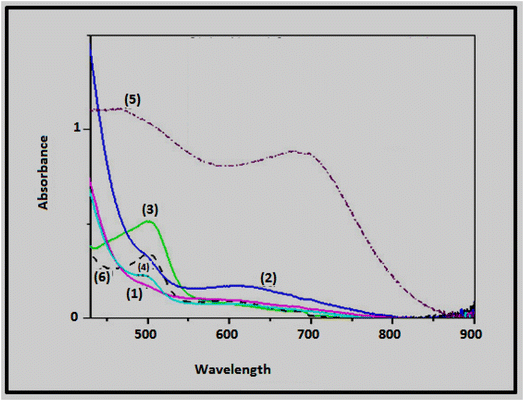
3.3. Magnetic moment measurements and electronic spectra
The electronic spectral data and magnetic moment values of the chromone ligand and its nickel(II) and cobalt(II) complexes are tabulated in Table 3; Fig. 2 presents the electronic spectra of some representative complexes. The chromone nickel(II) complexes (with the exception of 4–6) have magnetic moment values in the range of 2.80–3.20 B.M., demonstrating an octahedral geometry.38 However, the magnetic moment values of complexes 4–6 are in the range of 2.00–2.43 B.M., indicating an octahedral geometry with an antiferromagnetic interaction or an equilibrium between square and octahedral species.39 This explanation is further supported by the electronic spectra of the nickel(II) complexes showing two bands in the ranges of 493–504 and 605–693 nm which may be respectively assigned to the 3A2g(F) → 3T1g(P) and 3A2g(F) → 3T1g(F) electronic transitions in octahedral geometrical arrangements of Ni(II) ion.40The magnetic moment values of the chromone cobalt(II) complexes (except 9–11) are in the range 4.74–5.21 B.M., consistent with octahedral cobalt(II) complexes (4.8–5.2 B.M.).38 The lower values (Table 3) of complexes 9–11 could be attributed to antiferromagnetic interaction.41 The electronic spectra of cobalt(II) complexes revealed a band in the 488–532 nm range which might be attributed to the 4T1g → 4T1g(P) transition in an octahedral geometry.31,39 The negative slope of ΔEgab with λmax, as ΔEgab is known to increase with the increasing stability of complexes, leads to a decreasing in λmax, where ΔEgab = 8.35 (0.4251)–0.00443 (0.0007426) λmax, r = 92.2%, for complexes 1–4 and 11.
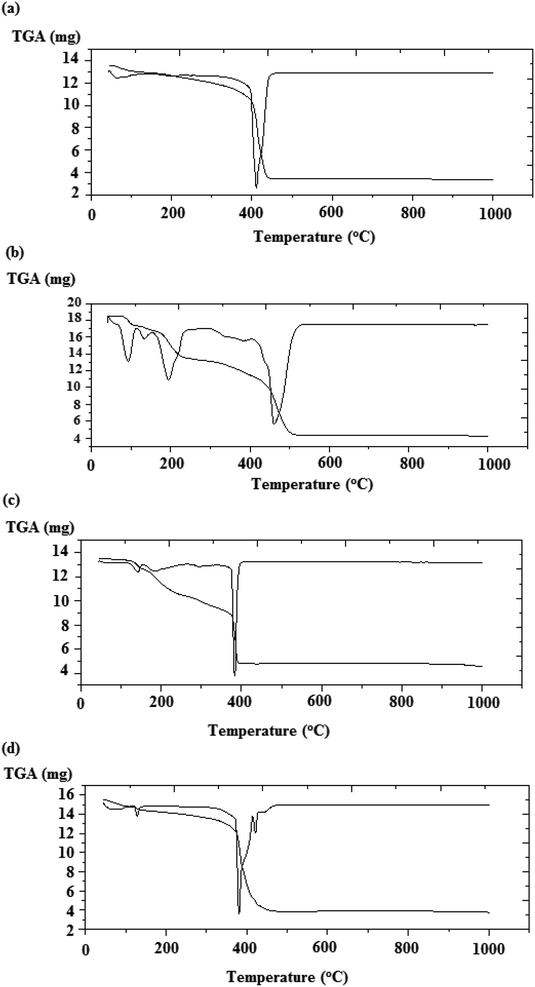
3.4. Thermal analysis
The TGA analysis of the chromone complexes, using 2, 5–7 and 10 as examples, were measured from room temperature to 800 °C with a heating rate of 10 °C min−1. Fig. 3a–d represent the thermal analysis data of 5–7 and 10 and the thermal data are summarized in Table 4.| Complex | Temperature range (°C) | % Wt loss | Lost fragment (no. of molecules) |
|---|---|---|---|
| Found/(calc.) | |||
| [Ni(L)3](NO3)2·0.5MeOH (2) | 43–132 | 1.67/(2.22) | 0.5MeOH (solv.) |
| [Ni(L)(8-HQ)2]·1.5H2O (5) | 45–142 | 4.27/(4.93) | 1.5H2O (hyd.) |
| [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 (6) | 42–116 | 5.93/(5.44) | 1MeOH (coord.) |
| 116–156 | 2.97/(3.06) | 1H2O (coord.) | |
| 156–336 | 21.1/(21.40) | 2HNO3 | |
| [Co(L)2(OAc)2]·1.5H2O (7) | 45–138 | 5.10/(4.89) | 1.5H2O (hyd.) |
| 138–309 | 21.6/(21.72) | 2AcOH | |
| [Co(L)(8-HQ)2]·1.5H2O (10) | 44–117 | 4.34/(4.92) | 1.5H2O (hyd.) |
The initial degradation step of the chromone complexes observed in the range 42–142 °C corresponds to the elimination of solvated molecules. The second degradation step in the range 116–309 °C agrees with the elimination of coordinated water or acetic acid molecules. In the case of complex 6, the third degradation step in the range 156–336 °C corresponds to the elimination of two nitric acid molecules. The decomposition processes of complex 6 are summarized in Scheme 4.
Furthermore, using the Coats–Redfern equations,42 the order n and activation parameters of the major degradation processes of the metal–chromone complexes were derived from the TG thermograms and the kinetic parameters are tabulated in Table 5. Based on the data in Table 5, it is possible to conclude the following points: (i) the decomposition processes are endothermic, as evidenced from the +ve ΔH values. (ii) The autocatalytic effect of nickel or cobalt ions on the thermal degradation of the complexes and the non–spontaneous processes is indicated by ΔG values that are comparatively low and have a +ve sign.43 (iii) ΔS values for complexes are -ve, demonstrating that the activated complex is more ordered than the reactants and/or the reactions are slow.44 (iv) In the ternary cobalt–phen complex (6), the value of activation energy E for the second stage of degradation is smaller than that of the first stage, demonstrating that the rate of decomposition of the second stage is higher than that of the first.45
| Complex | Step | n order | T (K) | A (S−1) | Δ E (kJ mol−1) | ΔH (kJ mol−1) | ΔS (kJ mol−1 K−1) | ΔG (kJ mol−1) |
|---|---|---|---|---|---|---|---|---|
| [Ni(L)3](NO3)2·0.5MeOH (2) | First | 1 | 349 | 2.13 × 106 | 1.0267 | −1.87 | −0.190 | 64.68 |
| [Ni(L)(8-HQ)2]·1.5H2O (5) | First | 1 | 356 | 9.75 × 103 | 29.160 | 26.20 | −0.290 | 130.50 |
| [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 (6) | First | 1 | 367 | 2.76 × 1013 | 106.09 | 103.0 | 2.37 × 10−3 | 102.16 |
| Second | 1 | 404 | 1.14 × 106 | 64.650 | 61.25 | −0.139 | 117.37 | |
| [Co(L)2(OAc)2]·1.5H2O (7) | First | 1 | 363 | 1.9 × 103 | 1.0816 | −1.94 | −0.192 | 67.76 |
| [Co(L)(8-HQ)2]·1.5H2O (10) | First | 0 | 353 | 2.57 × 109 | 84.160 | 81.20 | −0.074 | 107.37 |
3.5. Mass spectra
As illustrative examples, the mass spectra of complexes 4, 6, 7 and 10 were recorded to assign the most abundant molecular ion peaks. The mass spectra of complexes 4 and 6 are shown in Fig. 4a and b, respectively. For complexes 4 and 6, the observed highest mass peaks at m/z 478 and 589, respectively, are in good agreement with the formula weights of the complexes [Ni(L)2Cl2(H2O)2] (F. Wt = 477.93) and [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 (F. Wt = 588.13). For the mass spectra of complexes 7 and 10, the observed molecular ion peaks at m/z 525 and 521, respectively, are consistent with the formula weights of the anhydrous complexes [Co(L)2(OAc)2] (F. Wt = 525.36) and [Co(L)(8-HQ)2] (F. Wt = 521.42).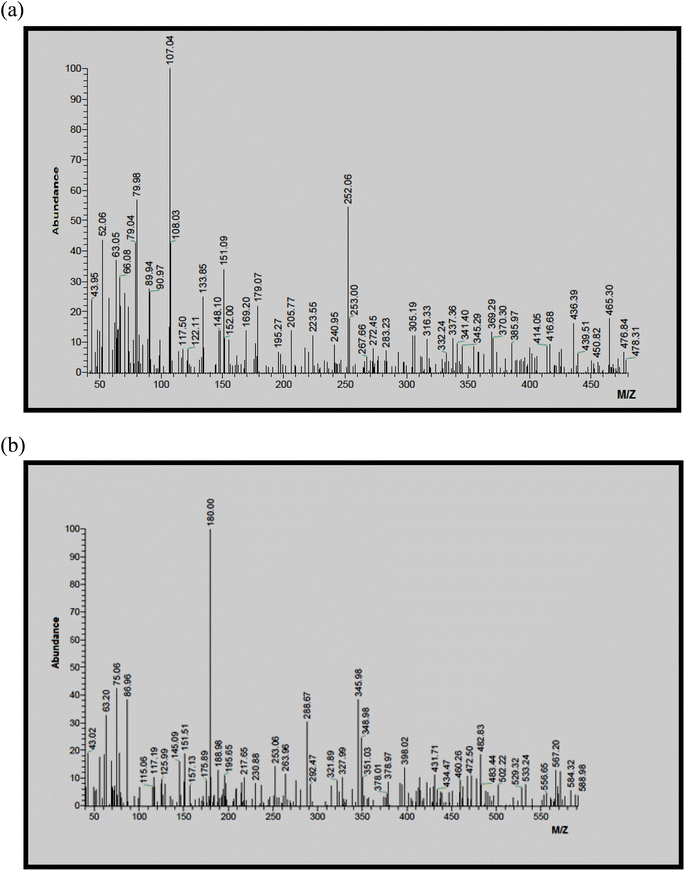 | ||
| Fig. 4 (a) Mass spectrum of the complex [Ni(L)2Cl2(H2O)2] (4). (b) Mass spectrum of the complex [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 (6). | ||
Based on the above interpretation, structures of the binary and ternary metal–chromone complexes are represented in Schemes 2 and 3.
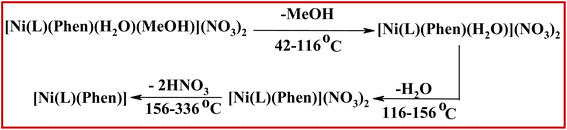 | ||
| Scheme 4 Thermal degradation pattern of complex 6, [Ni(L)(Phen)(H2O)(MeOH](NO3)2 in the range 42–336 °C. | ||
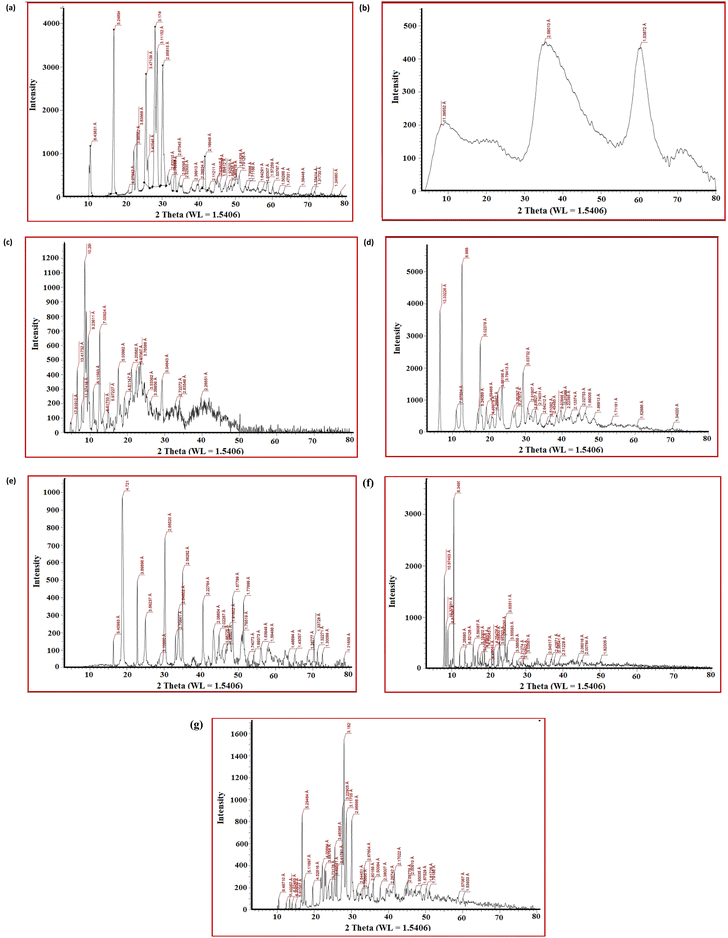
3.6. Powder XRD measurements
Powder XRD measurements of the ligand and complexes 2, 5, 6, 7, 10 and 11 (Fig. 5) showed several peaks with different intensities in the 2θ range from 0 to 80°, representing that the materials are crystalline, except for complex 2 which has an amorphous structure. The diffractograms and associated data depict the 2θ value for each peak, the relative intensity of the Co(II)- and Ni(II)-complexes to that of the ligand. This result suggests that the Co(II) and Ni(II) ions were coordinated to the ligand L to give new complexes. The main crystallite sizes of the most intense peak are 11.18, 27.59, 16.90, 102.09 and 11.11 nm for ligand complexes 5, 6, 7, 10 and 11, respectively.3.7. Antimicrobial activity
The antimicrobial effects of the obtained Ni(II)- and Co(II)-chromone complexes were assessed against Gram +ve bacteria Staphylococcus aureus and Bacillus subtilis, Gram −ve bacteria Escherichia coli and Salmonella typhimurium, the yeast Candida albicans and the fungus Aspergillus fumigatus (Table 6). From the data in Table 6, the following points are deduced: (i) all complexes are biologically active except complexes 5 and 10, which are inactive towards all organisms. (ii) Some complexes are inactive towards certain organisms, as follows: towards Staphylococcus aureus, complexes 3 and 9 are inactive, and towards Salmonella typhimurium, complex 6 is inactive. (iii) The bioactivity of the present complexes ranges from poor to a high nearly equivalent to that of the control. (iv) Against Staphylococcus aureus, all complexes exhibited poor activity except the ternary Co(II)–Phen complex 11, which showed high activity. On the other hand, most complexes exhibited high activity towards Bacillus subtilis and the highest activity was observed in the acetato- (1) and chloro- (4) nickel(II) complexes. (v) Against Salmonella typhimurium, most complexes exhibited poor to intermediate activity and the highest activity was given by the acetato-Ni(II) complex 1. The complexes exhibited intermediate to high activity towards Escherichia coli and the highest activity is given by the perchlorate-Ni(II) complex 3. (vi) Against Candida albicans and Aspergillus fumigates, the complexes exhibited intermediate to high activity and the highest activities are given by the acetato-Ni(II) complex 1 (in the case of Candida albicans) and the chloro-Co(II) complex 9 and the ternary Co(II)–Phen complex 11 (in the case of Aspergillus fumigates).| Organism | Meanb of zone diameter, nearest whole mm | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-positive bacteria | Gram-negative bacteria | Yeast and fungic | |||||||||||
| Staphylococcus aureus (ATCC 25923) | Bacillus subtilis (ATCC 6635) | Salmonella typhimurium (ATCC 14028) | Escherichia coli (ATCC 25922) | Candida albicans (ATCC 10231) | Aspergillus fumigatus | ||||||||
| Sample | Concentration | 1 mg ml−1 | 0.5 mg ml−1 | 1 mg ml−1 | 0.5 mg ml−1 | 1 mg ml−1 | 0.5 mg ml−1 | 1 mg ml−1 | 0.5 mg ml−1 | 1 mg ml−1 | 0.5 mg ml−1 | 1 mg ml−1 | 0.5 mg ml−1 |
| a — = no effect, L: low activity = mean of zone diameter ≤ 1/3 of mean zone diameter of control, I: intermediate activity = mean of zone diameter ≤ 2/3 of mean zone diameter of control, H: high activity = mean of zone diameter > 2/3 of mean zone diameter of control.b Calculated from 3 values.c Identified on the basis of routine cultural, morphological and microscopic characteristics.d Chloramphenicol in the case of Gram-positive bacteria, cephalothin in the case of Gram-negative bacteria and cycloheximide in the case of fungi. | |||||||||||||
| L | 22 I | 19H | 31H | 27H | 29H | 26H | 24 I | 20H | 43H | 38H | 25 I | 20H | |
| [Ni(L)2(OAc)2]·H2O (1) | 9 L | 8L | 26H | 23H | 15I | 12I | 17I | 15I | 26H | 23H | 25H | 22H | |
| [Ni(L)3](NO3)2·0.5MeOH (2) | 10L | 9L | 25H | 21H | 11L | 9L | 15I | 12I | 24H | 20H | 26H | 22H | |
| [Ni(L)2(H2O)2](ClO4)2·H2O (3) | — | — | 25H | 22H | 11L | 8L | 21I | 18H | 24H | 19H | 22I | 18I | |
| [Ni(L)2Cl2(H2O)2] (4) | 9L | 7L | 26H | 23H | 9L | 7L | 16I | 11I | 25H | 19H | 20I | 16I | |
| [Ni(L)(8-HQ)2]·1.5H2O (5) | — | — | — | — | — | — | — | — | — | — | — | — | |
| [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 (6) | 11L | 8L | 26H | 22H | — | — | 11L | 9L | 24H | 19H | 18I | 16I | |
| [Co(L)2(OAc)2]·1.5H2O (7) | 9 L | 8L | 20I | 18H | 11L | 9L | 12I | 10I | 24H | 22H | 16I | 14I | |
| [Co(L)3](NO3)2·MeOH (8) | 11L | 8L | 21I | 18H | 10L | 8L | 13I | 10I | 24H | 20H | 18I | 14I | |
| [Co(L)2Cl2] (9) | — | — | 22I | 18H | 8L | 7L | 14I | 10I | 20I | 16I | 27H | 23H | |
| [Co(L)(8-HQ)2]·1.5H2O (10) | — | — | — | — | — | — | — | — | — | — | — | — | |
| [Co(L)(Phen)(MeOH)2](NO3)2 (11) | 23I | 20H | 23I | 18H | 11L | 9L | 15I | 11I | 22I | 20H | 27H | 24H | |
| Controld | 35 | 26 | 35 | 25 | 36 | 28 | 38 | 27 | 35 | 28 | 37 | 26 | |
It has been reported that different parameters, including the characters of the chelating agent and its chelating donor atoms, the character of the metal ion, the geometrical arrangement of the complex, solubility, and other characteristics, have a major impact on the antibacterial activity exhibited by a compound.46
3.8. Molecular docking studies
The MOE 2014.0901 software was used for molecular docking studies for the ligand L and its complexes 2, 6, 8 and 11. Elena et al. studied molecular docking for chromone compounds using the S. aureus nucleoside diphosphate kinase (PDB ID: 3Q89) (Table 7).47| Compounds | Ligand | Receptor | Interaction | Distanceb | Ea | |||
|---|---|---|---|---|---|---|---|---|
| a kcal mol−1.b Å | ||||||||
| Ligand | O7 | NZ | LYS | 9 | (B) | H-Acceptor | 2.98 | −0.8 |
| O11 | MG | MG | 159 | (B) | Coordinate | 2.37 | −1.9 | |
| O14 | MG | MG | 159 | (B) | Coordinate | 2.29 | −1.7 | |
| 6-Ring | ND2 | ASN | 112 | (B) | Pi-H | 4.51 | −2.4 | |
| [Ni(L)3](NO3)2 · 0.5MeOH (2) | O28 | NZ | LYS | 9 | (B) | H-Acceptor | 2.95 | −18.0 |
| O43 | ND2 | ASN | 112 | (B) | H-Acceptor | 3.19 | −2.1 | |
| 6-Ring | NH1 | ARG | 102 | (B) | Pi-cation | 3.56 | −0.8 | |
| [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 (6) | O30 | NE2 | HIS | 52 | (B) | H-Acceptor | 2.89 | −14.0 |
| Ni29 | MG | MG | 159 | (B) | Ionic | 1.52 | −28.5 | |
| [Co(L)3](NO3)2·MeOH (8) | O28 | NZ | LYS | 9 | (B) | H-Acceptor | 2.97 | −18.6 |
| O43 | ND2 | ASN | 112 | (B) | H-Acceptor | 3.12 | −2.8 | |
| [Co(L)(Phen)(MeOH)2](NO3)2 (11) | O30 | NE2 | HIS | 52 | (B) | H-Acceptor | 3.40 | −1.7 |
| CO29 | MG | MG | 159 | (B) | Ionic | 1.47 | −30.3 | |
The ligand (Fig. 6a) formed three bonds with amino acid residues; one of them is the H-acceptor bond between O7 of L and LYS 9 (2.98 Å, −0.8 kcal mol−1). The second and third bonds are coordinate bonds between O11 and O14 of L and MG 159 (2.37 Å, −1.9 kcal mol−1 and 4.51 Å, −2.4 kcal mol−1, respectively).
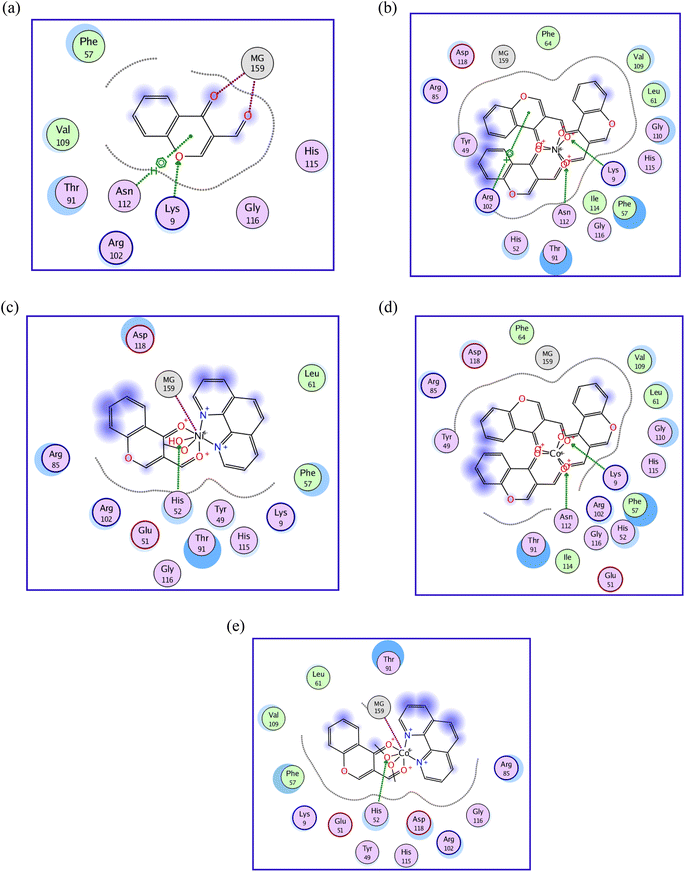 | ||
| Fig. 6 (a) 2D diagram of ligand interaction with S. aureus nucleoside diphosphate kinase, PDB ID: 3Q89. (b) 2D diagram of Ni(II)- complex 2 interaction with S. aureus nucleoside diphosphate kinase, PDB ID: 3Q89. (c) 2D diagram of Ni(II)- complex 6 interaction with S. aureus nucleoside diphosphate kinase, PDB ID: 3Q89. (d) 2D diagram of Co(II)- complex 8 interaction with S. aureus nucleoside diphosphate kinase, PDB ID: 3Q89. (e) 2D diagram of Co(II)- complex 11 interaction with S. aureus nucleoside diphosphate kinase, PDB ID: 3Q89. | ||
On the other hand, Ni(II)-complex 2 (Fig. 6b) formed three bonds. The first and second are H-acceptor bonds between O28 and O43 and LYS 9 and ASN 112 (2.95 Å, −18.0 kcal mol−1 and 3.19 Å, −2.1 kcal mol−1, respectively). The third bond is a pi-cation between the 6-ring and ARG 102 (3.56 Å, −0.8 kcal mol−1). Ni(II)-complex 6 (Fig. 6c) formed two bonds with the amino acid residues, the first being an H-acceptor bond between O30 and HIS 52 (2.89 Å, −14.0 kcal mol−1) and the second an ionic bond between Ni and MG 159 (1.52 Å, −28.5 kcal mol−1).
Co(II)-complex 8 (Fig. 6d) formed two bonds with the amino acid residues, H-acceptor bonds between O28 and O43 and LYS 9 and ASN 112 (2.97 Å, −18.6 kcal mol−1 and 3.12 Å, −2.8 kcal mol−1, respectively). Co(II)-complex 11 (Fig. 5e) formed two bonds with the amino acid residues in the active site of the protein. The first is an H-acceptor between O30 and HIS 52 (3.40 Å, −1.7 kcal mol−1). The second is an ionic bond between CO29 and MG 159 (1.47 Å, −30.3 kcal mol−1).
According to Hansch and Leo, highly lipophilic compounds will participate in the lipid interior of membranes and be retained there. A drug has low solubility with a higher log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and has difficulty penetrating lipid membranes with a lower log
P and has difficulty penetrating lipid membranes with a lower log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P.48 Lipophilicity (log
P.48 Lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) is a feature that affects absorption, solubility, distribution, excretion qualities, metabolism, and pharmacological activity; normalized B.S. and normalized C.A. rise as log
P) is a feature that affects absorption, solubility, distribution, excretion qualities, metabolism, and pharmacological activity; normalized B.S. and normalized C.A. rise as log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P increases.
P increases.
The docking data agreed with the antimicrobial activity data of the ligand and its complexes 2, 6, 8 and 11, where the ligand has the highest antimicrobial activity and formed 4 bonds, while its complexes formed 2 or 3 bonds.
Normalized B.S.a = 2.69 + 0.0592![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P − 0.0232 refractivity + 0.0628 polarizability + 0.481μ** + 0.289w + 1.44σ P − 0.0232 refractivity + 0.0628 polarizability + 0.481μ** + 0.289w + 1.44σ |
Normalized C.A.b = 0.307 + 0.0339![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P − 0.0297 refractivity + 0.0932 polarizability − 0.105μ** − 0.134w − 0.394σ P − 0.0297 refractivity + 0.0932 polarizability − 0.105μ** − 0.134w − 0.394σ |
3.9. Molecular modeling
Molecular modelling of the optimized compounds (Fig. 7) was carried out using the Hyperchem 7.52 program. The structures were fully optimized and geometric calculations were done to obtain structural parameters such as bond length, atomic charge distributions of the complexes, heat of formation, dipole moment, EHOMO, ELUMO, energy band gaps, and polarizabilities.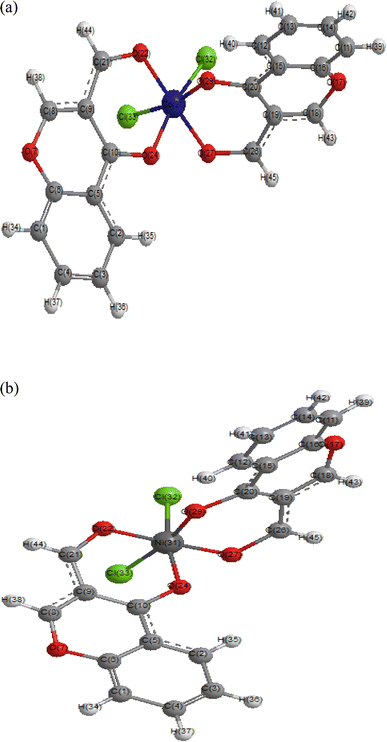 | ||
| Fig. 7 (a) Optimized structure of [(L)2CoCl2] (9) using Hyperchem 7.52 at PM3 level. (b) Optimized structure of [(L)2NiCl2] (4) using Hyperchem 7.52 at PM3 level. | ||
The values of EHOMO (eV), ELUMO (eV) and Egap (eV) of the binary complexes (Table 8) that include the acetate anions increase in the following order: Ni(II)- > Co(II)-complexes. Consequently, the band gap values of the complexes are less than that of the free chromone ligand, which means that, in any excitation process, the complexes need less energy than the free ligand. These orders reveal that the stability of the complexes is in the same order of increasing Egap. On the other hand, the Egap values of the binary and ternary chlorides show the order Ni- ≫ Co-complex, but the binary nitrate and ternary complexes of phenanthroline (phen) show the trend Co- > Ni-complex and the stability decreases in the same direction. These differences in the order of Egap might arise from the unlike hard–soft acid–base interactions (HSBA).
| No. | Complex | ELUMO | EHOMO | ΔEgab | Dipole moment | ΔH | T.E complex | η | μ | σ | ω |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | −0.880 | −9.770 | 8.890 | 4.080 | −63.830 | −49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 742.0 742.0 |
−4.4500 | 5.325 | −0.22 | −3.19 | |
| 1 | [Ni(L)2(OAc)2]·H2O | −1.360 | −7.528 | 6.168 | 8.494 | −550.090 | −163![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 798.7 798.7 |
3.0839 | −4.44415 | −0.32 | 3.2022 |
| 2 | [Ni(L)3](NO3)2·0.5MeOH | −0.724 | −6.387 | 5.663 | 2.627 | −380.629 | −173![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313.0 313.0 |
2.8315 | −3.5555 | −0.35 | 2.2323 |
| 3 | [Ni(L)2(H2O)2](ClO4)2·H2O | −0.802 | −6.566 | 5.764 | 2.677 | −444.901 | −138![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 578.4 578.4 |
2.8817 | −3.68395 | −0.35 | 2.3548 |
| 4 | [Ni(L)2Cl2(H2O)2] | −1.527 | −6.974 | 5.447 | 10.730 | −316.125 | −138![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 166.6 166.6 |
2.7236 | −4.25045 | −0.37 | 3.3167 |
| 5 | [Ni(L)(8-HQ)2]·1.5H2O | −0.744 | −7.910 | 7.166 | 3.730 | −190.970 | −148![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 218.6 218.6 |
3.5830 | −4.327 | −0.28 | 2.6127 |
| 6 | [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 | −1.628 | −5.614 | 3.986 | 10.700 | −299.620 | −135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 354.6 354.6 |
1.9932 | −3.62115 | −0.50 | 3.2894 |
| 7 | [Co(L)2(OAc)2]·1.5H2O | −2.042 | −3.420 | 1.378 | 12.070 | −592.470 | −157![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 975.3 975.3 |
0.6892 | −2.7308 | −1.45 | 5.4101 |
| 8 | [Co(L)3](NO3)2·MeOH | −1.035 | −7.514 | 6.479 | 2.612 | −482.560 | −67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 549.1 549.1 |
3.2395 | −4.2745 | −0.31 | 2.8201 |
| 9 | [Co(L)2Cl2] | −2.290 | −3.404 | 1.114 | 11.270 | −417.470 | −132![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 402.1 402.1 |
0.5570 | −2.847 | −1.80 | 7.2759 |
| 10 | [Co(L)(8-HQ)2]·1.5H2O | −1.208 | −3.712 | 2.504 | 4.733 | −373.310 | −142![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 935.1 935.1 |
1.2520 | −2.46 | −0.80 | 2.4168 |
| 11 | [Co(L)(Phen)(MeOH)2](NO3)2 | −0.783 | −6.853 | 6.070 | 6.132 | −396.250 | −133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 028.5 028.5 |
3.0352 | −3.81795 | −0.33 | 2.4013 |
The heats of formation of all complexes under study are exothermic. The [Co(L)2(OAc)2]·1.5H2O complex has the most negative heat of formation (−592.47 kcal mol−1), so it is more thermodynamically stable compared to the other complexes. The complex [Ni(L)2Cl2] has the most positive heat of formation and is the least stable.
Global chemical reactivity indices such as total energy, electronic chemical potential (μ), chemical hardness (η), electrophilicity (ω), softness (σ) and dipole moment (μ) are used to describe the reactivity and stability of any chemical compound.
The dipole moment in a molecule is an important property that is mainly used to study intermolecular interactions involving nonbonded type dipole–dipole interactions, because the higher the dipole moment, the stronger the expected intermolecular interactions. Increasing the stability of the complex formation is complemented by increasing the dipole moment value (more polarity) of the complex. Table 8 reveals that the highest polarity complex is [Co(L)2(OAc)2]·1.5H2O (12.07 Deby), while the least polar complex is [Co(L)3](NO3)2·MeOH (2.61 Deby).
The reactivity of complexes is Co(II)- > Ni(II)-complexes as indicated from the hardness (η) values in Table 8.43b
Electronic chemical potential (μ) describes the charge transfer within a system in the ground state. It is defined as the negative of electro-negativity. Physically, the electronic chemical potential (μ) is defined as the tendency of electrons to escape from the equilibrium state. Compounds having greater values of chemical potential are more reactive than those with smaller electronic chemical potential. The data in Table 8 reveals that the capability of an electron to leave the complex molecule based on the chemical potential (μ) values takes the order Ni(II)- > Co(II)-complexes. Therefore, [Ni(L)2(OAc)2]·H2O complex is the strongest electrophile while [Co(L)(8-HQ)2]·1.5H2O is the strongest nucleophile.
The electrophilicity index (ω) measures the stabilization of energy when the system acquires an additional electronic charge from the environment. The data in Table 8 reveal that the most powerful electrophile is the binary [Co(L)2Cl2] complex with ω = −7.28 eV, while the weakest electrophile is the binary [Ni(L)3](NO3)2 · 0.5MeOH complex with ω = −2.23 eV. The softness (σ) of the complexes ranges from −0.28 to −1.80 eV−1.
Based on the values in Table 9, it is observed that when these ligands are coordinated with the metal ion, there is an elongation of the bond length between the above-mentioned atoms which confirms the coordination of the carbonyl oxygen with the metal ion. When the atoms are coordinated with the metal ion by donating a lone pair of electrons, there is a decrease in electron density on the coordinating atoms, hence the bond length increases in the metal complexes. A comparison of the bond lengths of the ligand and its metal complexes is demonstrated in Table 9. The bond lengths from the PM3 method for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of the chromone after complexation are between 1.23 and 1.50 Å, longer than that of the free ligand (1.21 Å). Moreover, the M–O bond length of the metal ion with the carbonyl group was found to be in the range 1.86–1.97 Å.
O of the chromone after complexation are between 1.23 and 1.50 Å, longer than that of the free ligand (1.21 Å). Moreover, the M–O bond length of the metal ion with the carbonyl group was found to be in the range 1.86–1.97 Å.
| No. | Compound | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Oγ pyrone Oγ pyrone |
M–O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
M–O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cγ Cγ |
Other bond lengths |
|---|---|---|---|---|---|---|
| L | 1.21 | 1.22 | — | — | — | |
| 1 | [Ni(L)2(OAc)2]·H2O | 1.23 | 1.25 | 1.86 | 1.86 | M-OAC |
| 1.86–1.816 | ||||||
| 2 | [Ni(L)3](NO3)2·0.5MeOH | 1.50 | 1.50 | 1.88 | 1.89 | — |
| 3 | [Ni(L)2(H2O)2](ClO4)2·H2O | 1.50 | 1.50 | 1.876 | 1.88 | M-OH2 |
| 1.88–1.87 | ||||||
| 4 | [Ni(L)2Cl2(H2O)2] | 1.50 | 1.50 | 1.878 | 1.88 | M-Cl |
| 2.14–2.13 | ||||||
| 5 | [Ni(L)(8-HQ)2]·1.5H2O | 1.49 | 1.50 | 1.88 | 1.87 | M-8Hq |
| 1.90–1.89 | ||||||
| 6 | [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 | 1.24 | 1.24 | 1.92 | 1.97 | M-10Phen, 1.80 |
| M-OHCH3, 1.29 | ||||||
| M-OH2, 2.008 | ||||||
| 7 | [Co(L)2(OAc)2]·1.5H2O | 1.29 | 1.27 | 1.87 | 1.94 | M-OAC |
| 1.88–1.86 | ||||||
| 8 | [Co(L)3](NO3)2·MeOH | 1.28 | 1.28 | 1.89 | 1.93 | — |
| 9 | [Co(L)2Cl2] | 1.25 | 1.289 | 1.90 | 1.91 | M-Cl |
| 2.21 | ||||||
| 10 | [Co(L)(8-HQ)2]·1.5H2O | 1.29 | 1.26 | 1.915 | 1.94 | M-8Hq |
| 1.90 | ||||||
| 11 | [Co(L)(Phen)(MeOH)2](NO3)2 | 1.329 | 1.34 | 1.892 | 1.86 | M-10Phen, 1.90 |
| M-OHCH3, 1.993 | ||||||
| M-OH2, 2.023 |
The charge densities on the coordinating centres and the central metal ions are tabulated in Table 10. The results show that the range of charge density is lower in the case of Co(II)-complexes (0.011 to −0.142) than the electron density on the central metal ion of Ni(II)-complexes (−0.613 to −0.898).
| No. | Complex | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (1) O (1) |
CHO (1) | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (2) O (2) |
CHO (2) | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (3) O (3) |
CHO (3) | Metal | Other molecules |
|---|---|---|---|---|---|---|---|---|---|
| 1 | [Ni(L)2(OAc)2]·H2O | 0.013 | −0.016 | 0.003 | −0.013 | −0.623 | OAc = −0.168 | ||
| OAc = −0.171 | |||||||||
| 2 | [Ni(L)3](NO3)2 · 0.5MeOH | 0.013 | −0.034 | −0.010 | −0.169 | −0.001 | −0.029 | −0.731 | |
| 3 | [Ni(L)2(H2O)2](ClO4)2·H2O | −0.029 | −0.004 | 0.008 | −0.034 | −0.693 | OH2 = −0.079 | ||
| OH2 = −0.091 | |||||||||
| 4 | [Ni(L)2Cl2(H2O)2] | 0.036 | 0.003 | 0.030 | 0.001 | −0.613 | Cl = −0.199 | ||
| Cl = −0.202 | |||||||||
| 5 | [Ni(L)(8-HQ)2]·1.5H2O | −0.093 | −0.107 | −0.856 | N-8hy = 0.026 | ||||
| O-8hy = −0.129 | |||||||||
| N-8hy = 0.944 | |||||||||
| O-8hy = −0.110 | |||||||||
| 6 | [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 | −0.051 | −0.138 | −0.898 | N-phen = 0.766 | ||||
| N-phen = 0.831 | |||||||||
| MeOH = −0.210 | |||||||||
| OH2 = −0.105 | |||||||||
| 7 | [Co(L)2(OAc)2]·1.5H2O | −0.22 | −0.199 | −0.145 | −0.063 | 0.011 | OAc = −0.371 | ||
| OAc = −0.441 | |||||||||
| 8 | [Co(L)3](NO3)2·MeOH | −0.209 | −0.221 | −0.207 | −0.222 | −0.207 | −0.221 | −0.012 | |
| 9 | [Co(L)2Cl2] | −0.169 | −0.050 | −0.205 | −0.121 | −0.042 | Cl = −0.424 | ||
| Cl = −0.330 | |||||||||
| 10 | [Co(L)(8-HQ)2]·1.5H2O | −0.193 | −0.273 | −0.123 | O-8hy = −0.270 | ||||
| N-8hy = 0.378 | |||||||||
| O-8hy = −0.272 | |||||||||
| N-8hy = 0.42 | |||||||||
| 11 | [Co(L)(Phen)(MeOH)2](NO3)2 | −0.178 | −0.237 | −0.142 | MeOH = −0.225 | ||||
| MeOH = −0.196 | |||||||||
| N-phen = 0.175 | |||||||||
| N-phen = 0.171 |
The data in Table 11 show the following: (i) as ΔH increases, the lengths of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Oγ pyrone, C
Oγ pyrone, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and ν(M–O) increase, but the lengths of C
O and ν(M–O) increase, but the lengths of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Oγ–M pyrone, C
Oγ–M pyrone, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–M and ν(C
O–M and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) decrease. (ii) The positive slope of the relationship between ELUMO and the bond lengths of C
O) decrease. (ii) The positive slope of the relationship between ELUMO and the bond lengths of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Oγ pyrone indicates that increasing ELUMO leads to increasing stability and increasing bond lengths of C
Oγ pyrone indicates that increasing ELUMO leads to increasing stability and increasing bond lengths of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Oγ pyrone. On the other hand, the linear correlation between the calculated EHOMO with the frequency of C
Oγ pyrone. On the other hand, the linear correlation between the calculated EHOMO with the frequency of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and M–O indicates that increasing EHOMO leads to an increase in C
O and M–O indicates that increasing EHOMO leads to an increase in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and a decrease in M–O. (iii) However, increasing ΔEgab leads to increasing stability and ν(M–N) and decreasing ν(C
O and a decrease in M–O. (iii) However, increasing ΔEgab leads to increasing stability and ν(M–N) and decreasing ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and λmax.
O) and λmax.
| No. | Equation | r% | Complexes |
|---|---|---|---|
| 1 | ΔH = 8584(1327) − 4718(697.0) LM–O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cγ Cγ |
92.0% | 2, 4, 5, 7, 8 & 9 |
| 2 | ΔH = 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 486(5801) − 12 486(5801) − 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 684(3081) LM–O 684(3081) LM–O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
94.4% | 1, 5, 8 & 11 |
| 3 | ΔH = −2015(216.5) + 1215(159.8) LC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Oγ pyrone Oγ pyrone |
96.7% | 5, 8, 9 & 11 |
| 4 | ΔH = −2246 (74.06) + 1382(55.44) LC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
99.7% | 1, 5, 8 & 11 |
| 5 | ΔH = −1723(190.1) + 2.51(0.3557) ν(M–O) | 94.3% | 2, 3, 6, 9 & 10 |
| 6 | ΔH = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 442(933.4) − 6.73(0.5817) ν(C 442(933.4) − 6.73(0.5817) ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
97.8% | 2, 3, 5, 10 & 11 |
| 7 | ELUMO = −12.9(2.932) + 9.17(2.281) L(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
89.0% | 6, 8, 10 & 11 |
| 8 | ELUMO = −10.7(2.269) + 7.44(1.781) L(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Oγ pyrone) Oγ pyrone) |
85.3% | 1, 6, 8, 10 & 11 |
| 9 | EHOMO = −138(17.45) + 0.0812(0.01082) ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
98.3% | 1, 3 & 11 |
| 10 | EHOMO = 27.8(4.786) − 0.0589(0.008783) ν(M–O) | 97.8% | 6, 9 & 10 |
| 11 | ΔEgab = 8.35(0.4251) − 0.00443(0.0007426) λmax | 92.2% | 1–4 & 11 |
| 12 | ΔEgab = −69.2(9.483) + 0.170(0.02178) ν(M–N) | 96.8% | 1–4 & 10 |
| 13 | ΔEgab = 65.2(7.133) − 0.0367(0.004445) ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) O) |
95.8% | 1, 3, 5, 8 & 11 |
Table 12 shows that the QSAR properties of the complexes are higher than those of the ligand, including surface area (420.91–70, 558.50–738.27), octanol–water partition coefficient (lipophilicity, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, −1.84 to −0.04, 0.25–2.53), volume (982.79–1330.37), refractivity (103.68–150.72), polarizability (34.54–49.48) and molecular weight (443.05–581.40). The hydration energy for the ligand (7.40 kcal mol−1) changed to −1.2 to −26.15 kcal mol−1 for its complexes.
P, −1.84 to −0.04, 0.25–2.53), volume (982.79–1330.37), refractivity (103.68–150.72), polarizability (34.54–49.48) and molecular weight (443.05–581.40). The hydration energy for the ligand (7.40 kcal mol−1) changed to −1.2 to −26.15 kcal mol−1 for its complexes.
| No. | Compound | Surface area approx. | Surface area grid | Volume | Hydration energy (kcal mol−1) | -log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
Refractivity | Polarizability | MW |
|---|---|---|---|---|---|---|---|---|---|
| L | 277.25 | 329.02 | 508.16 | 7.40 | −0.61 | 50.43 | 17.62 | 174.16 | |
| 1 | [Ni(L)2(OAc)2]·H2O | 539.52 | 661.83 | 1184.05 | −5.40 | 1.15 | 123.91 | 42.56 | 525.11 |
| 2 | [Ni(L)3](NO3)2 · 0.5MeOH | 533.21 | 734.75 | 1317.94 | −2.24 | 1.89 | 150.72 | 49.48 | 581.18 |
| 3 | [Ni(L)2(H2O)2](ClO4)2·H2O | 420.91 | 558.50 | 982.79 | −26.15 | −0.04 | 103.68 | 34.54 | 443.05 |
| 4 | [Ni(L)2Cl2(H2O)2] | 458.21 | 585.96 | 1016.94 | −2.04 | 2.53 | 112.75 | 37.63 | 477.93 |
| 5 | [Ni(L)(8-HQ)2]·1.5H2O | 476.07 | 681.64 | 1214.83 | −7.16 | 0.52 | 143.98 | 48.95 | 521.17 |
| 6 | [Ni(L)(Phen)(H2O)(MeOH)](NO3)2 | 430.52 | 611.56 | 1085.19 | −8.78 | −1.84 | 120.70 | 41.40 | 463.13 |
| 7 | [Co(L)2(OAc)2]·1.5H2O | 535.92 | 670.61 | 1196.48 | −4.97 | 1.15 | 123.91 | 42.55 | 525.33 |
| 8 | [Co(L)3](NO3)2·MeOH | 552.44 | 738.27 | 1330.37 | −1.43 | 1.89 | 150.72 | 49.47 | 581.40 |
| 9 | [Co(L)2Cl2] | 514.62 | 604.21 | 1044.40 | −1.21 | 2.53 | 112.75 | 37.62 | 478.15 |
| 10 | [Co(L)(8-HQ)2]·1.5H2O | 647.70 | 560.06 | 1022.02 | −2.53 | 0.25 | 129.92 | 41.19 | 504.26 |
| 11 | [Co(L)(Phen)(MeOH)2](NO3)2 | 427.16 | 608.82 | 1115.62 | −4.16 | −1.59 | 125.59 | 43.23 | 477.38 |
4. Conclusion
The coordinating behavior of 3-formylchromone towards Ni(II) and Co(II) ions was investigated through the preparation of a series of binary and ternary metal complexes using metal ions with different anions (AcO−, NO3−, ClO4− and Cl−) and secondary ligands including N,O-donor 8-hydroxyquinoline or N,N-donor 1,10-phenanthroline. The prepared complexes were characterized by microanalytical, spectral, thermal, magnetic, and conductivity techniques. The analytical and spectroscopic techniques revealed the following: (i) all complexes exhibited octahedral geometry. (ii) The ligand behaves as a neutral bidentate, forming chelates that reflect the non-coordinating or weakly coordinating power of the ClO4− and NO3− anions as compared to the strongly coordinating power of the AcO− anion. (iii) The obtained complexes are formed with different molar ratios according to the anion. In the cases of the AcO−, ClO4− and Cl− anions, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 M
2 M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L complexes were obtained. In the case of the NO3− anion, 1
L complexes were obtained. In the case of the NO3− anion, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 M
3 M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L complexes were obtained. (iv) Ternary complexes are formed with two metal
L complexes were obtained. (iv) Ternary complexes are formed with two metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand
ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) secondary ligand molar ratios: 1
secondary ligand molar ratios: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in the case of 8-hydroxyquinoline and 1
2 in the case of 8-hydroxyquinoline and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in the case of 1,10-phenanthroline. The thermodynamic parameters of the metal complexes were calculated by the Coats–Redfern method and standard thermodynamic equations using the TGA curves from the thermal degradation of the solid complexes. Biological activity and molecular modelling showed good agreement, supported by the theoretical and experimental data.
2 in the case of 1,10-phenanthroline. The thermodynamic parameters of the metal complexes were calculated by the Coats–Redfern method and standard thermodynamic equations using the TGA curves from the thermal degradation of the solid complexes. Biological activity and molecular modelling showed good agreement, supported by the theoretical and experimental data.
Conflicts of interest
The authors of the article do not have any conflict of interest.References
- A. Husain, P. Ach and B. Anupama, J. Mol. Struct., 2022, 1254, 132341 CrossRef CAS.
- A. Innasiraj, B. Anandhi, Y. Gnanadeepam, N. Das, F. Paularokiadoss, A. V. Ilavarasi, C. D. Sheela, D. R. Ampasala and T. C. Jeyakumar, J. Mol. Struct., 2022, 1265, 133450 CrossRef CAS.
- M. B. Teimouri, Z. B. Savadjani, M. Shiri, R. Bikas and S. Naderi, Dyes Pigm., 2022, 199, 110106 CrossRef CAS.
- X. Yun, L. Chen, Y. Lv, Z. Lu, K. Huang and S. Yan, Green Synth. Catal., 2022 DOI:10.1016/j.gresc.2022.05.002 , in press..
- (a) S. B. A. Ghani, P. J. Mugisha, J. C. Wilcox, E. A. M. Gado, E. O. Medu, A. J. Lmb and R. C. D. Brown, Synth. Commun., 2013, 43, 1549–1556 CrossRef; (b) A. Ramesh, B. Srinivas, R. Pawar and A. Ramachandraiah, J. Mol. Struct., 2022, 1255, 132377 CrossRef CAS.
- K. M. Khan, N. Ambreen, U. R. Mughal, S. Jalil, S. Perveen and M. I. Choudhary, Eur. J. Med. Chem., 2010, 45, 4058–4064 CrossRef CAS PubMed.
- P. M. Kris-Etherton, K. D. Hecker, A. Bonanome, S. M. Coval, A. E. Binkoski, K. F. Hilpert, A. E. Griel and T. D. Etherton, Am. J. Med., 2002, 113, 71–88 CrossRef.
- (a) T. Zhou, Q. Shi, C.-H. Chen, H. Zhu, L. Huang, P. Ho and K.-H. Lee, Bioorg. Med. Chem., 2010, 18, 6678–6689 CrossRef CAS PubMed; (b) V. M. Patil, N. Masand, S. Verma and V. Masand, Chem. Biol. Drug Des., 2021, 98, 943–953 CrossRef CAS PubMed; (c) N. Balakrishnan, J. Haribabu, M. Dharmasivam, S. Swaminathan and R. Karvembu, Appl. Organomet. Chem., 2022, 36, e6750 CrossRef CAS; (d) I. Yousuf and M. Bashir, Appl. Organomet. Chem., 2021, 35, e6090 CrossRef CAS.
- (a) E. Venkateswararao, V. K. Sharma, M. Manickam, J. Yun and S.-H. Jung, Bioorg. Med. Chem. Lett., 2014, 24(22), 5256–5259 CrossRef CAS; (b) V. C. Patil, IJPSR, 2012, 3(12), 5006–5014 CAS; (c) A. Gomes, O. Neuwirth, M. Freitas, D. Couto, D. Ribeiro, A. G. P. R. Figueiredo, A. M. S. Silva, R. S. G. R. Seixas, D. C. G. A. Pinto, A. C. Tomé, J. A. S. Cavaleiro, E. Fernandes and J. L. F. C. Lima, Bioorg. Med. Chem., 2009, 17, 7218–7226 CrossRef CAS PubMed; (d) J. E. Philip, S. A. Antony, S. J. Eeettinilkunnathil, M. R. P. Kurup and M. P. Velayudhan, Inorg. Chim. Acta, 2018, 469, 87–97 CrossRef CAS; (e) P. Kavitha, M. Saritha and K. L. Reddy, Spectrochim. Acta, Part A, 2013, 102, 159–168 CrossRef CAS; (f) P. Mucha, A. Skoczyńska, M. Małecka, P. Hikisz and E. Budzisz, Molecules, 2021, 26(16), 4886 CrossRef CAS PubMed; (g) M. Gaber, K. El-Baradie, N. Elwakiel and S. Hafez, Appl. Organomet. Chem., 2020, 34(2), e5348 CrossRef CAS; (h) C. Chiruta, D. Schubert, R. Dargusch and P. Maher, J. Med. Chem., 2012, 55, 378–389 CrossRef CAS; (i) S. Selvamurugan, R. Ramachandran, P. Vijayan, R. Manikandan, G. Prakash, P. Viswanathamurthi, K. Velmurugan, R. Nandhakumar and A. Endo, Polyhedron, 2016, 107, 57–67 CrossRef CAS.
- S. Khursheed, S. Tabassum and F. Arjmand, Polyhedron, 2022, 214, 115638 CrossRef CAS.
- (a) A. Z. El-Sonbati, M. A. Diab, A. K. El-Sayed, M. I. Abou-Dobara and A. A. F. Gafer, Appl. Organomet. Chem., 2022, 36(5), e6651 CAS; (b) S. A. Sadeek, S. M. Abd El-Hamid, A. A. Mohamed, W. A. Zordok and H. A. El-Sayed, Appl. Organomet. Chem., 2019, 33, e4889 CrossRef.
- (a) H. F. Abd El-Halim, G. G. Mohamed and M. N. Anwar, Appl. Organomet. Chem., 2018, 32, e3899 CrossRef; (b) R. Neelaeni, S. Vasantha, R. Keerthana, S. Sivakolunthu and T. Angeline, Asian J. Pharm. Clin. Res., 2016, 9, 277–281 CrossRef.
- H. L. Mobley and R. P. Hausinger, Microbiol. Rev., 1989, 53, 85–108 CrossRef CAS PubMed.
- (a) D. Li and G.-Q. Zhong, Sci. World J., 2014, 2014, 641608 Search PubMed; (b) S. Altürk, D. Avci, B. Z. Kurt, Ö. Tamer, A. Başoğlu, F. Sِnmez, Y. Atalay and N. Dege, J. Inorg. Organomet. Polym. Mater., 2019, 29(4), 1265–1279 CrossRef.
- (a) A. Z. El-Sonbati, M. A. Diab and S. M. Morgan, J. Mol. Liq., 2017, 225, 195–206 CrossRef CAS; (b) M. Carcelli, S. Montalbano, D. Rogolino, V. Gandin, F. Miglioli, G. Pelosi and A. Buschini, Inorg. Chim. Acta, 2022, 533, 120779 CrossRef CAS; (c) A. Barma, D. Ghosh, P. Karmakar and P. Roy, J. Mol. Struct., 2022, 1250, 131687 CrossRef CAS.
- J. Shi, H. Ge, F. Song and S. Guo, J. Mol. Struct., 2022, 1253, 132263 CrossRef CAS.
- P. Patil and S. Zangade, J. Indian Chem. Soc., 2022, 99, 100274 CrossRef CAS.
- B. Kaya, D. Akyüz, T. Karakurt, O. Şahin, A. Koca and B. Ülküseven, Appl. Organomet. Chem., 2020, 34, e5930 CrossRef CAS.
- M. Echabaane, N. Mhadhbi, A. Rouis and H. Naïli, Optik, 2022, 256, 168770 CrossRef CAS.
- (a) S. Selvamurugan, R. Ramachandran, P. Vijayan, R. Manikandan, G. Prakash, P. Viswanathamurthi, K. Velmurugan, R. Nandhakumar and A. Endo, Polyhedron, 2016, 107, 57–67 CrossRef CAS; (b) P. Li, M. Niu, M. Hong, S. Cheng and J. Dou, J. Inorg. Biochem., 2014, 137, 101–108 CrossRef PubMed.
- A. Pui, C. Policar and J. P. Mahy, Inorg. Chim. Acta, 2007, 360, 2139–2144 CrossRef CAS.
- (a) M. Shebl, O. M. I. Adly, A. Taha and N. N. Elabd, J. Mol. Struct., 2017, 1147, 438–451 CrossRef CAS; (b) O. M. I. Adly and H. F. El-Shafiy, J. Coord. Chem., 2019, 72(2), 218–238 CrossRef CAS; (c) M. Shebl, S. M. E Khalil, M. A. A. kishk, D. M. EL-Mekkawi and M. Saif, Appl. Organomet. Chem., 2019, 33, e5147 CrossRef; (d) R. Fouad and O. M. I. Adly, J. Mol. Struct., 2021, 1225, 129158 CrossRef CAS.
- G. H. Jeffery, J. Bassett, J. Mendham and R. C. Denney, Vogel's Textbook of Quantitative Chemical Analysis, 5th edn, 1989 Search PubMed.
- F. E. Mabbs and D. I. Machin, Magnetism and Transition Metal Complexes, Chapman and Hall, London, 1973 Search PubMed.
- A. Nohara, T. Umetani and Y. Sanno, Tetrahedron, 1974, 30, 3553–3561 CrossRef CAS.
- A. U. Rahman, M. I. Choudhary and W. J. Thomsen, Bioassay Techniques for Drug Development, Harwood Academic Publishers, The Netherlands, 2001 Search PubMed.
- K. M. Khan, Z. S. Saify, A. K. Zeesha, M. Ahmed, M. Saeed, M. Schick, H. J. Kohlbau and W. Voelter, Arzneim. Forsch., 2000, 50, 915–922 CAS.
- HyperChem version 7.5, Hypercube, Inc, 2003 Search PubMed.
- (a) A. Dziewulska-Kulaczkowska, J. Therm. Anal. Calorim., 2010, 101(3), 1019–1026 CrossRef CAS; (b) Y. Li and Z. Yang, J. Coord. Chem., 2010, 63, 1960 CrossRef CAS; (c) D.-C. Ilies, E. Pahontu, S. Shova, R. Georgescu, N. Stanica, R. Olar, A. Gulea and T. Rosu, Polyhedron, 2014, 81, 123–131 CrossRef CAS; (d) S. J. Mohammed, A. M. Sheat, S. A. Abood and O. M. Yahya, Egypt. J. Chem., 2021, 64(11), 6423–6427 Search PubMed; (e) K. T. A. Al-Sultani and N. Al-Lami, Egypt. J. Chem., 2021, 64(6), 2953–2961 Search PubMed.
- E. M. Abdelrhman, B. A. El-Shetary, M. Shebl and O. M. I. Adly, Appl. Organomet. Chem., 2021, 35, e6183 CrossRef CAS.
- S. M. E. Khalil, M. Shebl and F. S. Al-Gohani, Acta Chim. Slov., 2010, 57, 716–725 CAS.
- (a) F. Samy, M. Shebl, A. Taha and H. S. Seleem, Egypt. J. Chem., 2022, 65(7), 85 Search PubMed; (b) F. Samy, A. Taha, M. Shebl, H. S. Seleem and F. I. Hanafy, Egypt. J. Chem., 2022 DOI:10.21608/EJCHEM.2022.138025.6079 , in press.
- N. T. Madhu and P. K. Radhakrichnan, Synth. React. Inorg. Met.-Org. Chem., 2001, 31, 315–330 CrossRef CAS.
- M. Shebl, A. A. Saleh, S. M. E. Khalil, M. Dawy and A. A. M. Ali, Inorg. Nano-Met. Chem., 2021, 51, 195–209 CrossRef CAS.
- F. Samy and M. Shebl, Appl. Organomet. Chem., 2020, 34, e5502 CrossRef CAS.
- (a) M. Shebl, O. M. I. Adly, H. F. El-Shafiy, S. M. E. Khalil, A. Taha and M. A. N. Mahdi, J. Mol. Struct., 2017, 1134, 649–660 CrossRef CAS; (b) M. Gaber, N. El-Wakiel, K. El-Baradie and S. Hafez, J. Iran. Chem. Soc., 2019, 16, 169–182 CrossRef CAS; (c) W. H. Mahmoud, R. G. Deghadi, M. M. I. El Desssouky and G. G. Mohamed, Appl. Organomet. Chem., 2019, 33, e4556 CrossRef; (d) F. Samy and M. Shebl, Appl. Organomet. Chem., 2022, 36(9), e6650 CAS.
- W. J. Geary, Coord. Chem. Rev., 1971, 7, 81–122 CrossRef CAS.
- F. A. Cotton and G. Wilkinson, Advanced Inorganic Chemistry. A Comprehensive Text, John Wiley and Sons, New York, 4th edn, 1986 Search PubMed.
- M. Shebl, J. Mol. Struct., 2017, 1128, 79–93 CrossRef CAS.
- (a) M. Shebl, Spectrochim. Acta, Part A, 2009, 73, 313–323 CrossRef PubMed; (b) S. M. E. Khalil, Synth. React. Inorg. Met.-Org. Chem., 2000, 30, 19 CrossRef CAS; (c) N. V. Kulkarni, M. P. Sathisha, S. Budagumpi, G. S. Kurdekar and V. K. Revankar, J. Coord. Chem., 2010, 63, 1451–1461 CrossRef CAS.
- (a) F. Samy and M. Shebl, Appl. Organomet. Chem., 2022, 36(5), e6650 CrossRef CAS; (b) K. A. R. Salib, S. L. Stefan, S. M. Abu El-Wafa and H. F. El-Shafiy, Synth. React. Inorg. Met.-Org. Chem., 2001, 31, 895–915 CrossRef CAS.
- A. W. Coats and J. P. Redfern, Nat, 1964, 201, 68–69 CrossRef CAS.
- (a) F. Samy, A. Taha and F. M. Omar, Appl. Organomet. Chem., 2022, 35(11), e6375 Search PubMed; (b) F. Samy and F. M. Omar, J. Mol. Struct., 2020, 1222, 128910 CrossRef CAS; (c) F. Samy, A. A. T. Ramadan, A. Taha and H. S. Seleem, Asian J. Chem., 2016, 28, 2650–2660 CrossRef CAS.
- C. R. Vinodkumar, M. K. M. Nair and P. K. Radhakrishnan, J. Therm. Anal. Calorim., 2000, 61, 143–149 CrossRef CAS.
- A. H. M. Siddaligaiah and S. G. Naik, J. Mol. Struct., 2002, 582, 129–136 CrossRef.
- B. Murukan and K. Mohanan, J. Enzyme Inhib. Med. Chem., 2007, 22(1), 65–70 CrossRef CAS PubMed.
- E. Pahonțu, M. Proks, S. Shova, G. Lupașcu, D.-C. Ilieș, Ș.-F. Bărbuceanu, L.-I. Socea, M. Badea, V. Păunescu, D. Istrati, A. Gulea, D. Drăgănescu and C. E. D. Pîrvu, Appl. Organomet. Chem., 2019, 33, e5185 CrossRef.
- M. A. Ibrahim, A. A. A. Emara, A. Taha, O. M. I. Adly, A. I. Nabeel and N. Salah, Appl. Organomet. Chem., 2022, 36, e6535 CAS.
| This journal is © The Royal Society of Chemistry 2022 |